Abstract
The Gram-negative Elizabethkingia express multiple antibiotic resistance and cause severe opportunistic infections. Vancomycin is commonly used to treat Gram-positive infections and has also been used to treat Elizabethkingia infections, even though Gram-negative organisms possess a vancomycin permeability barrier. Elizabethkingia anophelis appeared relatively vancomycin-susceptible and challenge with this drug led to morphological changes indicating cell lysis. In stark contrast, vancomycin growth challenge revealed that E. anophelis populations refractory to vancomycin emerged. In addition, E. anophelis vancomycin-selected mutants arose at high frequencies and demonstrated elevated vancomycin resistance and reduced susceptibility to other antimicrobials. All mutants possessed a SNP in a gene (vsr1 = vancomycin-susceptibility regulator 1) encoding a PadR family transcriptional regulator located in the putative operon vsr1-ORF551, which is conserved in other Elizabethkingia spp as well. This is the first report linking a padR homologue (vsr1) to antimicrobial resistance in a Gram-negative organism. We provide evidence to support that vsr1 acts as a negative regulator of vsr1-ORF551 and that vsr1-ORF551 upregulation is observed in vancomycin-selected mutants. Vancomycin-selected mutants also demonstrated reduced cell length indicating that cell wall synthesis is affected. ORF551 is a membrane-spanning protein with a small phage shock protein conserved domain. We hypothesize that since vancomycin-resistance is a function of membrane permeability in Gram-negative organisms, it is likely that the antimicrobial resistance mechanism in the vancomycin-selected mutants involves altered drug permeability.
1. Introduction
The Gram-negative Elizabethkingia are emerging opportunistic pathogens that cause severe disease (e.g., pneumonia, meningitis, and sepsis) in immunocompromised patients, the elderly, and neonates, that is associated with high mortality. To date, there are six characterized Elizabethkingia species that are known to cause human disease [1,2] and evidence suggests that most Elizabethkingia infections in humans are caused by Elizaethkingia anophelis [3]. Prior to a 2015–2016 E. anophelis community outbreak in the US Midwest involving 66 patients and 20 fatalities [4], outbreaks were primarily healthcare-related and often associated with water or a water source [5]. Three other species (Elizabethkingia umeracha, Elizabethkingia argenteiflava and an unnamed genomospecies) not yet associated with human infection were isolated from environmental and agricultural sources [6,7,8].
The Elizabethkingia exhibit intrinsic multiple antimicrobial resistance, which contributes to the high mortality attributed to infections caused by these organisms. Experimental research on the mechanisms of antimicrobial resistance in Elizabethkingia have focused on beta-lactamase mediated resistance [9] and fluoroquinolone resistance [10]. A unique aspect of these organisms is that they harbor multiple copies of putative beta-lactamase genes on their chromosome [11]. The Elizabethkingia also harbor multiple operons encoding putative Resistance-Nodulation-Division (RND) family intrinsic multidrug trans-envelope efflux pumps [12] which have been linked to clinically-relevant antimicrobial resistance [13].
The primary mechanism of action of the glycopeptide antibiotic vancomycin is the inhibition of peptidoglycan biosynthesis. Vancomycin binds to the terminal D-Ala-D-Ala residues of emerging muramyl-peptides on the membrane surface and prevents the linkage of new peptidoglycan monomers to the existing cell wall [14]. Vancomycin is used almost exclusively to treat Gram-positive infections [15] but it has also been used to treat Elizabethkingia infections with variable success [16]. The use of vancomycin to treat Elizabethkingia infections is highly unusual since Gram-negative organisms should be vancomycin-resistant due to the outer membrane permeability barrier and the molecular size exclusion (<600 Da and vancomycin is 1449 Da) of the aqueous filled outer membrane porins [17]. Drug efflux does not appear to play a role with resistance to vancomycin in Gram-negative organisms [17].
In the absence of Clinical and Laboratory Standards Institute (CLSI) guidelines for the Elizabethkingia, MIC breakpoints and Kirby Bauer zones of inhibition from the Gram-positive organism Staphylococcus aureus have been used to evaluate vancomycin susceptibility in Elizabethkingia [16], but several lines of evidence suggested that this is a mistake. Chiu et al., 2021 [18], provided evidence that disk diffusion and the E-test should not be employed to determine vancomycin susceptibility for E. anophelis. While most Gram-negative organisms exhibit high vancomycin MICs (64 to >1000 mg/L) [17,19,20], the Elizabethkingia demonstrate vancomycin MICs in the range of 1 to 64 mg/L [16,21,22,23] (this study). One study with 167 clinical Elizabethkingia isolates reported that 95.8% of E. anopheles and 100% of Elizabethkingia meningoseptica demonstrated intermediate vancomycin susceptibility [21]. Furthermore, it was reported that 108 Elizabethkingia spp. isolates demonstrated vancomycin resistance [22] while another study with 84 E. anophelis isolates also reported these strains were resistant to vancomycin as determined with agar dilution assays [18]. Collectively these findings support the notion that Elizabethkingia should be classified as demonstrating intermediate to full resistance to vancomycin.
To date there are no in vitro studies of the effects of vancomycin on Elizabethkingia yet considering the controversial use of this drug to treat infections caused by these Gram-negative organisms, thus such a study was warranted. The objectives of our research were to examine the effects of vancomycin on growth and the cell structure of Elizabethkingia, determine if vancomycin selection readily increases Elizabethkingia vancomycin-resistance, and determine the identity of mutations that alter vancomycin resistance in these organisms. This study provides more information on the nature of intrinsic antimicrobial resistance in these opportunistic pathogens.
