Progressive Design of a Ranatuerin-2 Peptide from Amolops wuyiensis: Enhancement of Bioactivity and In Vivo Efficacy
Abstract
1. Introduction
2. Results
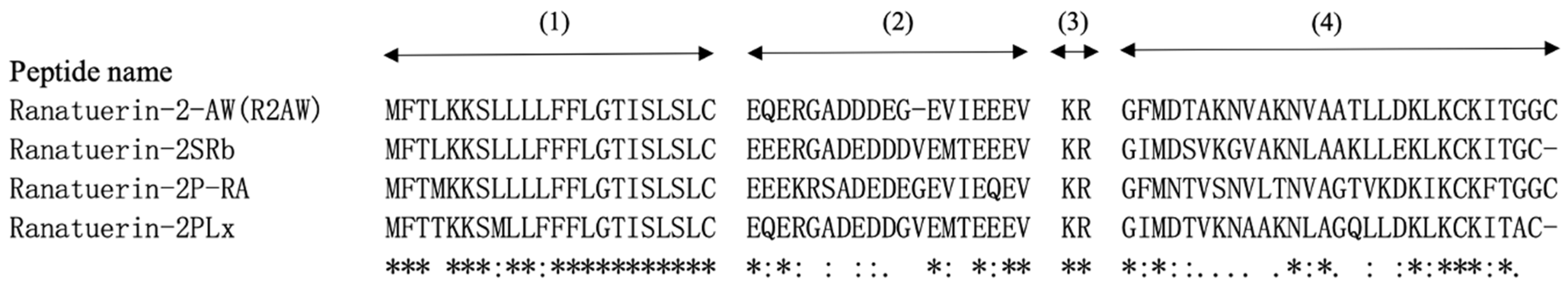
2.1. “Shotgun” Cloning of R2AW from Amolops Wuyiensis