Abstract
Foodborne infections pose a substantial global threat, causing an estimated 600 million illnesses and resulting in approximately 420,000 deaths annually. Among the diverse array of pathogens implicated in these infections, Escherichia coli (E. coli), specifically the O157 strain (E. coli O157), emerges as a prominent pathogen associated with severe outbreaks. This study employs a comprehensive bibliometric analysis and scholarly review focused on E. coli O157 research. The bibliometric analysis highlights the significant role played by the United States in the E. coli O157 research domain. Further exploration underscores the noteworthy contributions of the researcher Doyle MP, whose body of work, consisting of 84 documents and an impressive H-Index of 49, reflects their substantial impact in the field. Recent research trends indicate a discernible shift towards innovative detection methods, exemplified by the adoption of CRISPR-CAS and Loop-Mediated Isothermal Amplification. Moreover, high-throughput whole-genome sequencing techniques are gaining prominence for the expeditious analysis of pathogenic E. coli strains. Scientists are increasingly exploring antimicrobial agents, including phage therapy, to address the challenges posed by antibiotic-resistant E. coli strains, thereby addressing critical concerns related to multi-drug resistance. This comprehensive analysis provides vital insights into the dynamic landscape of E. coli O157 research. It serves as a valuable resource for researchers, policymakers, and healthcare professionals dedicated to mitigating E. coli O157 outbreaks and advancing global public health strategies.
1. Introduction
Foodborne infections persist as a considerable global health menace. According to data from the World Health Organization (WHO), nearly 600 million individuals suffer from illnesses resulting from the consumption of contaminated food each year, resulting in an estimated 420,000 deaths [1]. These infections not only pose immediate health hazards but also generate substantial economic consequences, manifested in escalating healthcare costs and diminished workforce productivity. Among the myriad of pathogens linked to foodborne diseases, E. coli occupies a distinctive position. This bacterium displays a captivating dual nature, functioning both as a benign inhabitant of the gut and a pernicious pathogen, rendering it a pivotal focus of scientific inquiry [2].
E. coli encompasses a diverse array of strains, with several exhibiting benign attributes essential to physiological processes such as vitamin K synthesis and the prevention of detrimental bacterial colonization. Notably, the O157 strain has drawn attention due to its association with severe foodborne outbreaks. Investigative studies have illuminated the multifaceted adaptive stress response of E. coli O157 when introduced into the human food chain, underscoring its distinctive survival characteristics that significantly influence its epidemiology and ecology. The consumption of the E. coli O157 strain can precipitate a spectrum of gastrointestinal complications. The particularly infamous subtype, E. coli O157:H7, is distinguished by its Shinga toxin production, which disrupts protein synthesis in human cells, ultimately culminating in cellular demise. This pathological process can manifest in a spectrum of symptoms ranging from bloody diarrhea to life-threatening conditions such as hemolytic uremic syndrome (HUS) [3].
Various research endeavors are underway to comprehensively elucidate the epidemiology, pathogenesis, transmission dynamics, and preventive strategies associated with E. coli O157. Concurrently, investigations into the role of external factors in the dissemination of E. coli O157 are being conducted. Notably, a study in Ohio, USA, explored the potential involvement of European starlings in the transmission of E. coli O157 among dairy farms, emphasizing the intricate web of transmission routes [4]. Similarly, a study in Syria utilized Random Amplified Polymorphic DNA (RAPD) markers to assess the genetic diversity of E. coli O157 in leafy greens irrigated by the Aleppo River, underscoring the significance of water sources in transmission dynamics [5]. Innovative approaches, such as Multiplex-PCR, are being employed for the rapid detection and comprehension of E. coli O157, as demonstrated in a study focusing on poultry, which simultaneously identified Salmonella enteritidis, Shigella flexneri, and E. coli O157: H7, emphasizing the importance of poultry as a potential source [6,7]. E. coli O157, particularly the H7 subtype, is predominantly identified in livestock, with cattle serving as significant carriers. This association underscores the frequent outbreaks associated with the consumption of raw or undercooked beef products, including ground beef. However, the spectrum of contamination extends beyond livestock to encompass fresh produce, unpasteurized dairy, and water sources. As a preventive measure, a comprehensive approach is imperative, integrating stringent agricultural and manufacturing practices, adherence to proper cooking temperatures, and public education on safe food handling [8].
In response to the escalating antibiotic resistance observed among various E. coli strains, the scientific community is actively exploring alternative therapeutic strategies [9,10]. Probiotics, such as Lactobacillus rhamnosus, have emerged as potential candidates offering protection against these pathogenic strains [11]. Simultaneously, nanotechnology is gaining prominence in the battle against bacterial infections. Nanoparticles, characterized by their minute size and expansive surface area, exhibit an enhanced interaction with microbial cells, augmenting their antimicrobial efficacy. Noteworthy examples include chitosan nanoparticles [12,13,14], especially when fortified with antimicrobial peptides, and metal oxide nanoparticles such as zinc oxide nanoparticles [NO_PRINTED_FORM], which are being investigated for their potent antibacterial properties against E. coli O157:H7 [15]. Additionally, lipid-based nanoparticles, known for their biocompatibility, are being tailored for targeted antibiotic therapy to mitigate the risk of resistance development [16].
In the face of the enduring global challenge posed by foodborne infections, gaining a nuanced understanding of pathogens such as E. coli O157 assumes paramount significance. The dual nature of E. coli, marked by its extensive strain diversity and the profound implications of its pathogenic variants, necessitates comprehensive research endeavors. Leveraging tools such as bibliometric analysis and innovative methodologies, the scientific community is dedicated to unraveling the epidemiology, transmission dynamics, and potential interventions associated with E. coli O157.
Bibliometric analysis, utilizing statistical and visualization tools, plays a pivotal role in decoding the extensive literature concerning this pathogen. Notably, researchers employing bibliometric analysis have successfully identified the key authors, tracked the evolution of research themes, pinpointed seminal papers, and elucidated research gaps [17]. In this paper, the amalgamation of bibliometric analysis and scholarly reviews aims to furnish a holistic perspective on E. coli O157 research, thereby bridging knowledge gaps and illuminating avenues for future exploration in alignment with the overarching objectives of this study.
2. Literature Review
2.1. Early Discoveries and Their Implications
Foundational research on E. coli O157 has paved the way for a deeper understanding of this pathogenic strain and its implications for public health. One of the seminal studies in this field was conducted by Morgan in their paper titled “Hemorrhagic Colitis Associated with a Rare E. coli Serotype”, published in 1988 [18]. This groundbreaking work identified E. coli O157 as the causative agent of a severe form of foodborne illness characterized by bloody diarrhea. Morgan and colleagues’ research not only established the association between E. coli O157 and hemorrhagic colitis but also highlighted the significance of this strain as a public health concern. Their findings underscored the need for enhanced surveillance and research efforts to understand the pathogenesis and epidemiology of E. coli O157 infections. Furthermore, this early research laid the foundation for subsequent investigations into the mechanisms of E. coli O157 virulence, its reservoirs in the environment, and strategies for preventing and managing outbreaks. The implications of this foundational work have reverberated through the years, influencing public health policies and interventions aimed at mitigating the risks associated with E. coli O157 infections.
2.2. Epidemiological Shifts
E. coli O157, commonly referred to as E. coli O157, is a significant public health concern due to its association with foodborne disease outbreaks. E. coli O157 is responsible for approximately 63,000 cases of hemorrhagic colitis annually in the United States. A comprehensive analysis of databases and studies from 10 out of 14 World Health Organization subregions revealed a global incidence of E. coli, totaling 2.8 million cases each year [19]. This pathogenic bacterium is often linked to the consumption of undercooked foods, particularly those contaminated with cattle manure harboring the bacterium. The epidemiological landscape of E. coli O157 is complex, with various factors influencing its prevalence and virulence. E. coli O157 primarily resides in cattle, especially those raised for beef, colonizing their intestines without noticeable symptoms. However, the bacteria from their fecal matter can contaminate food and water sources, causing human outbreaks. Understanding how E. coli O157 colonizes cattle is crucial for effective mitigation. Recent studies have highlighted gaps in knowledge regarding factors regulating its colonization, interactions with the host’s microbiota, and the host’s immune responses. Disturbances like stress during cattle production likely contribute to the bacterium’s survival and proliferation in the intestine [20]. Additionally, research has raised concerns about laboratory-acquired infections. A notable case involved a researcher contracting a severe E. coli O157 infection from nalidixic acid-resistant strains used in the lab, highlighting the bacterium’s virulence and emphasizing the need for strict safety protocols. The study also revealed that certain antibiotics, when given in sublethal doses, could enhance toxin expression, suggesting the potential exacerbation of infection severity with antibiotic treatment [21]. To gain insights into the epidemiological shifts and genetic aspects of E. coli O157, researchers have utilized murine models. These controlled environments enable the study of intestinal colonization, host responses, and the impact of host physiological status and microbiota on colonization and disease [20]. Such research offers a valuable understanding of host–pathogen–microbiota interactions, potentially leading to rational mitigation strategies for E. coli O157 in cattle.
2.3. Pathogenesis of E. coli O157
E. coli O157 stands out as a highly potent strain of enterohemorrhagic E. coli (EHEC). Primarily, cattle are a major natural reservoir of the E. coli O157 strain [22]. Furthermore, these cattle transmit E. coli O157 either through farm or food products to humans, where they survive in the gastrointestinal tract and cause infection. Its virulence lies in its ability to inflict extensive harm within the host, leading to various conditions, from mild gastroenteritis to severe hemorrhagic colitis and life-threatening hemolytic uremic syndrome (HUS). The pathogenicity of E. coli O157:H7 involves an intricate series of steps, incorporating bacterial adherence, toxin production, and strategies to evade the immune system [23]. Figure 1 visually represents the pathogenesis of E. coli O157 infection in humans.