2. Results
2.1. Vancomycin Susceptibility, Live Microscopy, and Vancomycin Survival Assays
The vancomycin MIC and MBC of E. anophelis R26 are shown in Table 1. Compared to other Gram-negative organisms [19], the vancomycin MIC and MBC for E. anophelis R26 appeared relatively low (Table 1), yet were in general, higher than the concentrations vancomycin can reach in certain tissues [24]. A representative sample of E. anophelis exposed to 1.5 X the vancomycin MIC for 4 h is shown in Figure 1. Exposure gave rise to a large percentage (78.44%) of lightly colored “empty” cells (white arrows in Figure 1) and we also noted numerous cells that demonstrated blebbing of the cell membrane (black arrow in Figure 1) which is indicative of bacterial cell wall degradation. Following 2 h of incubation, E. anophelis CFUs/mL began to decline precipitously in the 1.5 X MIC and the MBC culture up to 8 h (Figure 2). These cultures then rebounded to reach similar CFUs/mL as the control culture by the 24 h timepoint (Figure 2).
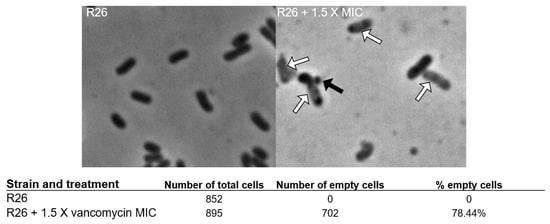
Figure 1.
Representative light microscopy pictures of R26 controls and R26 challenged with 1.5 X the vancomycin MIC. Black arrow points to representative membrane bleb, white arrows point to empty cells.
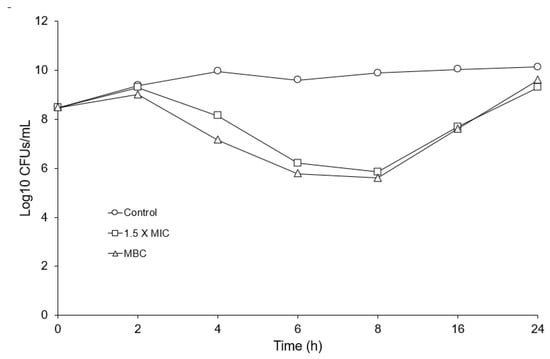
Figure 2.
Vancomycin growth survival assay of R26 challenged with 1.5 X MIC and the vancomycin MBC, showing surviving Log10 CFUs/mL over time.
2.2. Isolation of Vancomycin-Selected Mutants
In studies with Escherichia coli and Staphylococcus aureus, the mutation frequency for antibiotic resistance was reported to be between 10−5 and 10−9 [25,26,27], which means that one cell in population of 105 to 109 cells possessed spontaneous mutation(s) that granted antibiotic resistance. Vancomycin-selected mutants of E. anophelis appeared on media containing 16 mg/L vancomycin at a frequency of 10−4, which represented an unusually high frequency (Table 1). Three randomly chosen vancomycin-selected R26 mutants (R26VS1, R26VS2 and R26VS3) demonstrated higher vancomycin MICs and MBCs compared to parent strain R26 (Table 1). Gradient plate analysis (Table 2) also revealed that all R26 vancomycin-selected mutants demonstrated reduced susceptibility to ciprofloxacin, clindamycin, and rifampin. Under the microscope, we noted that the R26 parent cells had a longer rod length (2.37 ± 0.59 mm, n = 1652) compared to R26VS1 (2.09 mm ± 0.49 mm, n = 3417, p < 0.001) and R26VS2 (2.11 ± 0.47 mm, n = 2138, p < 0.001).
2.3. Mutations Associated with Vancomycin Resistance
Genome sequencing revealed that all three randomly picked R26 vancomycin-selected mutants possess a single cytosine insertion in a PadR-family helix-turn-helix transcriptional regulator gene [28,29,30,31] encoding a 112 aa product (WP_009089502) we have termed the “vancomycin-susceptibility regulator 1” or vsr1 (Figure 3). vsr1 overlaps by 7 bps and forms a putative bicistronic operon with ORF551 (Figure 3) which encodes a 551 aa protein (WP_009089500.1) that possesses five transmembrane-spanning regions and an internal small phage shock protein (58 aa)-conserved domain. Bioinformatic analysis of the upstream vsr1 promoter region did not reveal a PadR binding consensus site, but it did uncover a putative binding site for a helix-turn-helix SoxS regulatory protein, a RpoD sigma factor binding site, and -35 and -10 consensus sequences (Figure 3).
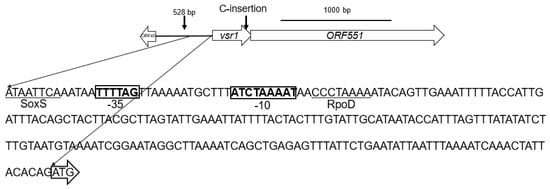
Figure 3.
E. anopheles vsr1-ORF551 operon showing location of cytosine (C) insertion at the end of vsr1 in R26 vancomycin-selected mutants and the 233 base pair promoter sequence upstream of vsr1. The vsr1 start codon is surrounded by an open arrow, the SoxS and RpoD sigma factor consensus binding sites are indicated (underlined) as are −35 and −10 regions (bolded in boxes) in the vsr1 promoter region.
The insertion of the cytosine in vsr1 (Figure 3) resulted in a frameshift mutation that caused three amino acid substitutions (R75 → T75, Y77 → I77, and Y78 → L78) and introduced a premature stop codon at aa position 79 in Vsr1 (Figure 4). This nonsense mutation leads to the deletion of the C-terminal 34 amino acids in Vsr1 which contains an important PadR dimerization domain [32]. To determine conserved motifs within Elizabethkingia PadR homologues, the amino acid sequences for 293 Elizabethkingia PadRs were downloaded from NCBI and compared via multiple alignments with the program mafft [33], and the data was visualized using WebLogo [34]. This analysis indicated that all Elizabethkingia PadRs share 41 conserved amino acids (or demonstrate 35% amino acid identity) with 100% conservation of a RKYY motif that is the beginning of the deletion observed in the vancomycin selected E. anophelis mutants (Figure 4).