Skin Secretion-Derived cDNA Library
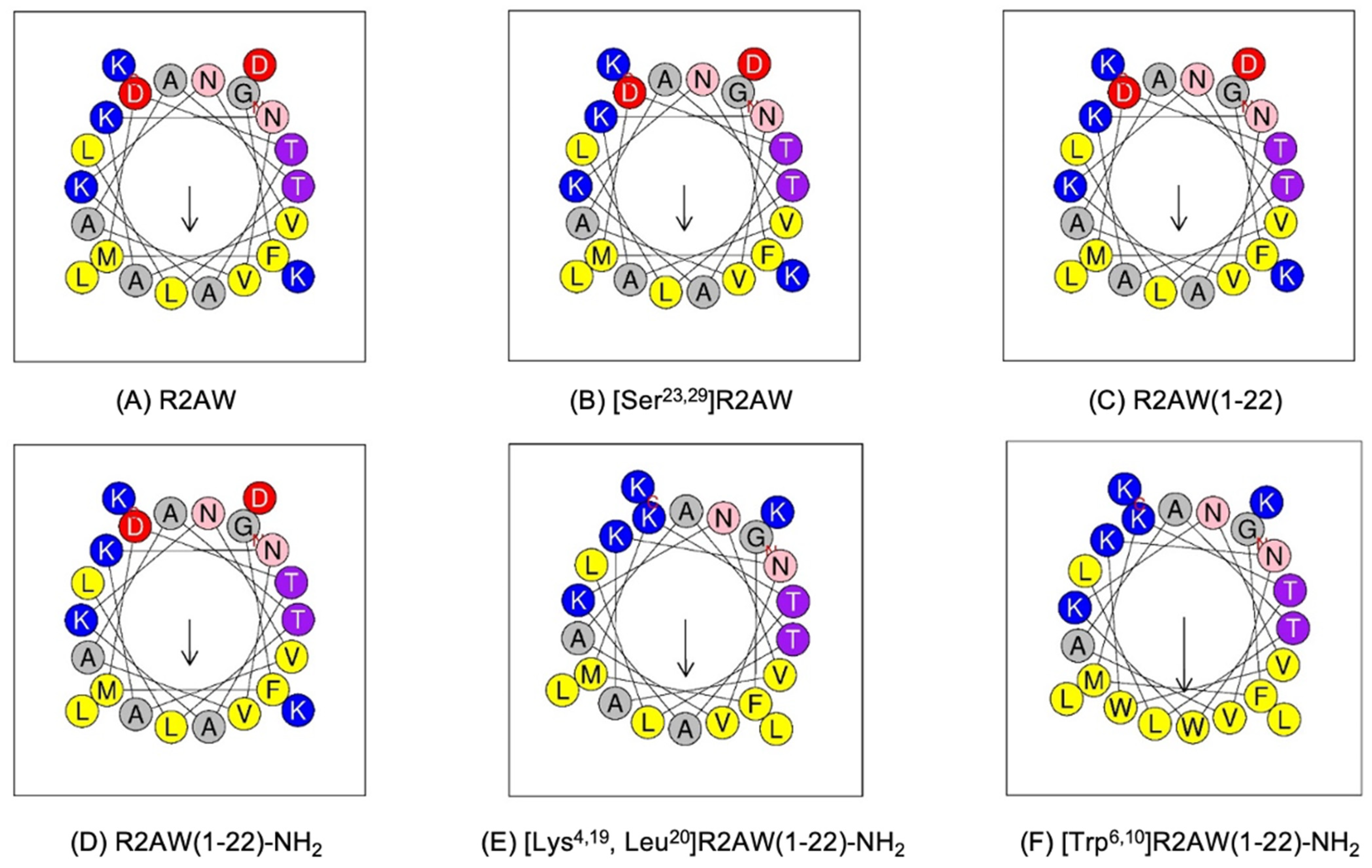
2.2. Peptide Design
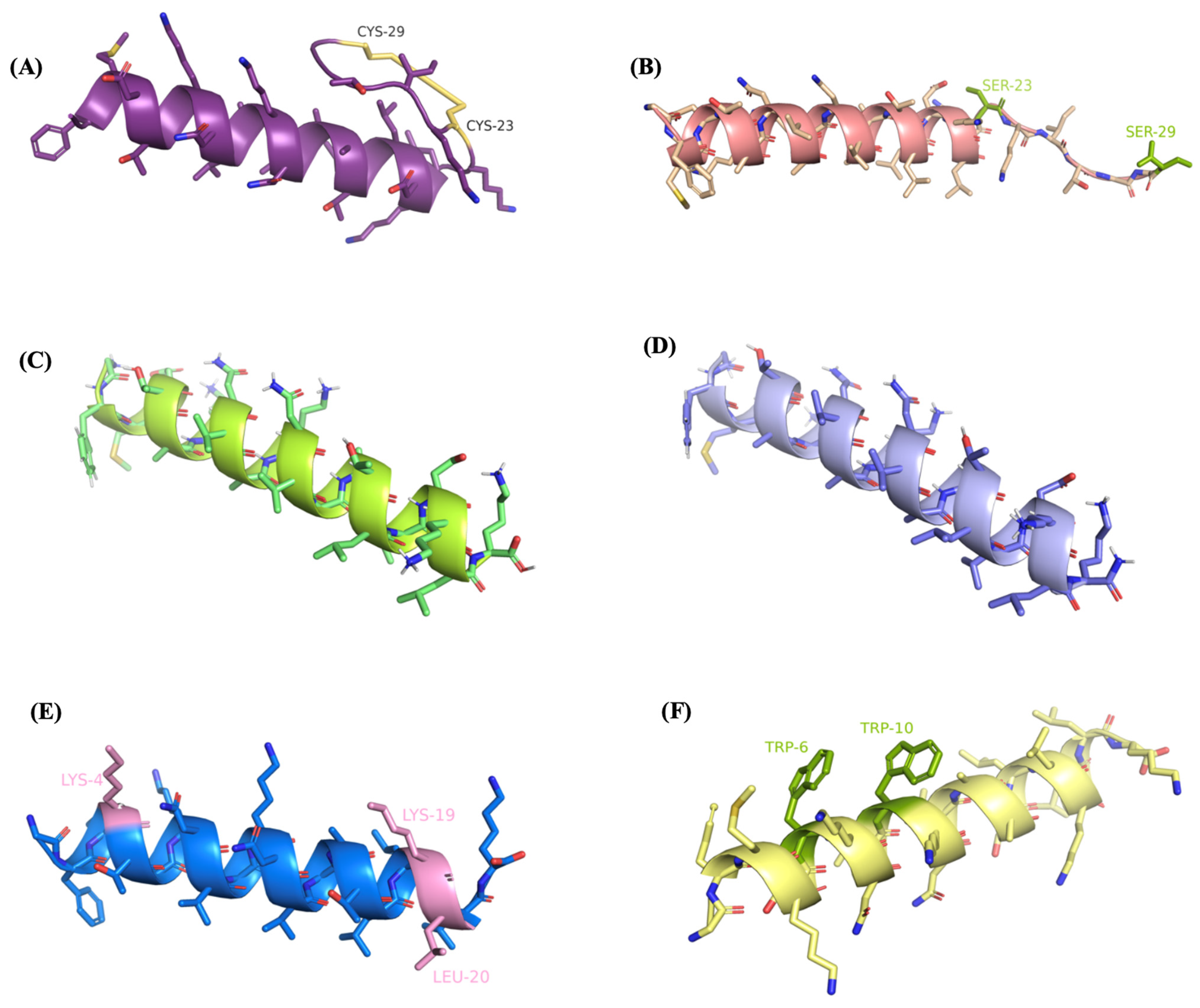
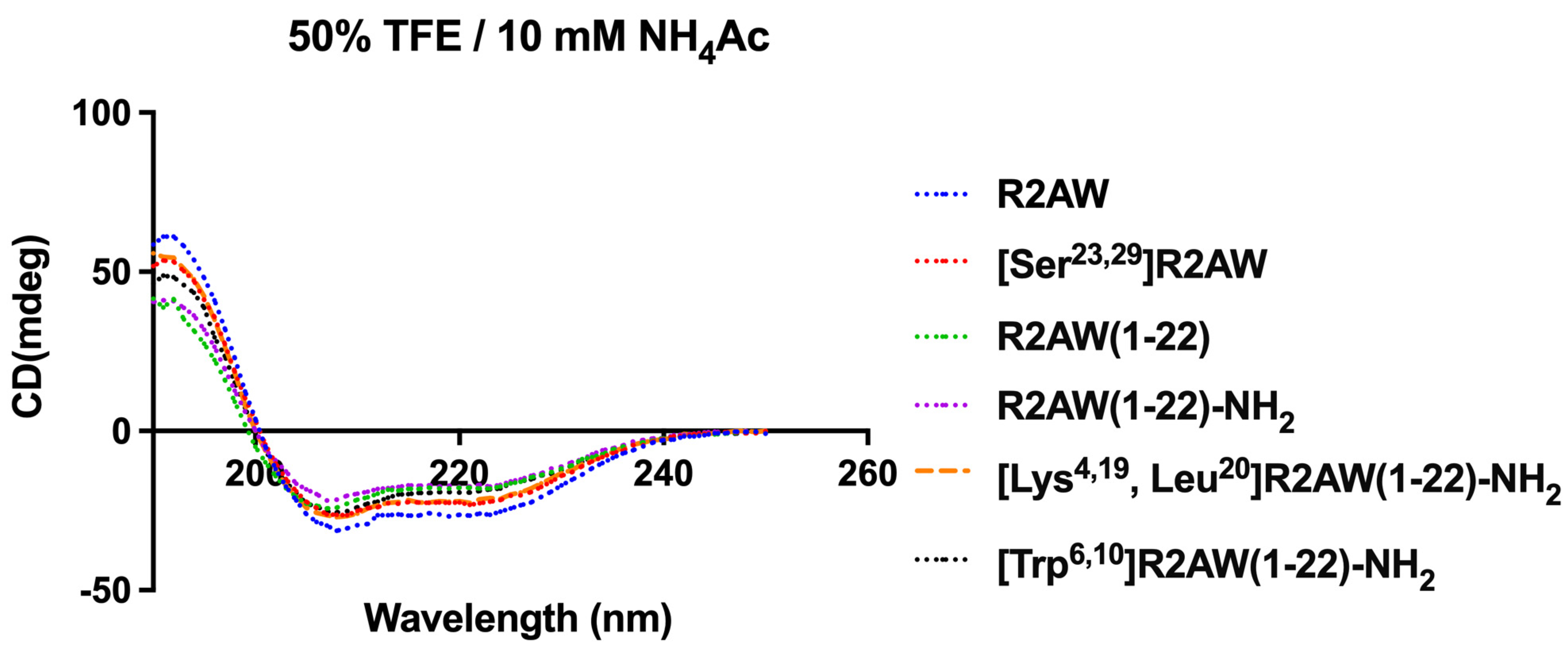
2.3. Conformational Analysis of the Designed Analogues of R2AW