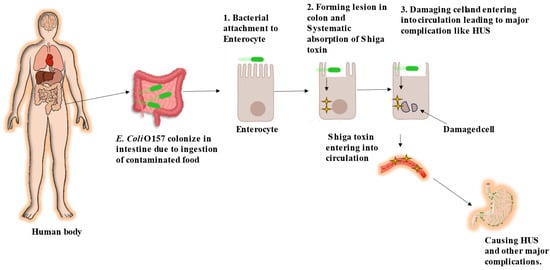
Figure 1.
Pathogenesis of E. coli O157 in human [19].
Various virulence factors play a role in the pathogenesis of E. coli O157 infection. Figure 2 visually represents the different virulence factors involved in each stage of E. coli O157 infection.
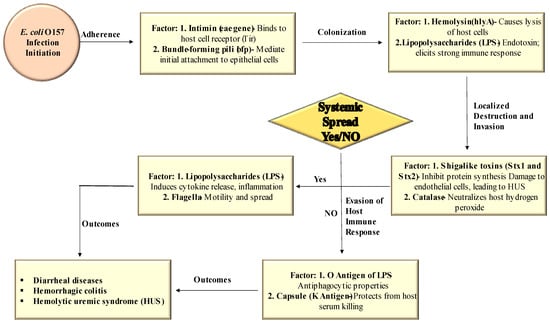
Figure 2.
Role of different virulence factors at each stage of E. coli O157 infection.
2.4. Antimicrobial Strategies against Escherichia coli O157: Traditional and Emerging Approaches
Escherichia coli O157 has become a notable public health issue, particularly in the realm of foodborne illnesses. Dealing with infections caused by this pathogen has proven to be difficult due to its virulence and escalating resistance to conventional antimicrobials [24]. This section concentrates on a variety of antimicrobial approaches employed against E. coli O157, encompassing both established treatments and inventive methods that have arisen in response to concerns about antibiotic resistance.
In the management of E. coli O157 infections, traditional treatment approaches have heavily relied on antibiotics such as ciprofloxacin, tetracycline, sulfamethoxazole, and penicillin [25]. However, the escalating resistance to these antibiotics has raised significant concerns [26]. Notably, E. coli O157 has demonstrated substantial resistance to antibiotics like tetracycline, sulfamethoxazole, and erythromycin, particularly in strains isolated from various sources, including pigs, cattle, and humans [27]. This increasing resistance pattern complicates existing treatment strategies and emphasizes the need for a more in-depth understanding of the mechanisms of antibiotics and the dynamics of resistance development. In response to the challenges posed by antibiotic resistance, research efforts have shifted towards alternative antimicrobial strategies. One promising avenue involves the exploration of natural compounds and probiotics [28]. For example, a specific combination of natural antimicrobials has exhibited effectiveness against E. coli O157 in both in vitro studies and animal models, suggesting its potential use in controlling this pathogen within the animal gut. Additionally, combinations of plant extracts and organic acids have been found to diminish the virulence of E. coli O157, presenting an innovative approach to managing this pathogen effectively [29].
Beyond natural compounds, the scientific community is also investigating the potential of bacteriophages in controlling E. coli O157:H7. These bacteriophages can be administered through various means, such as oral or rectal administration, to ruminants or through spraying or immersion treatments for fruits and vegetables. This versatility positions them as a significant tool in combating E. coli O157:H7 [30]. Moreover, mucoadhesive chitosan microparticles have been identified as potent bactericidal agents capable of disrupting cell membranes, making them a promising option for treating infections caused by E. coli O157:H7 [7,31] and highlighting their ongoing innovation in antimicrobial research.
The subsequent Table 1 provides a comprehensive overview of both the traditional and novel antimicrobials used against E. coli O157. It outlines their mechanisms of action, effectiveness, applications, and notes on resistance, offering a holistic perspective on current and prospective antimicrobial strategies. This comparison not only illuminates the changing landscape of antimicrobial resistance but also underscores the innovation in antimicrobial research, which is crucial for addressing the challenges posed by E. coli O157.

Table 1.
Overview of traditional and novel antimicrobials used against E. coli O157.
2.5. Innovations in Detection and Prevention
In recent times, significant progress has been made in enhancing the detection and prevention methods related to E. coli O157, leading to notable improvements in food safety and public health. The introduction of innovative technologies has transformed our capabilities in identifying and combating this bacterium effectively. These advancements include a wide range of techniques, from rapid molecular assays to sophisticated biosensors, all aimed at improving the sensitivity, specificity, and efficiency of E. coli O157 detection [52].
Within our extensive review, we compiled a table featuring the latest 20 state-of-the-art detection techniques (as mentioned in Table 2). Each method represents a distinct approach to diagnosing E. coli O157 infections, incorporating cutting-edge developments in molecular biology, immunological assays, nanotechnology, and other relevant fields. Our exploration of this diverse array of innovations aims to provide a comprehensive understanding of the current landscape of E. coli O157 detection strategies. This analysis sheds light on the progress achieved thus far and offers insights into potential directions for future research and development in this crucial area.

Table 2.
Recent detection technique for the diagnosis of E. coli O157 infection.
3. Methodology
3.1. Data Sources and Selection
Data were taken from the Scopus database on 20 October 2023. Scopus has the most extensive databases containing abstracts and citations from the peer-reviewed literature, spanning diverse subjects and offering a comprehensive overview of the research landscape. Utilizing Scopus provided several advantages for our study which included broad coverage as a vast array of journals across multiple disciplines were considered, ensuring a comprehensive perspective on E. coli O157 research, high-quality data as the Scopus database maintains fixed selection criteria indexing only reputable and impactful journals, interdisciplinary insights, and lastly, advanced search and analytical tools, which provide streamlining data retrieval and preliminary analysis. Furthermore, during data collection, we set certain exclusion criteria, like studies encompassing only peer-reviewed articles directly related to the topic. Articles in languages other than English, those lacking abstracts, and duplicates were meticulously excluded from our dataset [72].
3.2. Bibliometric Tools and Metrics
Bibliometric analysis offers a quantitative approach to understanding the patterns, networks, and impact within specific research areas. In our study on E. coli O157, we utilized various advanced bibliometric tools for a comprehensive analysis:
- ➢
- VOSviewer: This tool facilitated the visualization and analysis of bibliometric networks, mapping co-authorship patterns and revealing collaborative clusters among researchers and institutions. Its keyword co-occurrence analysis unveiled central themes and emerging trends in the field [73].
- ➢
- R Studio: We employed R Studio along with tailored bibliometric packages and scripts to conduct in-depth analyses, including co-citation assessments. Its flexibility and statistical capabilities enabled us to visualize complex bibliometric data effectively [74].
- ➢
- Thematic Mapping: To visually represent the thematic evolution in E. coli O157 research, we employed thematic mapping techniques. This involved clustering-related keywords and topics to identify distinct thematic areas and tracing their emergence, convergence, and divergence over time. This mapping highlighted dominant research themes, emerging areas, and potential gaps in the literature [75].We focused on pivotal metrics for a comprehensive bibliometric assessment:
- ➢
- Publication Count: This metric provided insights into research volume and growth, revealing periods of heightened activity [76].
- ➢
- Citation Analysis: Examining citation patterns pinpointed influential papers, authors, and institutions, indicating the impact and recognition of specific works within the research community [77].
- ➢
- Co-authorship Networks: These networks illuminated collaborations, showcasing influential research groups and their contributions, as well as the interconnectedness of the global research community [78].
- ➢
- Keyword Analysis: By examining keyword frequency and co-occurrence, we traced the evolution of research themes, identifying dominant topics, emerging interests, and potential gaps [79].
Through these tools, metrics, and thematic mapping techniques, our bibliometric analysis aimed to provide a detailed, data-driven overview of the E. coli O157 research landscape, capturing its complexities and evolution.
4. Results and Discussion
Data were collected from the Scopus database on 20 October 2023 using a specific search string: TITLE-ABS-KEY (“E. coli O157”) combined with filters to limit the results to the English language and document type “AR”. To ensure the integrity and quality of the data, an initial cleaning process was undertaken using Zotero Version 6.0.27, which effectively removed duplicate entries. Following this, a rigorous data processing phase was executed, wherein a total of 669 publications were excluded based on the following specific criteria: 250 review articles, 233 conference papers, 86 book chapters, 43 letters, 51 other types of articles, and 6 editorials. After these exclusions, a total of 7855 publications remained. These were then subjected to quantitative analysis using R Studio (Version R 3.3.0, 2023) with the bibliometrix R package. For visualization purposes, Vos Viewer Version 1.6.19 was employed. Any subsequent data preparation and adjustments were carried out using Microsoft Excel 2021 to ensure the data were primed for in-depth analysis. Figure 3 shows an overview of collected data from the Scopus database.

Figure 3.
Visualization of collected data from Scopus database.
4.1. Evolution of Publications over Time
The research landscape on E. coli O157 has seen a dynamic evolution since its inception. The foundational paper in this domain was published in 1973 [80], bearing the title “Isolation of E. coli O157 from pigs with colibacillosis in Canada and the United States.” Following this groundbreaking work, a noticeable gap emerged with no publications from 1974 to 1981. A modest revival was observed between 1982 and 1989, with less than 10 papers. However, the subsequent years, particularly post-1989, marked a significant uptick in research interest, culminating in peak numbers of publications of 377 and 368 in 2015 and 2020, respectively. As of 2023, the momentum remains robust, with 230 papers already published.
This trend analysis provides valuable insights into the shifting priorities and focus of the scientific community. The hiatus posts that the 1973 publication might reflect the challenges faced in the nascent stages of research, potentially due to technological constraints, funding limitations, or the preliminary nature of the topic. The modest activity between 1982 and 1989 suggests a budding but cautious interest. The exponential growth post-1989, however, underscores the escalating significance of E. coli O157 in both clinical and research contexts.