Figure 4.
Sequence logo produced by alignment of 293 PadR sequences from all known Elizabethkingia species revealing highly conserved areas (positions represented by single large letters). The conserved RKYY motif where the truncation of Vsr1 occurs in all vancomycin-selected mutants is indicated by the black bar above amino acids.

Table 1.
Strains utilized in study.
Table 1.
Strains utilized in study.
| Strain | Parent Strain | Vancomycin Selection Concentration (mg/L) | Mutation Frequency | Vancomycin MIC (mg/L) | Vancomycin MBC (mg/L) | Ref. |
|---|---|---|---|---|---|---|
| E. anophelis R26 | 8 | 16 | [34] | |||
| R26VS1 | R26 | 16 | 4.33 × 10−4 | 128 | >256 | This study |
| R26VS2 | R26 | 16 | 4.33 × 10−4 | 64 | 128 | This study |
| R26VS3 | R26 | 16 | 4.33 × 10−4 | 64 | 128 | This study |

Table 2.
Gradient plate antibiotic susceptibility analysis.
Table 2.
Gradient plate antibiotic susceptibility analysis.
| Strain | Ciprofloxacin 0 → 0.5 mg/L | Clindamycin 0 → 1 mg/L | Rifampin 0 → 0.25 mg/L | Vancomycin 0 → 64 mg/L |
|---|---|---|---|---|
| R26 | 3.67 ± 0.33 A | 31.00 ± 1.15 A | 41.33 ± 1.76 A | 6.33 ± 0.67 |
| R26 VS1 | 7.67 ± 0.67 BC | 65.67 ± 1.45 B | 63.67 ± 2.60 B | 90.00 ± 0.00 |
| R26 VS2 | 7.00 ± 0.58 C | 52.67 ± 1.20 C | 70.33 ± 2.03 B | 89.00 ± 1.00 |
| R26 VS3 | 9.67 ± 0.33 B | 61.67 ± 2.19 B | 80.33 ± 1.45 C | 90.00 ± 0.00 |
One-way ANOVA with groups (A, B, and or C) displaying significant differences in susceptibility to the individual antibiotics ciprofloxacin, clindamycin, and rifampin (p < 0.05). Because strains grew to the top of the vancomycin gradient, statistical analyses could not be performed.
We then completed a phylogeny of the PadRs found in the genomes of five Elizabethkingia-type strains compared to three PadRs (PadR1, PadR2 and PadR3) found within a high quality Burkholderia cepacia complete genome (Figure 5). This analysis revealed the presence of two structurally distinct PadR subfamilies (PadR1 and PadR2) in the Elizabethkingia and Vsr1 belongs to the PadR1 subfamily. The PadR1 subfamily is characterized by the separation of the DNA binding domain in the N-terminal from the dimerization domain in the C-terminal region. An ORF551 gene lies upstream of all padR1s in all five Elizabethkingia genomes examined (see materials methods Section 4.4). The gene products encoded for by these genes exhibit 95.54 to 100% aa identity for Vsr1 and 89.67% to 100% aa identity for ORF155, demonstrating that the putative vsr1-ORF551 operon is conserved in the Elizabethkingia.
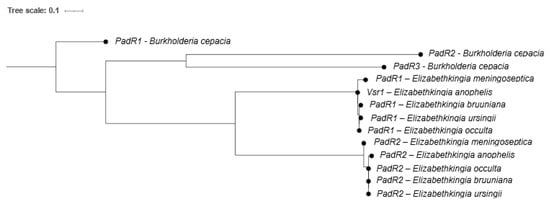
Figure 5.
Maximum likelihood phylogeny of PadR amino acid sequences from Elizabethkingia spp. PadR genes from Burkholderia cepacia BC16 are included as an outgroup.
2.4. qPCR Analysis of vrs1 Expression
Compared to parent strain R26, the expression levels of both vsr1 and ORF551 were increased in R26VS1 (224.4 ± 0.3 and 99.73 ± 0.1, respectively) and R26VS2 (272.4 ± 0.7 and 80.44 ± 0.1, respectively; N = 3, ±standard error) in drug-free cultures. These genes were also upregulated following vancomycin induction in R26VS1 and R26VS2 (Table 3), and only vsr1 was upregulated by vancomycin induction in R26 (Table 3).

Table 3.
Relative gene expression in vancomycin-induced cultures compared to uninduced.
3. Discussion
The vancomycin MIC and MBC for E. anophelis appeared to be relatively low for Gram-negative organisms and this organism responded to a growth inhibitory vancomycin concentration in a fashion that indicates the inhibition of cell wall synthesis. The formation of empty R26 cells with associated membrane blebbing following vancomycin challenge was similar to those reported by Huang et al. [35] who challenged vancomycin-susceptible E. coli mutants with vancomycin and noted similar abnormalities. The vancomycin growth survival challenge, however, indicated that E. anophelis cell populations were selected that were refractory to the action of vancomycin and E. anophelis vancomycin-selected mutants appeared on media containing a growth inhibitory vancomycin concentration at unusually high mutation frequencies. These findings demonstrate the ineffectiveness of vancomycin action against a population of E. anophelis and lends support to studies demonstrating that the Elizabethkingia display intermediate to full resistance to vancomycin [18,21,22]. We provide additional evidence that the vancomycin-selected mutants also demonstrated reduced susceptibility to other antimicrobials. The relative ease of selection for elevated vancomycin resistance and multiple antibiotic susceptibility mechanism should raise concern since vancomycin has been used to treat Elizabethkingia infections.
All three randomly chosen vancomycin-selected R26 mutants examined had the same mutation within vsr1, which encodes a PadR homologue that is located in the putative bicistronic operon vsr1-ORF551, an operon that is present in all Elizabethkinigia analyzed. Sequence analysis of 293 Elizabethkingia putative PadR proteins revealed these proteins exhibit a great deal of amino acid conservation and a highly conserved RKYY motif in the C-terminal domain of Elizabethkingia PadRs, which represents the beginning of the deletion in Vsr1 observed in all vancomycin-selected mutants of E. anophelis. Based on this bioinformatic analysis, we hypothesized that the truncated Vsr1 missing the C-terminal 34 amino acids and the conserved RKYY motif in vancomycin selected mutants cannot bind DNA and Vsr1 activity is compromised in these mutants. Our data also demonstrated that the vsr1-ORF551 operon was upregulated in vancomycin-selected mutants, suggesting that Vsr1 acts in one way or another as a repressor of the vsr1-ORF551 operon. However, no PadR binding site was identified upstream of the vsr1 start codon which was not wholly unexpected given the diversity of PadR regulators and therefore the sequences they bind [28,29,30,31], and the documented difficulties in promoter prediction for less well-studied organisms. Analysis of the promoter region upstream of vsr1-ORF551 did however identify a potential binding site for another helix-turn-helix SoxS type regulatory protein, and it should be noted that soxS genes have been implicated to play a role in the regulation of genes associated with intrinsic multiple antimicrobial resistance in multiple Gram-negative bacterial species [36,37,38,39].
The Vsr1 and ORF155 proteins of Elizabethkingia spp. are highly conserved and the genes encoding these proteins demonstrated synteny among the species analyzed. Additional experimentation is however required to determine if vsr1-ORF551 operons are associated with intrinsic antimicrobial resistance in other Elizabethkingia. In addition, we identified two distinct Elizabethkingia PadR subfamilies suggesting that these diverged gene families likely provide distinct functions to the cell.
PadR homologues in Gram-positive bacteria control genes encoding multidrug efflux pumps [28,29,30,31] and a padR operon in Streptococcus pneumoniae expressed several membrane proteins with unknown functions that control vancomycin tolerance [40]. PadR homologues have a structure that is similar to the MarR family of proteins (which includes SoxS), which have been reported to control intrinsic multiple antimicrobial resistance in the Enterobacteriaceae. In E. coli, MarR regulates the expression of MarA, which controls the production and activity of outer membrane porins and RND efflux pumps such as AcrAB-TolC, leading to a reduction in antimicrobial accumulation [41,42,43]. To the best of our knowledge, this is the first description of a PadR homologue playing a role in antimicrobial resistance in a Gram-negative organism.
The vancomycin-selected mutants were shorter than the parent strain and since cell morphology is maintained by peptidoglycan structure [44], we suggest that cell wall biosynthesis is affected in these mutants. ORF551 encodes a protein with membrane spanning regions and possesses a small phage shock protein-conserved domain. In Escherichia coli, it has been proposed that the phage shock protein system can detect and mitigate issues that affect inner membrane permeability [45]. Since Gram-negative organisms do not efflux vancomycin and are reported to be vancomycin-resistant due to an inherent outer membrane permeability barrier [17], we hypothesize that the vancomycin-selected mechanism affects membrane permeability in E. anophelis.
We now intend to determine the effects of the inactivation and complementation of the vsr1-ORF551 operon on intrinsic antimicrobial resistance and antimicrobial accumulation.
4. Materials and Methods
4.1. Bacterial Strains, Growth Conditions, Vancomycin-Selected Mutants, Antibiotic Susceptibility Testing, and Live Cell Microscopy
The type strain Elizabethkingia anophelis R26 isolated from the midgut of Anopheles gambiae [46] was used for this study. A complete list of bacterial strains used in this study can be found in Table 1. All freezer stocks were maintained in heart infusion broth (HIB) containing 20% v/v glycerol (final concentration) at −80 °C. Working stocks were maintained on heart infusion agar (HIA; Remel, San Diego, CA, USA) supplemented with 5% defibrinated rabbit blood (Hemostat Laboratories, Dixon, CA, USA). All overnight cultures were prepared by inoculating a single colony into HIB or Mueller-Hinton (MHB) broth, followed by overnight incubation (37 °C, 200 rpm, 18 h). All chemicals and antibiotics were purchased from MilliporeSigma (St. Louis, MO, USA).
Vancomycin-selected mutants of E. anophelis R26 were isolated by plating diluted HIB overnight cultures onto HIA plates supplemented with 16 mg/L of vancomycin (MilliporeSigma, St. Louis, MO, USA). Following overnight incubation (37 °C) single isolated colonies were picked, passaged three times on HIA, and HIB glycerol freezer archive stocks were prepared. Mutation frequencies (Table 1) were defined as the proportion of the colonies growing on vancomycin selection plates to the total viable colony count.
Vancomycin MIC and MBC concentrations were determined by broth dilution following standard CLSI guidelines [47] and relative antibiotic susceptibility was compared utilizing the antimicrobial gradient plate analysis as described previously [48,49]. Distances grown on antimicrobial gradient plates (Table 1) were analyzed by one-way ANOVA with groups displaying significant differences (p < 0.05) and subsequently differentiated using Tukey’s Honestly Significant Differences post hoc testing [50]. All analysis was performed using JMP Pro (version 14; SAS Institute Inc., Carey, NC).