The peak years of 2015 and 2020 stand as a testament to the culmination of research endeavors and the global emphasis on understanding this bacterium. The sustained engagement in 2023 indicates not only the continued relevance but also hints at emerging challenges or innovations that keep researchers intrigued (please refer to Figure 4). In summary, the evolution of publication trends offers a comprehensive view of the historical progression of E. coli O157 research. It serves as both a reflection of past endeavors and a beacon, highlighting potential avenues and gaps for future exploration.
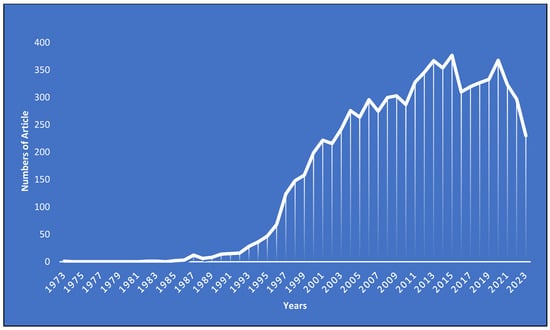
Figure 4.
Yearly published research articles on E. coli 0157 as the research domain.
4.2. Publication Distribution Based on Region
The regional distribution of research on E. coli O157 offers a compelling insight into global research dynamics [81]. The USA distinctly leads the forefront, followed by China, Canada, South Korea, and the UK. The USA’s dominance in this research domain might be attributed to its robust research infrastructure, substantial funding opportunities, and a long-standing tradition of scientific inquiry. With a frequency of 13,972, it significantly outpaces other nations in its contributions. China, holding the second position with a frequency of 5027, underscores its rapid ascent in the global research arena. This reflects China’s growing scientific capabilities and its emphasis on health-related research. Canada’s contribution, with a frequency of 2803, might be influenced by its proximity to the USA and shared concerns regarding public health. South Korea, with a frequency of 2240, showcases its commitment to scientific advancements in health and microbiology. The UK, with a rich history of scientific research, registers a frequency of 2161, emphasizing its continued role in global health research. In essence, this geographic distribution underscores the universal significance of research on E. coli O157 (as shown in Figure 5). While certain regions lead in terms of sheer volume, the collective effort from various countries highlights the global commitment to understanding this bacterium.
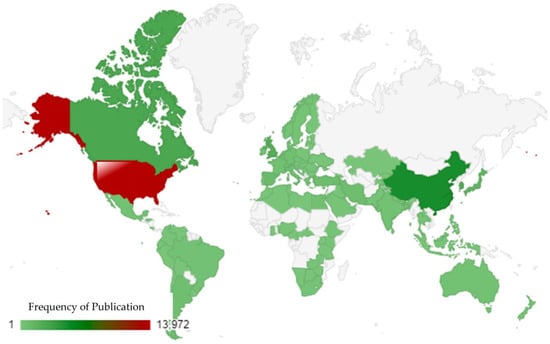
Figure 5.
Visualization of region-based distribution of publications using a world map.
Additionally, Vosviewer was employed to visualize collaboration networks among countries, specifically concentrating on those nations with at least 5 documents out of the total 164 countries. Seventy-three countries met this criterion and were chosen for analysis, as depicted in Figure 6.
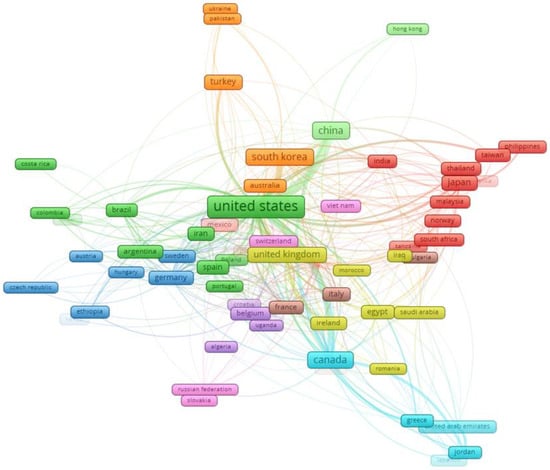
Figure 6.
Visualization of country collaboration network using Vosviewer.
4.3. Co-Authorship Analysis
The utilization of bibliometric analysis to examine co-authorship patterns provides valuable insights into the collaborative dynamics inherent within the scientific community [82]. Our comprehensive data analysis encompassed a vast pool of 7456 authors who have made significant contributions to the field of E. coli O157 research. To facilitate a more detailed investigation, we applied the criterion of selecting authors with a substantial body of work, specifically those with more than five publications in this domain, ensuring that we focused on individuals with a considerable impact in the field.
Within this cohort of prolific authors, five individuals emerged as the foremost contributors to E. coli O157 research, each distinguished by their exceptional publication records. Notably, Doyle MP, affiliated with the Department of Food Microbiology and Toxicology and the Food Research Institute at the University of Wisconsin, Madison, USA, stands out prominently. Doyle’s remarkable portfolio comprises an impressive 84 documents and a substantial H-Index of 49, firmly establishing him as a preeminent figure in the realm of E. coli O157 research. Furthermore, an exploration of his collaborative network reveals a robust total link strength, indicative of the extensive connections he has forged within the research community. Table 2 below shows the top 10 most prolific authors in the field of the E. coli O157 research domain.
Examining collaboration patterns among countries using the Multiple Countries Publications (MCP) and Single Country Publications (SCP) metrics has yielded crucial information about international partnerships in E. coli O157 research [83]. The analysis reveals that the USA leads both single-country publications and extensive collaborations with multiple countries, indicating its prominent role in this research area. Following closely behind are China and Korea, emphasizing their active involvement in both single-country and collaborative publications within this domain, as shown in Figure 7.
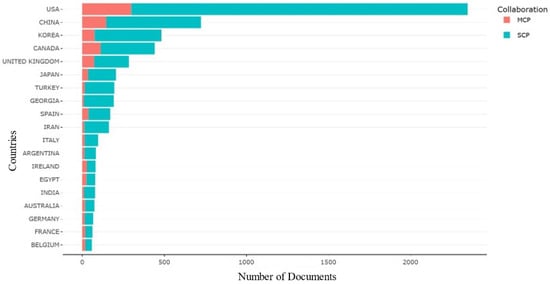
Figure 7.
Countries collaboration based on MCP and SCP.
Vosviewer is used to visualize the collaboration network of authors based on total linkage strength, as shown in Figure 8. Criteria were set to authors with a minimum of five research articles, which were selected for analysis out of 7456 authors.
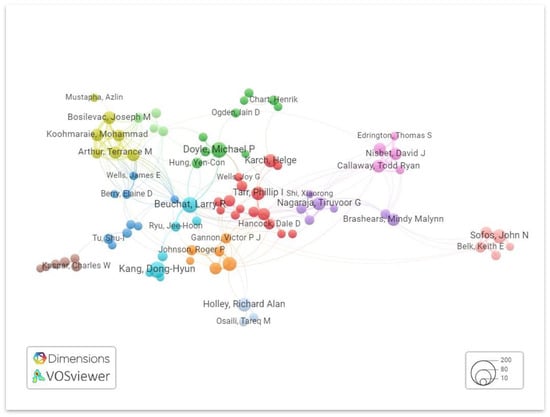
Figure 8.
Network visualization of the collaboration network of authors. Different clusters are denoted by a different color.
Turning our attention to institutions, the University of Georgia and the Eastern Regional Research Centre have assumed leadership positions as the top institutional contributors in the domain of E. coli O157 research, as shown in Figure 9. The University of Georgia, renowned for its research excellence, has made a substantial impact with 568 published documents. Meanwhile, the Eastern Regional Research Centre has made significant strides with 486 publications, reflecting their dedication to advancing knowledge in this crucial area.
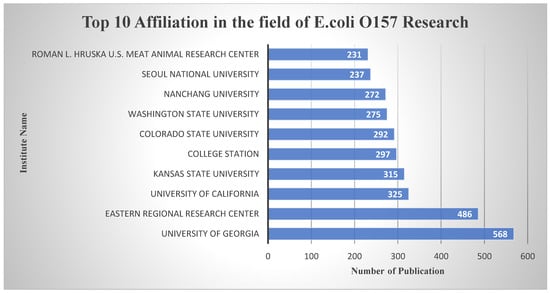
Figure 9.
Top 10 affiliations in the field of E. coli O157 Research domain based on publication number.
4.4. Keywords Analysis
Examining keywords in bibliometric research has proven to be a robust approach for revealing and comprehending the fundamental themes, trends, and intellectual structure within a particular research area [84]. In our investigation, we employed a dataset obtained from the Scopus database and conducted a comprehensive analysis of keyword co-occurrence.
Among the 28,269 identified keywords, 9899 were author keywords, while the rest were index keywords. We focused on author keywords that appeared more than five times, categorizing them as high-frequency keywords for our analysis. As a result, out of the 9899 author keywords, 704 met this criterion and were included in our study.
The most frequently occurring author keyword was “Escherichia coli O157:H7”, noted 1065 times, with a substantial total linkage strength of 2218. Following closely were the keywords “Escherichia coli” and “Escherichia coli O157,” appearing 328 and 213 times, respectively, with total linkage strengths of 761 and 435, as detailed in Table 3.

Table 3.
Most prolific author in the field of E. coli O157 research domain.
To visually depict the co-occurrence network of these keywords, we employed VOSviewer, as illustrated in Figure 10. Furthermore, Figure 11 displays a word cloud created using R Studio, emphasizing the most frequently appearing keywords based on their size.
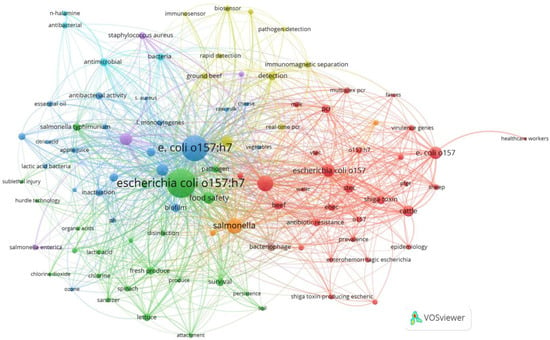
Figure 10.