Vancomycin growth survival assays were performed in 50 mL flasks containing MHB cultures initiated with overnight culture (beginning OD600nm = 0.01) containing no addition or vancomycin (1.5 X MIC and MBC). These flasks were then incubated with shaking (200 rpm, 37 °C) and surviving
CFUs/mL were determined over time by plating dilutions onto Mueller-Hinton agar (MHA) followed by overnight incubation (37 °C).
For live cell microscopy, overnight cultures were diluted in fresh MHB to reach an OD600nm = 0.01 and incubated for 3 h (37 °C, 200 rpm). Following incubation, vancomycin was added to a final concentration of 1.5 X the MIC for each isolate and a 1 µL aliquot was transferred to a sterile 1% agar pad at 25 °C for visualization. Vancomycin-challenged cultures were then incubated for 4 h (37 °C, 200 rpm), with 1 µL aliquots removed for imaging at 2 h and 4 h post-challenge. Phase contrast images were collected on a NikonNi-E epifluorescent microscope equipped with a 100X/1.45 NA objective (Nikon, Tokyo, Japan), Zyla 4.2 plus cooled sCMOS camera (Andor, Belfast, Northern Ireland), and NIS Elements software (Nikon).
4.2. Whole Genome Sequencing and Identification of Mutations Associated with Enhanced Vancomycin Resistance
Upon arrival at CDC, strains were grown on HIA (Difco, Tokyo, Japan) supplemented with 5% rabbit blood (Hemostat Laboratories) at 35 °C. Genomic DNA was extracted using the CTAB protocol provided by the Department of Energy’s Joint Genome Institute [51]. E. anophelis vancomycin-selected mutant libraries were prepared using the NexteraTM DNA Flex kit according to manufacturer’s instructions, and genomes were sequenced using a 2 × 150 paired end protocol using an Illumina iSeq 100 Sequencing System (Illumina, Inc., San Diego, CA, USA).
Paired raw reads were trimmed to remove adapter sequences and for quality control using a quality threshold of 0.02 and 0 allowable ambiguous nucleotides. Trimmed reads were then mapped to the complete R26 genome using the default options and the consensus sequence for each isolate was extracted. All reported mutations were verified by inspection of the raw reads. All trimming and mapping steps were performed using CLC Genomics Workbench v11.0.1. Consensus sequences were annotated using the Rapid Annotations Using Subsystems Technology (RAST) server [52]. Regulatory elements were predicted using the BPROM program [53] while the identity and putative functional domains of hypothetical proteins were investigated using nucleotide and protein Basic Local Alignment Search Tool (BLAST) [54].
4.3. Quantitative Real-Time qPCR Analysis
Overnight HIB cultures (in triplicate) were used to inoculate (0.1% v/v inoculum) fresh HIB which was incubated (37 °C, 200 rpm) until an OD600nm = 0.5. These cultures were then divided into equal portions, and one portion of the culture was induced with 4 mg/L vancomycin. All cultures were then incubated (37 °C, 200 rpm) and the cells were harvested after 30 min growth. Total RNA was isolated utilizing Trizol (Invitrogen, ThermoFisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The isolated RNA was then solubilized in the RNase-free water and treated with DNA-free(tm) (Ambion, Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. RNA quantity and quality were then measured on a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). cDNA synthesis was carried out with isolated RNA (25 ng) using the High-Capacity cDNA Reverse Transcription Kit according to the manufacturer’s proto-cols (Applied Biosystems™, Thermo Fisher Scientific, Vilnius, Lithuania). The cDNA samples were then diluted five-fold and the relative quantification of the target genes in triplicate was carried out using the LightCycler® 96 real-time PCR system (Roche Diagnostics, Mannheim, Germany) and iQ SYBR® Green Supermix as per the manufacturer’s recommended protocols (Bio-Rad, Hercules, CA, USA). Gene expression was then normalized using species-specific rpoB primers, expression levels were calculated using the 2−ΔΔCT method [55], and results are presented as the means and standard errors of the data. All the primers used for RT-qPCR are shown in Table 4.

Table 4.
List of primers used in qRT-PCR analysis.
4.4. Promoter, Structural Sequence and Phylogenetic Analyses
All vsr1 promoter sequences and potential DNA binding protein binding sites were identified with a previously described program [53]. Amino acid sequences for 293 putative Elizabethkingia PadR genes were downloaded from NCBI, aligned by mafft [56], and then visualized using WebLogo [34]. To further investigate the differences between the two PadR families in Elizabethkingia, PadR amino acid sequences were downloaded from the NCBI RefSeq for each Elizabethkingia species (E. anophelis R26, accession # GCF_002023665.2; E. bruuniana FDAARGOS_1031, accession # GCF_016599835.1; E. meningoseptica G4120, accession # GCF_002022145.1; E. occulta G4070, accession # GCF_002023715.1; and E. ursingii CSID_3000516135, accession # GCF_002023405.1), along with three PadR sequences from Burkholderia cepacia (strain BC16, accession # GCF_009586235.1) to serve as a comparator. Amino acid sequences were aligned by mafft, and maximum likelihood phylogenies were created by IQ-Tree using the Q.pfam + I model, with the -bb and -alrt options set for 10,000 bootstraps each.
5. Conclusions
In the laboratory, vancomycin was readily selected for elevated vancomycin resistance, intrinsic multiple antimicrobial reduced susceptibility, and altered cell wall morphology in E. anopheles. A mutation within a padR homologue (vsr1) within a putative bicistronic operon vsr1-ORF551 and vsr1-ORF551 upregulation, was observed in vancomycin-selected mutants. The vsr1-ORF551 operon is conserved in the Elizabethkingia and we identified two PadR genes within genomes of this genus. This work represents the first time a padR homologue and a gene like ORF551 have been associated with intrinsic antimicrobial resistance in a Gram-negative organism. Overall, this work adds to the knowledge of mutation-mediated intrinsic antimicrobial resistance mechanisms of the Elizabethkingia and adds credence to the literature that demonstrated that these organisms are vancoycin-resistant.