Keyword co-occurrence network visualization using Vosviewer. The size of the nodes signifies the importance or prominence of items in the dataset, with larger nodes indicating greater importance. The color of the nodes categorizes items based on characteristics, helping to identify clusters or groups within the network, with each color representing a different category or group.

Figure 11.
Word cloud prepared using R-studio. The size of word indicates the number of co- occurrences of the keyword. Text size in Figure 11 depicts the number of occurrences of the keyword, providing a visual representation of its frequency.
Additionally, utilizing the R-studio R package bibliometrix, we generated a word frequency graph based on the occurrence of keywords across different years. This graph highlights the evolving landscape of research within the E. coli O157 research domain over time, as shown in Figure 12.
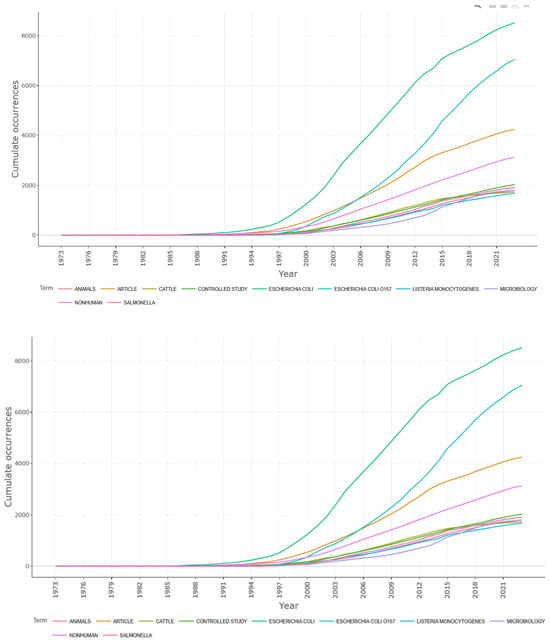
Figure 12.
Word frequency over the time prepared using R-studio.
Analyzing trending topics using the R-Studio Bibliometrix package has provided insights into emerging themes in E. coli O157 research based on keyword co-occurrence analysis, as shown in Figure 13. This analysis indicates a recent shift in focus within the last five years. There has been increased emphasis on the development of detection techniques, particularly those utilizing CRISPR-CAS and Loop-Mediated Isothermal Amplification methods. Additionally, there is a growing interest in advancing high-throughput whole-genome sequencing techniques for the rapid sequencing of pathogenic E. coli strains. The research spotlight has also turned towards the development of antimicrobial agents to combat antibiotic-resistant E. coli strains, including innovative therapies such as phage therapy, aiming to address the challenges posed by multi-drug resistance in E. coli strains.
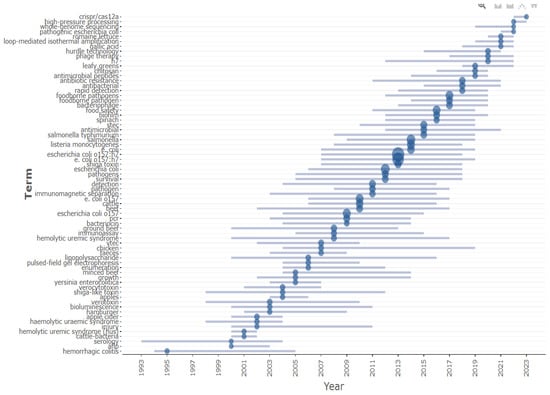
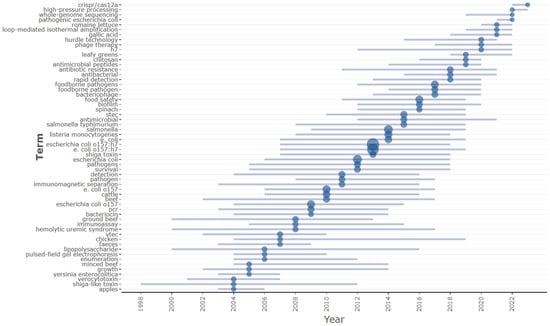
Figure 13.
Keyword arrangement year wise; the line represents the corresponding year and the blue circle size indicates the number of occurrences. Trend topic analysis using R-studio.
4.5. Citation Analysis
This segment shows the substantial contributions made by prior research and scholarly works, which have greatly influenced and enhanced the content of our study [85]. The subsequent citation analysis presents a comprehensive compilation of references utilized throughout our research, spanning academic papers, books, articles, and other credible sources. These references form a robust basis for our exploration of the E. coli O157 research domain. Table 4 below lists the top 10 most-cited documents in the field of the E. coli O157 research domain.

Table 4.
Most frequent author keyword in the research domain of E. coli O157 research.
After the most cited documents come the most impactful journals published; Table 5 below provides an overview of the most cited journals, accompanied by their H, G, and M indexes, offering a complete perspective on the highly influential journals within this research area.

Table 5.
Most cited document in the research domain of E. coli O157 research.
Bradford’s Law, a pivotal notion in bibliometric research, provides valuable perspectives on the unequal dispersion of information and scholarly publications within particular academic domains. This concept was formulated during the 1930s by Samuel C. Bradford, a British librarian. It suggests that within any given field of study, the scholarly literature frequently exhibits a discernible pattern: a nucleus of exceptionally prolific journals or sources, succeeded by a region where returns on information tend to diminish, and ultimately, an area characterized by dispersed or less central sources. Bradford’s Law has played a crucial role in enhancing our comprehension of the clustering of scientific knowledge and assisting researchers in refining their approaches to searches made in the literature [96]. Using this approach, Figure 14 shows core sources based on the number of articles using Bradford’s Law.
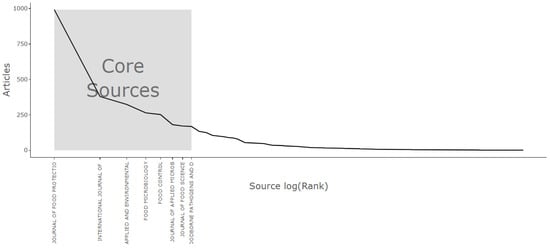
Figure 14.
Core source analysis based on Bradford’s Law.
Further Table 6 shows most cited journal in the research domain of E. coli O157. A deeper analysis of citations highlights the most frequently cited countries, with the USA leading significantly with a total citation count of 105,195 and an average of 44.80 citations per article. Table 7 below illustrates this dominance, showing the USA as the top country in terms of citation numbers. China, listed second, lags far behind with 17,814 citations, indicating a substantial difference compared to the USA’s citation count.

Table 6.
Most Cited Journal in the research domain of E. coli O157.

Table 7.
Most cited countries.
4.6. Three-Field Plot
This visual representation is a three-field plot, commonly known as a Sankey diagram or Alluvial diagram, depicting relationships between various data dimensions [97]. Specifically, it illustrates the interconnections among authors’ countries (AU_CO), individual authors (AU), and the topics or keywords of their research (ID) in the realm of E. coli O157 research, as shown in Figure 15.
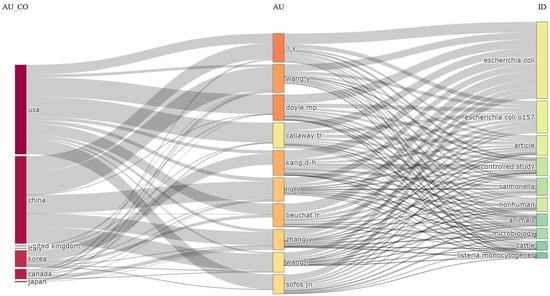
Figure 15.
Three-field plots (Sankey diagram) based on the author’s country (Left), author (Middle), and keyword (Right).
In the leftmost column (AU_CO), different countries are represented by dark bars indicating their respective contributions to the research domain. The United States (USA) exhibits the most substantial presence, followed by China, the United Kingdom, Korea, Canada, and Japan. This suggests that these nations have active research communities dedicated to E. coli O157.
The middle column (AU) displays individual authors, with color-coding that likely signifies publication count or contribution weight, although this aspect is not clarified in the image. The width of the lines connecting authors to their countries and research topics reflects the volume of work or the strength of their association. Authors such as “Li Y” and “Wang Y” have extensive collaborations connections, suggesting a significant number of publications or a strong association with the listed topics.
The rightmost column (ID) showcases topics, identifiers, or keywords related to research publications. “Escherichia coli” and “Escherichia coli O157” emerge as the most prevalent topics, indicating a substantial focus on these bacteria. Additional linked topics encompass the following: “article”, “controlled study”, “salmonella”, “nonhuman”, “animals”, “microbiology”, “cattle”, and “listeria monocytogenes”. This diversity signifies varied yet interconnected areas of interest within E. coli O157 research. The inclusion of keywords like “cattle” and “listeria monocytogenes” implies research spanning beyond E. coli O157 to encompass related topics in food safety and zoonotic diseases. In summary, this three-field plot provides a visual summary of the E. coli O157 research landscape, showcasing active countries, influential authors, and frequently studied topics. It offers insights into the collaborative network and thematic focus within the research community studying E. coli O157.
4.7. Thematic Mapping and Factorial Analysis
A thematic map is commonly employed in bibliometric studies to visually depict the structure of a research field [98]. Here, it focuses on research related to E. coli O157, a specific strain of the E. coli bacterium known for causing foodborne illnesses, as shown in Figure 16. The map is segmented into four quadrants based on two axes: the vertical axis representing the development degree or density (indicating internal connections between themes), and the horizontal axis representing the relevance degree or centrality (indicating interactions with other themes). The following provides a breakdown of each quadrant:
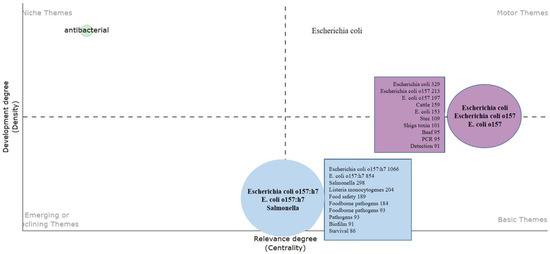
Figure 16.
Theme map based on the E. coli O157 research domain.
- 1.
- Motor Themes (upper right): These themes are both well-developed and central, signifying their maturity and strong interconnections with other research topics. The prominent bubble in this quadrant is labeled “Escherichia coli”, “E. coli O157”, and “E. coli O157”, highlighting it as a core topic in the field with extensive research. Smaller bubbles also focus on E. coli O157 and related subtopics such as “cattle”, which is significant due to cattle being a primary reservoir for this bacterium.
- 2.
- Niche Themes (upper left): These themes are less developed but still represent niche areas of emerging or specialized interest. The single theme “antibacterial” suggests a concentration on treatments or resistance issues related to E. coli O157.
- 3.
- Basic Themes (lower right): These themes are well-developed but less central, foundational to the field, but not at the forefront of current research. This quadrant includes themes like “food safety”, indicating substantial research but is potentially less connected to other themes.
- 4.
- Emerging or Declining Themes (lower left): These themes have lower density and centrality, indicating that they are either new areas of interest yet to be fully explored (“emerging”) or areas losing focus within the research community (“declining”). Themes like “E. coli O157:H7” and “salmonella” fall into this category, suggesting some focus on these pathogens, though not necessarily as primary subjects or possibly newly emerging subjects within the field.
Each bubble within the quadrant represents a cluster of themes, with its size denoting the volume of research or attention given to that topic. The numbers alongside each term could indicate the number of publications or the weight of the theme in the analysis.
Researchers can use this map to identify active, emerging, and underexplored areas in E. coli O157 research. Funding agencies can utilize it to allocate resources effectively, while new researchers can identify gaps in the existing literature.
Figure 17 below is a visual representation resulting from a multiple correspondence analysis (MCA), portraying research topics associated with E. coli O157 in a spatial arrangement. MCA, a statistical method, aims to position each object in a multidimensional space, preserving the distances between objects as accurately as possible. In this context, each point on the plot represents a specific research topic or keyword, and their proximity signifies their closeness in the realm of research in the literature [99].
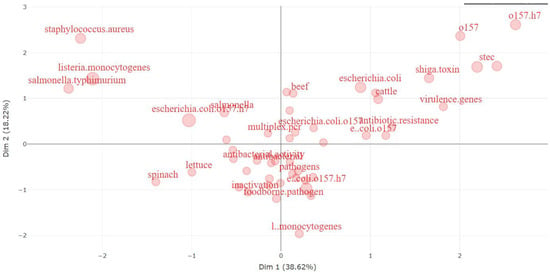
Figure 17.
Visual representation of multiple correspondence analysis (MCA) based on the E. coli O157 research domain.
Breaking down the image:
- ➢
- Dimension 1 (horizontal axis, explaining 38.62% of the variance): This dimension likely signifies a continuum in research topics, ranging from general E. coli-related subjects on the left to more specific subtopics or issues concerning E. coli on the right. Topics of higher specificity or those currently prominent in research tend to cluster towards the right end of this axis.
- ➢
- Dimension 2 (vertical axis, explaining 13.29% of the variance): This dimension captures another aspect of variation among research topics, potentially related to the nature of the research, such as the study type (e.g., clinical, environmental, food safety) or the research approach (e.g., genetic, epidemiological).This plot reveals distinct clusters of keywords:
- ➢
- Upper Right Quadrant: This area encompasses terms associated with genetic aspects and types of E. coli O157, including “stec” (shiga toxin-producing E. coli), “virulence genes”, and strain differentiations such as “O157” and “O157:H7”.
- ➢
- Lower Right Quadrant: Terms in this section pertain to food sources and resistance, such as “beef”, “cattle”, and “antibiotic resistance”, highlighting a focus on the origins of E. coli O157 infections and the challenges in treating them.
- ➢
- Left Quadrant: This portion includes broader terms like “spinach” and “lettuce”, linked to E. coli outbreaks, as well as general pathogens. It also encompasses terms like “E. coli” without specifying the O157 strain, indicating a more general research focus.
- ➢
- Upper Left Quadrant: Here, other bacterial species like “Staphylococcus aureus” and “Salmonella typhimurium” are located, likely part of comparative studies in food safety or pathogenicity.
The relative distances between terms offer insights into the interconnectedness of research themes. For example, the central positioning of “E. coli” on the horizontal axis implies it is a primary focus, with research branching into specific areas like “O157”, “Shiga toxin”, and “foodborne pathogen”. This visualization proves invaluable for researchers, aiding their comprehension of the E. coli O157 research landscape. It helps identify closely related topics, potential research gaps, and emerging trends within the field.
5. Limitations
Database Limitation (Scopus): Our research exclusively relied on Scopus as the main database, owing to its extensive coverage of the scholarly literature across diverse fields. As Scopus is a reputable and widely acknowledged database, relying solely on it might result in overlooking pertinent studies available in other databases, potentially constraining the comprehensiveness of our analysis [100].
Language Restriction (English articles): We limited our analysis to English-language articles, introducing a potential language bias. However, by excluding non-English publications, we did not miss any valuable contributions from researchers in regions where English is not the primary language, as while using this filter, we ensured not to miss any potentially relevant studies on E. coli O157 research domain. Therefore, it can be said that this limitation has not led to a partial representation of the global research landscape on this topic [101].
Publication Bias: The analysis is vulnerable to publication bias since it primarily focuses on published articles. Published works often emphasize positive results, possibly neglecting studies with neutral or negative outcomes. This bias has not shown any potential to influence the overall interpretation of research patterns and themes [102].
6. Conclusions
The conducted bibliometric analysis has yielded insightful revelations within the domain of E. coli research, unraveling noteworthy patterns and advancements that delineate the historical evolution of investigations in this field. A pivotal milestone was the inaugural paper in the E. coli O157 research sphere in 1973, which triggered a notable upswing in publications commencing from 1993 onward. A striking observation is the pronounced influence of the United States in E. coli O157 research, as evidenced by a substantial output of 13,972 publications, surpassing other nations. This dominance extends to citations, totaling 105,195, and is accompanied by an impressive average article citation rate of 44.80.
Furthermore, the analysis highlights the “Journal of Food Protection” as the preeminent publication venue in this area, boasting 992 published articles and accruing a total of 41,238 citations. Eminent contributors to this domain include Li Y and Doyle Mp, with 102 and 84 publications, respectively, underscoring their significant contributions. Additionally, this study delves into the intellectual framework using techniques such as theme mapping, trend topic analysis, and multiple correspondence analysis (MCA) analysis. These methodologies shed light on emerging research themes and identify existing research gaps, thereby offering a comprehensive perspective for researchers, policymakers, and industries engaged in E. coli O157 research. Such insights hold considerable value for steering future research initiatives, shaping policies, and guiding industry practices within this critical domain.
Author Contributions
Conceptualization, H.J. and G.K.; methodology, H.J. and D.K.; software, H.J.; validation, G.K., R.K., and N.M.; writing—original draft preparation, H.J. and D.K.; writing—review and editing, H.J., D.K., and R.K.; supervision, G.K. and N.M. All authors have read and agreed to the published version of the manuscript.
Funding
This study was not supported by any external funding agency.
Institutional Review Board Statement
An institutional review board statement and approval number is not relevant to this study.
Informed Consent Statement
Not applicable.
Data Availability Statement
Raw data are available from the corresponding author on request.
Acknowledgments
The authors are thankful to the management of Lovely Professional University, the University of Nebraska Medical Centre, and the University of Wisconsin-Madison for providing the necessary facilities to carry out this study.
Conflicts of Interest
All authors have declared no conflicts of interest.
References
- WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007–2015. Available online: https://iris.who.int/handle/10665/199350 (accessed on 28 December 2023).