Author Contributions
J.E.G., W.L.J., S.K.G., S.M. and R.M.M. designed the study. W.L.J., S.K.G., S.M., A.C.N. and J.R.M. carried out all the experimentation. J.E.G. led manuscript development. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by funding from the Oklahoma Agricultural Experiment Station to John E. Gustafson and by an Institutional Development Award from the National Institute of General Medical Sciences of the National Institutes of Health to Randy Morgenstein (P20 GM134973).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Sequence data for strains R26-VER1, R26-VER2, and R26-VER3 have been deposited to NCBI BioProjects PRJNA875105, PRJNA875110, and PRJNA875117 respectively, and released with the accession numbers JANZKF000000000, JANZKG000000000, and JANZKH000000000.
Acknowledgments
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention (CDC). The mention of company names or products does not constitute endorsement by CDC.
Conflicts of Interest
The authors declare of conflicst of interest.
References
- King, E.O. Studies on a group of previously unclassified bacteria associated with meningitis in infants. Am. J. Clin. Pathol. 1959, 31, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, A.C.; Gulvik, C.A.; Whitney, A.M.; Humrighouse, B.W.; Graziano, J.; Emery, B.; Bell, M.; Loparev, V.; Juieng, P.; Gartin, J.; et al. Revisiting the taxonomy of the genus Elizabethkingia using whole-genome sequencing, optical mapping, and MALDI-TOF, along with proposal of three novel Elizabethkingia species: Elizabethkingia bruuniana sp. nov., Elizabethkingia ursingii sp. nov., and Elizabethkingia occulta sp. nov. Antonie Leeuwenhoek 2018, 111, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Chew, K.L.; Cheng, B.; Lin, R.T.P.; Teo, J.W.P. Elizabethkingia anophelis Is the Dominant Elizabethkingia Species Found in Blood Cultures in Singapore. J. Clin. Microbiol. 2018, 56, e01445-17. [Google Scholar] [CrossRef] [PubMed]
- Perrin, A.; Larsonneur, E.; Nicholson, A.C.; Edwards, D.J.; Gundlach, K.M.; Whitney, A.M.; Gulvik, C.A.; Bell, M.E.; Rendueles, O.; Cury, J.; et al. Evolutionary dynamics and genomic features of the Elizabethkingia anophelis 2015 to 2016 Wisconsin outbreak strain. Nat. Commun. 2017, 8, 15483. [Google Scholar] [CrossRef] [PubMed]
- Kyritsi, M.A.; Mouchtouri, V.A.; Pournaras, S.; Hadjichristodoulou, C. First reported isolation of an emerging opportunistic pathogen (Elizabethkingia anophelis) from hospital water systems in Greece. J. Water Health 2018, 16, 164–170. [Google Scholar] [CrossRef]
- Hem, S.; Jarocki, V.M.; Baker, D.J.; Charles, I.G.; Drigo, B.; Aucote, S.; Donner, E.; Burnard, D.; Bauer, M.J.; Harris, P.N.A.; et al. Genomic analysis of Elizabethkingia species from aquatic environments: Evidence for potential clinical transmission. Curr. Res. Microb. Sci. 2022, 3, 100083. [Google Scholar] [CrossRef]
- Hwang, J.H.; Kim, J.; Kim, J.H.; Mo, S. Elizabethkingia argenteiflava sp. nov., isolated from the pod of soybean, Glycine max. Int. J. Syst. Evol. Microbiol. 2021, 71, 004767. [Google Scholar] [CrossRef]
- Liu, K.M.; Chang, H.L.; Hsu, M.H.; Lin, Y.Z.; Lee, Y.L.; Chen, Y.T. Complete Genome Sequencing of Elizabethkingia sp. Strain 2-6. Microbiol. Resour. Announc. 2019, 8. [Google Scholar] [CrossRef]
- Colapietro, M.; Endimiani, A.; Sabatini, A.; Marcoccia, F.; Celenza, G.; Segatore, B.; Amicosante, G.; Perilli, M. BlaB-15, a new BlaB metallo-beta-lactamase variant found in an Elizabethkingia miricola clinical isolate. Diagn. Microbiol. Infect. Dis. 2016, 85, 195–197. [Google Scholar] [CrossRef]
- Jian, M.J.; Cheng, Y.H.; Chung, H.Y.; Cheng, Y.H.; Yang, H.Y.; Hsu, C.S.; Perng, C.L.; Shang, H.S. Fluoroquinolone resistance in carbapenem-resistant Elizabethkingia anophelis: Phenotypic and genotypic characteristics of clinical isolates with topoisomerase mutations and comparative genomic analysis. J. Antimicrob. Chemother. 2019, 74, 1503–1510. [Google Scholar] [CrossRef]
- Matyi, S.A.; Hoyt, P.R.; Ayoubi-Canaan, P.; Hasan, N.A.; Gustafson, J.E. Draft Genome Sequence of Strain ATCC 33958, Reported to Be Elizabethkingia miricola. Genome Announc. 2015, 3, e00828-15. [Google Scholar] [CrossRef]
- Zgurskaya, H.I.; Weeks, J.W.; Ntreh, A.T.; Nickels, L.M.; Wolloscheck, D. Mechanism of coupling drug transport reactions located in two different membranes. Front. Microbiol. 2015, 6, 100. [Google Scholar] [CrossRef]
- Mazzariol, A.; Tokue, Y.; Kanegawa, T.M.; Cornaglia, G.; Nikaido, H. High-level fluoroquinolone-resistant clinical isolates of Escherichia coli overproduce multidrug efflux protein AcrA. Antimicrob. Agents Chemother. 2000, 44, 3441–3443. [Google Scholar] [CrossRef]