- Kornacki, J.L.; Marth, E.H. Foodborne Illness Caused by Escherichia coli: A Review. J. Food Prot. 1982, 45, 1051–1067. [Google Scholar] [CrossRef] [PubMed]
- Singha, S.; Thomas, R.; Viswakarma, J.N.; Gupta, V.K. Foodborne Illnesses of Escherichia coli O157origin and Its Control Measures. J. Food Sci. Technol. 2023, 60, 1274–1283. [Google Scholar] [CrossRef] [PubMed]
- LeJeune, J.; Homan, J.; Linz, G.; Pearl, D.L. Role of the European Starling in the Transmission of E. coli O157 on Dairy Farms. Proc. Vertebr. Pest Conf. 2008, 23, 31–34. [Google Scholar] [CrossRef]
- Darkazanli, M.; Kiseleva, I.; Darkazanli, K. Genetic Diversity of E. coli O157:H7 Isolated from Some Leafy Greens, Irrigated by Aleppo River, Using Random Amplified Polymorphic DNA (RAPD) Marker. Russ. Agric. Sci. 2018, 44, 146–152. [Google Scholar] [CrossRef]
- Sadeqi, S.; Heidariyeh, P.; Qorbani, M.; Nikkhahi, F.; Amin Marashi, S.M. Evaluation of Multiplex-PCR for Simultaneous Identification of Salmonella Enteritidis, Shigella Flexneri, and Escherichia coli O157: H7 in Poultry. Infect. Epidemiol. Microbiol. 2019, 5, 13–18. [Google Scholar]
- Kumar, R.; Islam, T.; Nurunnabi, M. Mucoadhesive Carriers for Oral Drug Delivery. J. Control Release 2022, 351, 504–559. [Google Scholar] [CrossRef] [PubMed]
- Laing, C.; Pegg, C.; Yawney, D.; Ziebell, K.; Steele, M.; Johnson, R.; Thomas, J.E.; Taboada, E.N.; Zhang, Y.; Gannon, V.P.J. Rapid Determination of Escherichia coli O157:H7 Lineage Types and Molecular Subtypes by Using Comparative Genomic Fingerprinting. Appl. Environ. Microbiol. 2008, 74, 6606–6615. [Google Scholar] [CrossRef]
- Dutt, S.; Gupta, A.K.; Aadil, K.R.; Bunekar, N.; Mishra, V.K.; Kumar, R.; Gupta, A.; Chaudhary, A.; Kumar, A.; Chawla, M.; et al. Nanomaterials of Metal and Metal Oxides for Optical Biosensing Application. In Metal Oxides for Biomedical and Biosensor Applications; Elsevier: Amsterdam, The Netherlands, 2022; pp. 321–352. [Google Scholar] [CrossRef]
- Kumar, R.; Pulikanti, G.R.; Shankar, K.R.; Rambabu, D.; Mangili, V.; Kumbam, L.R.; Sagara, P.S.; Nakka, N.; Yogesh, M. Surface Coating and Functionalization of Metal and Metal Oxide Nanoparticles for Biomedical Applications. In Metal Oxides for Biomedical and Biosensor Applications; Elsevier: Amsterdam, The Netherlands, 2022; pp. 205–231. [Google Scholar] [CrossRef]
- Karimi, S.; Azizi, F.; Nayeb-Aghaee, M.; Mahmoodnia, L. The Antimicrobial Activity of Probiotic Bacteria Escherichia coli Isolated from Different Natural Sources against Hemorrhagic E. coli O157:H7. Electron. Physician 2018, 10, 6548–6553. [Google Scholar] [CrossRef]
- Divya, K.; Vijayan, S.; George, T.K.; Jisha, M.S. Antimicrobial Properties of Chitosan Nanoparticles: Mode of Action and Factors Affecting Activity. Fibers Polym. 2017, 18, 221–230. [Google Scholar] [CrossRef]
- Hou, T.; Sankar Sana, S.; Li, H.; Wang, X.; Wang, Q.; Boya, V.K.N.; Vadde, R.; Kumar, R.; Kumbhakar, D.V.; Zhang, Z.; et al. Development of Plant Protein Derived Tri Angular Shaped Nano Zinc Oxide Particles with Inherent Antibacterial and Neurotoxicity Properties. Pharmaceutics 2022, 14, 2155. [Google Scholar] [CrossRef]
- Sana, S.S.; Vadde, R.; Kumar, R.; Arla, S.K.; Somala, A.R.; Krishna Rao, K.S.V.; Zhijun, Z.; Boya, V.K.N.; Mondal, K.; Mamidi, N. Eco-Friendly and Facile Production of Antibacterial Zinc Oxide Nanoparticles from Grewia flavescens (G. flavescens) Leaf Extract for Biomedical Applications. J. Drug Deliv. Sci. Technol. 2023, 80, 104186. [Google Scholar] [CrossRef]
- Hassanien, A.A.; Shaker, E.M. Investigation of the Effect of Chitosan and Silver Nanoparticles on the Antibiotic Resistance of Escherichia coli O157:H7 Isolated from Some Milk Products and Diarrheal Patients in Sohag City, Egypt. Vet. World 2020, 13, 16471653. [Google Scholar] [CrossRef]
- Kumar, R. Lipid-Based Nanoparticles for Drug-Delivery Systems. In Nanocarriers for Drug Delivery: Nanoscience and Nanotechnology in Drug Delivery; Elsevier: Amsterdam, The Netherlands, 2019; pp. 249–284. [Google Scholar] [CrossRef]
- Deng, R.; Wang, M.; Song, Y.; Shi, Y. A Bibliometric Analysis on the Research Trend of Exercise and the Gut Microbiome. Microorganisms 2023, 11, 903. [Google Scholar] [CrossRef]
- Morgan, G.M.; Newman, C.; Palmer, S.R.; Allen, J.B.; Shepherd, W.; Rampling, A.M.; Warren, R.E.; Gross, R.J.; Scotland, S.M.; Smith, H.R. First Recognized Community Outbreak of Haemorrhagic Colitis Due to Verotoxin-Producing Escherichia coli O 157.H7 in the UK. Epidemiol. Infect. 1988, 101, 83–91. [Google Scholar] [CrossRef]
- Ameer, M.A.; Wasey, A.; Salen, P. Escherichia coli (e Coli 0157 H7). In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Lange, M.E.; Uwiera, R.R.E.; Inglis, G.D. Enteric Escherichia coli O157:H7 in Cattle, and the Use of Mice as a Model to Elucidate Key Aspects of the Host-Pathogen-Microbiota Interaction: A Review. Front. Vet. Sci. 2022, 9, 937866. [Google Scholar] [CrossRef]
- Eppinger, M.; Almería, S.; Allué-Guardia, A.; Bagi, L.K.; Kalalah, A.A.; Gurtler, J.B.; Fratamico, P.M. Genome Sequence Analysis and Characterization of Shiga Toxin 2 Production by Escherichia coli O157:H7 Strains Associated With a Laboratory Infection. Front. Cell Infect. Microbiol. 2022, 12, 888568. [Google Scholar] [CrossRef]
- Dunn, J.R.; Keen, J.E.; Thompson, R.A. Prevalence of Shiga-Toxigenic Escherichia coli O157:H7 in Adult Dairy Cattle. J. Am. Vet. Med. Assoc. 2004, 224, 1151–1158. [Google Scholar] [CrossRef]
- Gambushe, S.M.; Zishiri, O.T.; El Zowalaty, M.E. Review of Escherichia coli O157:H7 Prevalence, Pathogenicity, Heavy Metal and Antimicrobial Resistance, African Perspective. Infect. Drug Resist. 2022, 15, 4645–4673. [Google Scholar] [CrossRef]
- Oluwarinde, B.O.; Ajose, D.J.; Abolarinwa, T.O.; Montso, P.K.; Du Preez, I.; Njom, H.A.; Ateba, C.N. Safety Properties of Escherichia coli O157:H7 Specific Bacteriophages: Recent Advances for Food Safety. Foods 2023, 12, 3989. [Google Scholar] [CrossRef]
- Kumar, R.; Siril, P.F. Drop-by-Drop Solvent Hot Antisolvent Interaction Method for Engineering Nanocrystallization of Sulfamethoxazole to Enhanced Water Solubility and Bioavailability. J. Drug Deliv. Sci. Technol. 2020, 55, 101359. [Google Scholar] [CrossRef]
- Fair, R.J.; Tor, Y. Antibiotics and Bacterial Resistance in the 21st Century. Perspect. Med. Chem. 2014, 6, 25–64. [Google Scholar] [CrossRef] [PubMed]
- Ateba, C.N.; Bezuidenhout, C.C. Characterisation of Escherichia coli O157 Strains from Humans, Cattle and Pigs in the North-West Province, South Africa. Int. J. Food Microbiol. 2008, 128, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Ngo, H.; Wu, C. Natural and Bio-Based Antimicrobials: A Review. ACS Symp. Ser. 2018, 1287, 1–24. [Google Scholar] [CrossRef]
- Stratakos, A.C.; Linton, M.; Ward, P.; Campbell, M.; Kelly, C.; Pinkerton, L.; Stef, L.; Pet, I.; Stef, D.; Iancu, T.; et al. The Antimicrobial Effect of a Commercial Mixture of Natural Antimicrobials Against Escherichia coli O157:H7. Foodborne Pathog. Dis. 2019, 16, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Qu, K.; Li, X.; Cao, Z.; Wang, X.; Li, Z.; Song, Y.; Xu, Y. Use of Bacteriophages to Control Escherichia coli O157:H7 in Domestic Ruminants, Meat Products, and Fruits and Vegetables. Foodborne Pathog. Dis. 2017, 14, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Li, Y.; Liu, Y.; Li, N.; Zhang, X.; Yan, C. Antimicrobial Properties of Chitosan and Chitosan Derivatives in the Treatment of Enteric Infections. Molecules 2021, 26, 7136. [Google Scholar] [CrossRef] [PubMed]
- Shariati, A.; Arshadi, M.; Khosrojerdi, M.A.; Abedinzadeh, M.; Ganjalishahi, M.; Maleki, A.; Heidary, M.; Khoshnood, S. The Resistance Mechanisms of Bacteria against Ciprofloxacin and New Approaches for Enhancing the Efficacy of This Antibiotic. Front. Public Health 2022, 10, 1025633. [Google Scholar] [CrossRef]
- Grossman, T.H. Tetracycline Antibiotics and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025387. [Google Scholar] [CrossRef]
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and Resistance Mechanisms of Antibiotics: A Guide for Clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300–305. [Google Scholar] [CrossRef]