- Nieto, M.; Perkins, H.R. Modifications of the acyl-D-alanyl-D-alanine terminus affecting complex-formation with vancomycin. Biochem. J. 1971, 123, 789–803. [Google Scholar] [CrossRef]
- Bruniera, F.R.; Ferreira, F.M.; Saviolli, L.R.; Bacci, M.R.; Feder, D.; da Luz Goncalves Pedreira, M.; Sorgini Peterlini, M.A.; Azzalis, L.A.; Campos Junqueira, V.B.; Fonseca, F.L. The use of vancomycin with its therapeutic and adverse effects: A review. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 694–700. [Google Scholar]
- Jean, S.S.; Hsieh, T.C.; Ning, Y.Z.; Hsueh, P.R. Role of vancomycin in the treatment of bacteraemia and meningitis caused by Elizabethkingia meningoseptica. Int. J. Antimicrob. Agents 2017, 50, 507–511. [Google Scholar] [CrossRef]
- Krishnamoorthy, G.; Leus, I.V.; Weeks, J.W.; Wolloscheck, D.; Rybenkov, V.V.; Zgurskaya, H.I. Synergy between Active Efflux and Outer Membrane Diffusion Defines Rules of Antibiotic Permeation into Gram-Negative Bacteria. mBio 2017, 8, e01172-17. [Google Scholar] [CrossRef]
- Chiu, C.T.; Lai, C.H.; Huang, Y.H.; Yang, C.H.; Lin, J.N. Comparative Analysis of Gradient Diffusion and Disk Diffusion with Agar Dilution for Susceptibility Testing of Elizabethkingia anophelis. Antibiotics 2021, 10, 450. [Google Scholar] [CrossRef]
- Dorr, T.; Delgado, F.; Umans, B.D.; Gerding, M.A.; Davis, B.M.; Waldor, M.K. A Transposon Screen Identifies Genetic Determinants of Vibrio cholerae Resistance to High-Molecular-Weight Antibiotics. Antimicrob. Agents Chemother. 2016, 60, 4757–4763. [Google Scholar] [CrossRef]
- Fass, R.J.; Barnishan, J. In vitro susceptibilities of nonfermentative gram-negative bacilli other than Pseudomonas aeruginosa to 32 antimicrobial agents. Rev. Infect. Dis. 1980, 2, 841–853. [Google Scholar] [CrossRef]
- Chang, T.Y.; Chen, H.Y.; Chou, Y.C.; Cheng, Y.H.; Sun, J.R. In vitro activities of imipenem, vancomycin, and rifampicin against clinical Elizabethkingia species producing BlaB and GOB metallo-beta-lactamases. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 2045–2052. [Google Scholar] [CrossRef]
- Kuo, S.C.; Tan, M.C.; Huang, W.C.; Wu, H.C.; Chen, F.J.; Liao, Y.C.; Wang, H.Y.; Shiau, Y.R.; Lauderdale, T.L. Susceptibility of Elizabethkingia spp. to commonly tested and novel antibiotics and concordance between broth microdilution and automated testing methods. J. Antimicrob. Chemother. 2021, 76, 653–658. [Google Scholar] [CrossRef]
- Lin, X.H.; Xu, Y.H.; Sun, X.H.; Huang, Y.; Li, J.B. Genetic diversity analyses of antimicrobial resistance genes in clinical Chryseobacterium meningosepticum isolated from Hefei, China. Int. J. Antimicrob. Agents 2012, 40, 186–188. [Google Scholar] [CrossRef]
- Rybak, M.J. The pharmacokinetic and pharmacodynamic properties of vancomycin. Clin. Infect. Dis. 2006, 42 (Suppl. S1), S35–S39. [Google Scholar] [CrossRef]
- Baquero, M.R.; Nilsson, A.I.; Turrientes Mdel, C.; Sandvang, D.; Galan, J.C.; Martinez, J.L.; Frimodt-Moller, N.; Baquero, F.; Andersson, D.I. Polymorphic mutation frequencies in Escherichia coli: Emergence of weak mutators in clinical isolates. J. Bacteriol. 2004, 186, 5538–5542. [Google Scholar] [CrossRef]
- Price, C.T.; Gustafson, J.E. Increases in the mutation frequency at which fusidic acid-resistant Staphylococcus aureus arise with salicylate. J. Med. Microbiol. 2001, 50, 104–106. [Google Scholar] [CrossRef][Green Version]
- Gustafson, J.E.; Candelaria, P.V.; Fisher, S.A.; Goodridge, J.P.; Lichocik, T.M.; McWilliams, T.M.; Price, C.T.; O’Brien, F.G.; Grubb, W.B. Growth in the presence of salicylate increases fluoroquinolone resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1999, 43, 990–992. [Google Scholar] [CrossRef]
- Lubelski, J.; de Jong, A.; van Merkerk, R.; Agustiandari, H.; Kuipers, O.P.; Kok, J.; Driessen, A.J. LmrCD is a major multidrug resistance transporter in Lactococcus lactis. Mol. Microbiol. 2006, 61, 771–781. [Google Scholar] [CrossRef]
- Huillet, E.; Velge, P.; Vallaeys, T.; Pardon, P. LadR, a new PadR-related transcriptional regulator from Listeria monocytogenes, negatively regulates the expression of the multidrug efflux pump MdrL. FEMS Microbiol. Lett. 2006, 254, 87–94. [Google Scholar] [CrossRef]
- Madoori, P.K.; Agustiandari, H.; Driessen, A.J.; Thunnissen, A.M. Structure of the transcriptional regulator LmrR and its mechanism of multidrug recognition. EMBO J. 2009, 28, 156–166. [Google Scholar] [CrossRef]
- Hauf, S.; Moller, L.; Fuchs, S.; Halbedel, S. PadR-type repressors controlling production of a non-canonical FtsW/RodA homologue and other trans-membrane proteins. Sci. Rep. 2019, 9, 10023. [Google Scholar] [CrossRef]
- Fibriansah, G.; Kovacs, A.T.; Pool, T.J.; Boonstra, M.; Kuipers, O.P.; Thunnissen, A.M. Crystal structures of two transcriptional regulators from Bacillus cereus define the conserved structural features of a PadR subfamily. PLoS ONE 2012, 7, e48015. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Huang, K.C.; Mukhopadhyay, R.; Wen, B.; Gitai, Z.; Wingreen, N.S. Cell shape and cell-wall organization in Gram-negative bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 19282–19287. [Google Scholar] [CrossRef]
- Aly, S.A.; Boothe, D.M.; Suh, S.J. A novel alanine to serine substitution mutation in SoxS induces overexpression of efflux pumps and contributes to multidrug resistance in clinical Escherichia coli isolates. J. Antimicrob. Chemother. 2015, 70, 2228–2233. [Google Scholar] [CrossRef]
- Bratu, S.; Landman, D.; George, A.; Salvani, J.; Quale, J. Correlation of the expression of acrB and the regulatory genes marA, soxS and ramA with antimicrobial resistance in clinical isolates of Klebsiella pneumoniae endemic to New York City. J. Antimicrob. Chemother. 2009, 64, 278–283. [Google Scholar] [CrossRef]
- McMurry, L.M.; Oethinger, M.; Levy, S.B. Overexpression of marA, soxS, or acrAB produces resistance to triclosan in laboratory and clinical strains of Escherichia coli. FEMS Microbiol. Lett. 1998, 166, 305–309. [Google Scholar] [CrossRef]
- Perez, A.; Poza, M.; Aranda, J.; Latasa, C.; Medrano, F.J.; Tomas, M.; Romero, A.; Lasa, I.; Bou, G. Effect of transcriptional activators SoxS, RobA, and RamA on expression of multidrug efflux pump AcrAB-TolC in Enterobacter cloacae. Antimicrob. Agents Chemother. 2012, 56, 6256–6266. [Google Scholar] [CrossRef]
- Liu, X.; Li, J.W.; Feng, Z.; Luo, Y.; Veening, J.W.; Zhang, J.R. Transcriptional Repressor PtvR Regulates Phenotypic Tolerance to Vancomycin in Streptococcus pneumoniae. J. Bacteriol. 2017, 199, e00054-17. [Google Scholar] [CrossRef]
- Cohen, S.P.; Hachler, H.; Levy, S.B. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J. Bacteriol. 1993, 175, 1484–1492. [Google Scholar] [CrossRef]
- Hachler, H.; Cohen, S.P.; Levy, S.B. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 1991, 173, 5532–5538. [Google Scholar] [CrossRef]
- Sharma, P.; Haycocks, J.R.J.; Middlemiss, A.D.; Kettles, R.A.; Sellars, L.E.; Ricci, V.; Piddock, L.J.V.; Grainger, D.C. The multiple antibiotic resistance operon of enteric bacteria controls DNA repair and outer membrane integrity. Nat. Commun. 2017, 8, 1444. [Google Scholar] [CrossRef]
- Young, K.D. Approaching the physiological functions of penicillin-binding proteins in Escherichia coli. Biochimie 2001, 83, 99–102. [Google Scholar] [CrossRef]
- Flores-Kim, J.; Darwin, A.J. The Phage Shock Protein Response. Annu. Rev. Microbiol. 2016, 70, 83–101. [Google Scholar] [CrossRef]
- Kampfer, P.; Matthews, H.; Glaeser, S.P.; Martin, K.; Lodders, N.; Faye, I. Elizabethkingia anophelis sp. nov., isolated from the midgut of the mosquito Anopheles gambiae. Int. J. Syst. Evol. Microbiol. 2011, 61, 2670–2675. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, Approved Standard, 11th ed.; M07-A11; CLSI: Wayne, PA, USA, 2018. [Google Scholar]
- Johnson, W.L.; Ramachandran, A.; Torres, N.J.; Nicholson, A.C.; Whitney, A.M.; Bell, M.; Villarma, A.; Humrighouse, B.W.; Sheth, M.; Dowd, S.E.; et al. The draft genomes of Elizabethkingia anophelis of equine origin are genetically similar to three isolates from human clinical specimens. PLoS ONE 2018, 13, e0200731. [Google Scholar] [CrossRef]
- O’Leary, J.O.; Langevin, M.J.; Price, C.T.; Blevins, J.S.; Smeltzer, M.S.; Gustafson, J.E. Effects of sarA inactivation on the intrinsic multidrug resistance mechanism of Staphylococcus aureus. FEMS Microbiol. Lett. 2004, 237, 297–302. [Google Scholar] [CrossRef]
- Tukey, J.W. Comparing individual means in the analysis of variance. Biometrics 1949, 5, 99–114. [Google Scholar] [CrossRef]
- JGI Bacterial DNA Isolation CTAB Protocol. Available online: https://jgi.doe.gov/user-programs/pmo-overview/protocols-sample-preparation-information/jgi-bacterial-dna-isolation-ctab-protocol-2012/ (accessed on 26 September 2023).
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Solayev, V.; Salamov, A. Automatic Annotation of Microbial Genomes and Metagenomic Sequences. In Metagenomics and Its Applications in Agriculture, Biomedicine and Environmental Studies; Li, R.W., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2011; pp. 61–78. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).