- Krause, K.M.; Serio, A.W.; Kane, T.R.; Connolly, L.E. Aminoglycosides: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a027029. [Google Scholar] [CrossRef]
- Arumugham, V.B.; Gujarathi, R.; Cascella, M. Third-Generation Cephalosporins. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Garneau-Tsodikova, S.; Labby, K.J. Mechanisms of Resistance to Aminoglycoside Antibiotics: Overview and Perspectives. Medchemcomm 2016, 7, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Ovung, A.; Bhattacharyya, J. Sulfonamide Drugs: Structure, Antibacterial Property, Toxicity, and Biophysical Interactions. Biophys. Rev. 2021, 13, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Mączka, W.; Twardawska, M.; Grabarczyk, M.; Wińska, K. Carvacrol—A Natural Phenolic Compound with Antimicrobial Properties. Antibiotics 2023, 12, 824. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ashby, R.D.; Solaiman, D.K.Y.; Liu, Y.; Fan, X. Antimicrobial Activity and Inactivation Mechanism of Lactonic and Free Acid Sophorolipids against Escherichia coli O157:H7. Biocatal. Agric. Biotechnol. 2017, 11, 176–182. [Google Scholar] [CrossRef]
- Sváb, D.; Falgenhauer, L.; Papp, V.; Rohde, M.; Chakraborty, T.; Tóth, I. Characterisation of New Anti-O157 Bacteriophages of Bovine Origin Representing Three Genera. Arch. Microbiol. 2022, 204, 231. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.; Yin, H.B.; Bauchan, G.; Mowery, J. Inhibition of Escherichia coli O157:H7 and Salmonella Enterica Virulence Factors by Benzyl Isothiocyanate. Food Microbiol. 2020, 86, 103303. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, Y.G.; Cho, H.S.; Ryu, S.Y.; Cho, M.H.; Lee, J. Coumarins Reduce Biofilm Formation and the Virulence of Escherichia coli O157:H7. Phytomedicine 2014, 21, 1037–1042. [Google Scholar] [CrossRef]
- Nannapaneni, R.; Muthaiyan, A.; Crandall, P.G.; Johnson, M.G.; O’Bryan, C.A.; Chalova, V.I.; Callaway, T.R.; Carroll, J.A.; Arthington, J.D.; Nisbet, D.J.; et al. Antimicrobial Activity of Commercial Citrus-Based Natural Extracts Against Escherichia coli O157:H7 Isolates and Mutant Strains. Foodborne Pathog. Dis. 2008, 5, 695–699. [Google Scholar] [CrossRef]
- Ye, R.; Xu, H.; Wan, C.; Peng, S.; Wang, L.; Xu, H.; Aguilar, Z.P.; Xiong, Y.; Zeng, Z.; Wei, H. Antibacterial Activity and Mechanism of Action of ε-Poly-l-Lysine. Biochem. Biophys. Res. Commun. 2013, 439, 148–153. [Google Scholar] [CrossRef]
- Vuthikunchai, S.P.V.; Suwalak, S. Antibacterial Activities of Semipurified Fractions of Quercus Infectoria against Enterohemorrhagic Escherichia coli O157:H7 and Its Verocytotoxin Production. J. Food Prot. 2008, 71, 1223–1227. [Google Scholar] [CrossRef]
- Rahal, E.A.; Kazzi, N.; Nassar, F.J.; Matar, G.M. Escherichia coli O157:H7-Clinical Aspects and Novel Treatment Approaches. Front. Cell Infect. Microbiol. 2012, 2, 138. [Google Scholar] [CrossRef] [PubMed]
- Bhavaniramya, S.; Vishnupriya, S.; Al-Aboody, M.S.; Vijayakumar, R.; Baskaran, D. Role of Essential Oils in Food Safety: Antimicrobial and Antioxidant Applications. Grain Oil Sci. Technol. 2019, 2, 49–55. [Google Scholar] [CrossRef]
- Wang, X.; Lee, S.Y.; Akter, S.; Huq, M.A. Probiotic-Mediated Biosynthesis of Silver Nanoparticles and Their Antibacterial Applications against Pathogenic Strains of Escherichia coli O157:H7. Polymers 2022, 14, 1834. [Google Scholar] [CrossRef] [PubMed]
- Bhagwat, A.; Mixon, M.; Collins, C.H.; Dordick, J.S. Opportunities for Broadening the Application of Cell Wall Lytic Enzymes. Appl. Microbiol. Biotechnol. 2020, 104, 9019–9040. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Liu, Y.; Li, C.; Hua, S.; Sun, C.; Huang, L. Antibacterial Mechanism of Vanillin against Escherichia coli O157: H7. Heliyon 2023, 9, e19280. [Google Scholar] [CrossRef] [PubMed]
- Lian, F.; Wang, D.; Yao, S.; Ge, L.; Wang, Y.; Zhao, Y.; Zhao, J.; Song, X.; Zhao, C.; Li, J.; et al. A Detection Method of Escherichia coli O157:H7 Based on Immunomagnetic Separation and Aptamers-Gold Nanoparticle Probe Quenching Rhodamine B’s Fluorescence: Escherichia coli O157:H7 Detection Method Based on IMS and Apt-AuNPs Probe Quenching Rho B’ s Fluorescence. Food Sci. Biotechnol. 2021, 30, 1129–1138. [Google Scholar] [CrossRef] [PubMed]
- Bazsefidpar, S.; Serrano-Pertierra, E.; Gutiérrez, G.; Calvo, A.S.; Matos, M.; Blanco-López, M.C. Rapid and Sensitive Detection of E. coli O157:H7 by Lateral Flow Immunoassay and Silver Enhancement. Microchim. Acta 2023, 190, 264. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Li, D.; Jiang, Q.; Gan, N. A Paper-Chip-Based Phage Biosensor Combined with a Smartphone Platform for the Quick and On-Site Analysis of E. coli O157:H7 in Foods. Chemosensors 2023, 11, 151. [Google Scholar] [CrossRef]
- Banerjee, T.; Panchal, N.; Sutton, C.; Elliott, R.; Patel, T.; Kajal, K.; Arogunyo, E.; Koti, N.; Santra, S. Tunable Magneto-Plasmonic Nanosensor for Sensitive Detection of Foodborne Pathogens. Biosensors 2023, 13, 109. [Google Scholar] [CrossRef]
- Chen, C.; Coronel-Aguilera, C.P.; Applegate, B.M.; Gehring, A.G.; Bhunia, A.K.; Paoli, G.C. Studies on Simultaneous Enrichment and Detection of Escherichia coli O157:H7 during Sample Shipment. Foods 2022, 11, 3653. [Google Scholar] [CrossRef]
- Liu, D.; Lv, X.; Zhao, C.; Li, J.; Huang, J.; Weng, L.; He, L.; Liu, S. NaBiF4 Upconversion Nanoparticle-Based Electrochemiluminescent Biosensor for E. coli O157:H7 Detection. RSC Adv. 2022, 12, 30174–30180. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Li, P.; Feng, X.; Liu, D.; Wang, X.; Wang, L. Detection of 13 Foodborne Pathogens in Aquatic Products Using Visual Chromogenic Chips Based on Asymmetric Multiplex Polymerase Chain Reaction and Nucleic Acid Hybridization. Food Control 2024, 155, 110100. [Google Scholar] [CrossRef]
- Feng, X.; Zhou, D.; Mu, D.; Xu, H. An Esterase Activity-Based Biosensor for Rapid and Sensitive Detection of Viable Escherichia coli O157:H7 in Milk. Int. Dairy J. 2023, 147, 105769. [Google Scholar] [CrossRef]
- Yinur, D.; Moges, B.; Hassen, A.; Tessema, T.S. Loop Mediated Isothermal Amplification as a Molecular Diagnostic Assay: Application and Evaluation for Detection of Enterohaemorrhagic Escherichia coli (O157:H7). Pract. Lab. Med. 2023, 37, e00333. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Wang, Z.; St-Pierre, J.P.; Dan, H.; Lin, M.; Zou, S.; Cao, X. Multivalent Aptamer Meshed Open Pore Membrane and Signal Amplification for High-Flux and Ultra Sensitive Whole Cell Detection of E. coli O157:H7 in Complex Food Matrices. Sens. Actuators B Chem. 2023, 394, 134378. [Google Scholar] [CrossRef]
- Shao, Y.; Wang, Z.; Xie, J.; Zhu, Z.; Feng, Y.; Yu, S.; Xue, L.; Wu, S.; Gu, Q.; Zhang, J.; et al. Dual-Mode Immunochromatographic Assay Based on Dendritic Gold Nanoparticles with Superior Fluorescence Quenching for Ultrasensitive Detection of E. coli O157:H7. Food Chem. 2023, 424, 136366. [Google Scholar] [CrossRef]
- Li, J.; Yun, W.; Zhang, H.; Chen, L.; Ho, H.P.; Pu, X.; Huang, Y.; Shen, Y.; Cao, H. MoS2 Nanosheets Based Label-Free Colorimetric Aptasensor for Escherichia coli O157: H7 Detection. Colloids Surf. A Physicochem. Eng. Asp. 2023, 674, 131955. [Google Scholar] [CrossRef]
- Xiao, S.; Cao, C.; Ming, T.; Cao, Y.; Yu, Z.; Gan, N. Simultaneous and Rapid Screening of Live and Dead E. coli O157:H7 with Three Signal Outputs: An All-in-One Biosensor Using Phage-Apoferritin@CuO2 Signal Tags on MXenes-Modified Electrode Platform. J. Hazard. Mater. 2023, 458, 131875. [Google Scholar] [CrossRef]
- Liu, J.; Huang, T.; Xu, R.; Xiang, Z.; Soteyome, T.; Chen, X.; Zhang, Q.; Huang, Q.; Wu, Z.; Huang, Y.; et al. A Propidium Monoazide-Polymerase Spiral Reaction (PMA-PSR) Designed for Direct Detection of Escherichia coli O157:H7 Viable Cell. LWT 2023, 186, 115212. [Google Scholar] [CrossRef]
- Liu, Y.; Wei, C.; Wan, H.; Sarengaowa; Liang, X.; Jiang, T.; Dong, Y.; Zhao, X.; Zhong, T. Preliminary Study on Rapid and Simultaneous Detection of Viable Escherichia coli O157:H7, Staphylococcus Aureus, and Salmonella by PMA-MPCR in Food. Molecules 2023, 28, 5835. [Google Scholar] [CrossRef]
- You, H.; Wang, M.; Wang, S.; Xu, J.; Hu, S.; Li, T.; Yu, Z.; Tang, D.; Gan, N. Ultrasensitive and Specific Phage@DNAzyme Probe-Triggered Fluorescent Click Chemistry for On-Site Detection of Foodborne Pathogens Using a Smartphone. Anal. Chem. 2023, 95, 11211–11218. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Toushik, S.H.; Park, H.J.; Kim, S.A.; Shim, W.B. Rapid Detection of Shiga-Toxin-Producing Escherichia coli O157:H7 Based on a Colorimetric Loop-Mediated Isothermal Amplification (CLAMP) Assay Using a Molecular Beacon Paired with HRPzyme. Anal. Bioanal. Chem. 2023, 415, 4973–4984. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bu, T.; Cao, Y.; Wu, H.; Xi, J.; Feng, Q.; Xuan, C.; Wang, L. A Versatile PdRu Bimetallic Nanoenzyme-Integrated Enzyme-Linked Immunosorbent Assay for Highly Sensitive Escherichia coli O157:H7 Detection. Anal. Chem. 2023, 95, 9237–9243. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Liang, Z.; Xu, Y.; Chen, Z.; Wang, J.; Zhou, L. Ultrasensitive and Rapid Visual Detection of Escherichia coli O157:H7 Based on RAA-CRISPR/Cas12a System. Biosensors 2023, 13, 659. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Bu, S.; Xue, L.; Li, X.; Zhou, H.; Wang, X.; Wan, J. Ultrasensitive Detection of Pathogenic Bacteria by Primer Exchange Reaction Coupled with PGM. Microchem. J. 2023, 187, 108401. [Google Scholar] [CrossRef]
- Donthu, N.; Kumar, S.; Mukherjee, D.; Pandey, N.; Lim, W.M. How to Conduct a Bibliometric Analysis: An Overview and Guidelines. J. Bus. Res. 2021, 133, 285–296. [Google Scholar] [CrossRef]
- Perianes-Rodriguez, A.; Waltman, L.; van Eck, N.J. Constructing Bibliometric Networks: A Comparison between Full and Fractional Counting. J. Informetr. 2016, 10, 1178–1195. [Google Scholar] [CrossRef]
- Aria, M.; Cuccurullo, C. Bibliometrix: An R-Tool for Comprehensive Science Mapping Analysis. J. Informetr. 2017, 11, 959–975. [Google Scholar] [CrossRef]
- Alkhammash, R. Bibliometric, Network, and Thematic Mapping Analyses of Metaphor and Discourse in COVID-19 Publications from 2020 to 2022. Front Psychol 2023, 13, 1062943. [Google Scholar] [CrossRef]
- Szomszor, M.; Adams, J.; Fry, R.; Gebert, C.; Pendlebury, D.A.; Potter, R.W.K.; Rogers, G. Interpreting Bibliometric Data. Front. Res. Metr. Anal. 2021, 5, 628703. [Google Scholar] [CrossRef]
- Mejia, C.; Wu, M.; Zhang, Y.; Kajikawa, Y. Exploring Topics in Bibliometric Research Through Citation Networks and Semantic Analysis. Front. Res. Metr. Anal. 2021, 6, 742311. [Google Scholar] [CrossRef] [PubMed]
- Ullah, M.; Shahid, A.; ud Din, I.; Roman, M.; Assam, M.; Fayaz, M.; Ghadi, Y.; Aljuaid, H. Analyzing Interdisciplinary Research Using Co-Authorship Networks. Complexity 2022, 2022, 2524491. [Google Scholar] [CrossRef]
- Pesta, B.; Fuerst, J.; Kirkegaard, E.O.W. Bibliometric Keyword Analysis across Seventeen Years (2000–2016) of Intelligence Articles. J. Intell. 2018, 6, 46. [Google Scholar] [CrossRef] [PubMed]
- Glantz, P.J.; Gyles, C.L.; Greenfield, J.; Otskov, F. Isolation of Escherichia coli O157 from Pigs with Colibacillosis in Canada and the United States. Can. J. Comp. Med. 1973, 37, 200. [Google Scholar] [PubMed]
- Zacca-González, G.; Chinchilla-Rodríguez, Z.; Vargas-Quesada, B.; De Moya-Anegón, F. Bibliometric Analysis of Regional Latin America’s Scientific Output in Public Health through SCImago Journal & Country Rank. BMC Public Health 2014, 14, 632. [Google Scholar] [CrossRef]
- Tian, W.; Cai, R.; Fang, Z.; Geng, Y.; Wang, X.; Hu, Z. Understanding Co-Corresponding Authorship: A Bibliometric Analysis and Detailed Overview. J. Assoc. Inf. Sci. Technol. 2024, 75, 3–23. [Google Scholar] [CrossRef]
- Simopoulou, M.; Sfakianoudis, K.; Maziotis, E.; Rapani, A.; Giannelou, P.; Pantou, A.; Anifandis, G.; Bakas, P. Assessing Clinical Embryology Research: A Global Bibliometric Analysis. Medicina 2020, 56, 210. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Xiao, L. Selecting Publication Keywords for Domain Analysis in Bibliometrics: A Comparison of Three Methods. J. Informetr. 2016, 10, 212–223. [Google Scholar] [CrossRef]
- Simko, I. Analysis of Bibliometric Indicators to Determine Citation Bias. Palgrave Commun. 2015, 1, 15011. [Google Scholar] [CrossRef]
- Perna, N.T.; Plunkett, G.; Burland, V.; Mau, B.; Glasner, J.D.; Rose, D.J.; Mayhew, G.F.; Evans, P.S.; Gregor, J.; Kirkpatrick, H.A.; et al. Genome Sequence of Enterohaemorrhagic Escherichia coli O157:H7. Nature 2001, 409, 529–533. [Google Scholar] [CrossRef]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria Can Protect from Enteropathogenic Infection through Production of Acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Makino, K.; Ohnishi, M.; Kurokawa, K.; Ishii, K.; Yokoyama, K.; Han, C.G.; Ohtsubo, E.; Nakayama, K.; Murata, T.; et al. Complete Genome Sequence of Enterohemorrhagic Eschelichia Coli O157:H7 and Genomic Comparison with a Laboratory Strain K-12. DNA Res. 2001, 8, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Mcdaniel, T.K.; Jarvis, K.G.; Donnenberg, M.S.; Kaper, J.B. A Genetic Locus of Enterocyte Effacement Conserved among Diverse Enterobacterial Pathogens. Proc. Natl. Acad. Sci. USA 1995, 92, 1664–1668. [Google Scholar] [CrossRef] [PubMed]
- Rangel, J.M.; Sparling, P.H.; Crowe, C.; Griffin, P.M.; Swerdlow, D.L. Epidemiology of Escherichia coli O157:H7 Outbreaks, United States, 1982–2002-Volume 11, Number 4—April 2005-Emerging Infectious Diseases Journal-CDC. Emerg. Infect. Dis. 2005, 11, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.S.; Jelacic, S.; Habeeb, R.L.; Watkins, S.L.; Tarr, P.I. The Risk of the Hemolytic–Uremic Syndrome after Antibiotic Treatment of Escherichia coli O157:H7 Infections. N. Engl. J. Med. 2000, 342, 1930–1936. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, B.; Barrett, T.J.; Hunter, S.B.; Tauxe, R.V. PulseNet: The Molecular Subtyping Network for Foodborne Bacterial Disease Surveillance, United States-Volume 7, Number 3—June 2001-Emerging Infectious Diseases Journal-CDC. Emerg. Infect. Dis. 2001, 7, 382–389. [Google Scholar] [CrossRef] [PubMed]
- Besser, R.E.; Lett, S.M.; Weber, J.T.; Doyle, M.P.; Barrett, T.J.; Wells, J.G.; Griffin, P.M. An Outbreak of Diarrhea and Hemolytic Uremic Syndrome From Escherichia coli O157:H7 in Fresh-Pressed Apple Cider. JAMA 1993, 269, 2217–2220. [Google Scholar] [CrossRef]
- Surette, M.G.; Miller, M.B.; Bassler, B.L. Quorum Sensing in Escherichia coli, Salmonella Typhimurium, and Vibrio Harveyi: A New Family of Genes Responsible for Autoinducer Production. Proc. Natl. Acad. Sci. USA 1999, 96, 1639–1644. [Google Scholar] [CrossRef]
- Liu, Y.; He, L.; Mustapha, A.; Li, H.; Hu, Z.Q.; Lin, M. Antibacterial Activities of Zinc Oxide Nanoparticles against Escherichia coli O157:H7. J. Appl. Microbiol. 2009, 107, 1193–1201. [Google Scholar] [CrossRef]
- Brookes, B.C. Bradford’s Law and the Bibliography of Science. Nature 1969, 224, 953–956. [Google Scholar] [CrossRef]
- Koo, M. Systemic Lupus Erythematosus Research: A Bibliometric Analysis over a 50-Year Period. Int. J. Environ. Res. Public Health 2021, 18, 7095. [Google Scholar] [CrossRef] [PubMed]
- Khare, A.; Jain, R. Mapping the Conceptual and Intellectual Structure of the Consumer Vulnerability Field: A Bibliometric Analysis. J. Bus. Res. 2022, 150, 567–584. [Google Scholar] [CrossRef]
- Tzeng, J.; Lu, H.; Li, W.H. Multidimensional Scaling for Large Genomic Data Sets. BMC Bioinform. 2008, 9, 179. [Google Scholar] [CrossRef] [PubMed]
- Belter, C.W. Bibliometric Indicators: Opportunities and Limits. J. Med. Libr. Assoc. 2015, 103, 219. [Google Scholar] [CrossRef]
- Kumar, D.; Kumar, G.; Kumar, R.; Mamidi, N.; Jangid, H.; Kumar, D.; Kumar, G.; Kumar, R.; Mamidi, N. Bibliometric Examination of Global Scientific Research about Carbapenem-Resistant Acinetobacter Baumannii (CRAB). Antibiotics 2023, 12, 1593. [Google Scholar] [CrossRef]
- Chuang, C.W.; Chang, A.; Chen, M.; Selvamani, M.J.P.; Shia, B.C. A Worldwide Bibliometric Analysis of Publications on Artificial Intelligence and Ethics in the Past Seven Decades. Sustainability 2022, 14, 11125. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).