2.4. Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) of Five Analogues of R2AW
2.5. Prevention and Eradication of Biofilm by Five Designed Analogues of R2AW
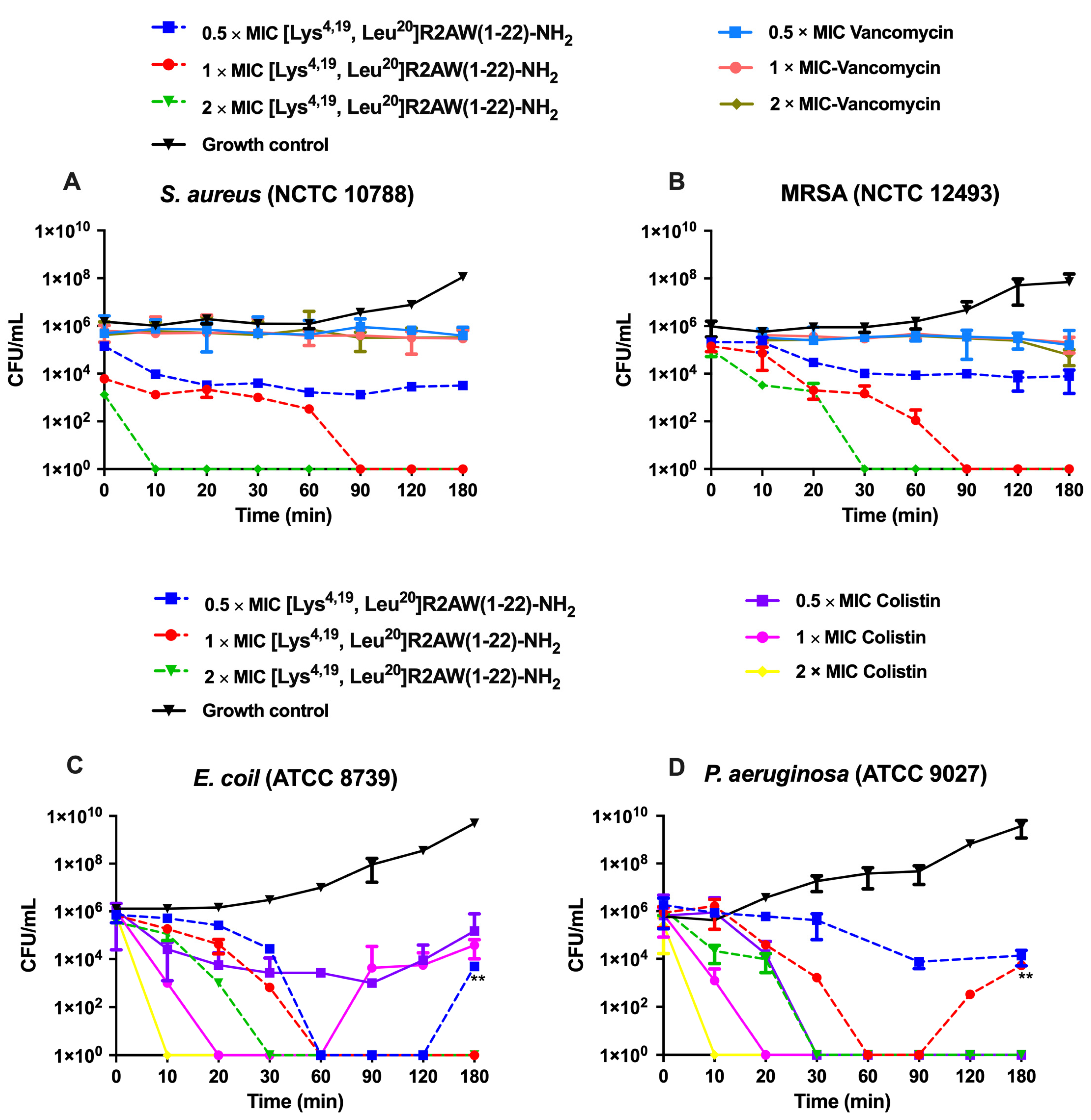
2.6. Killing Kinetics against S. aureus, E. coli, MRSA, and P. aeruginosa by [Lys4,19, Leu20]R2AW(1-22)-NH2
2.7. Membrane Permeability of S. aureus, E. coli, MRSA and P. aeruginosa by [Lys4,19, Leu20]R2AW(1-22)-NH2
2.8. Treatment of MRSA-Infected Waxworms with [Lys4,19, Leu20]R2AW(1-22)-NH2
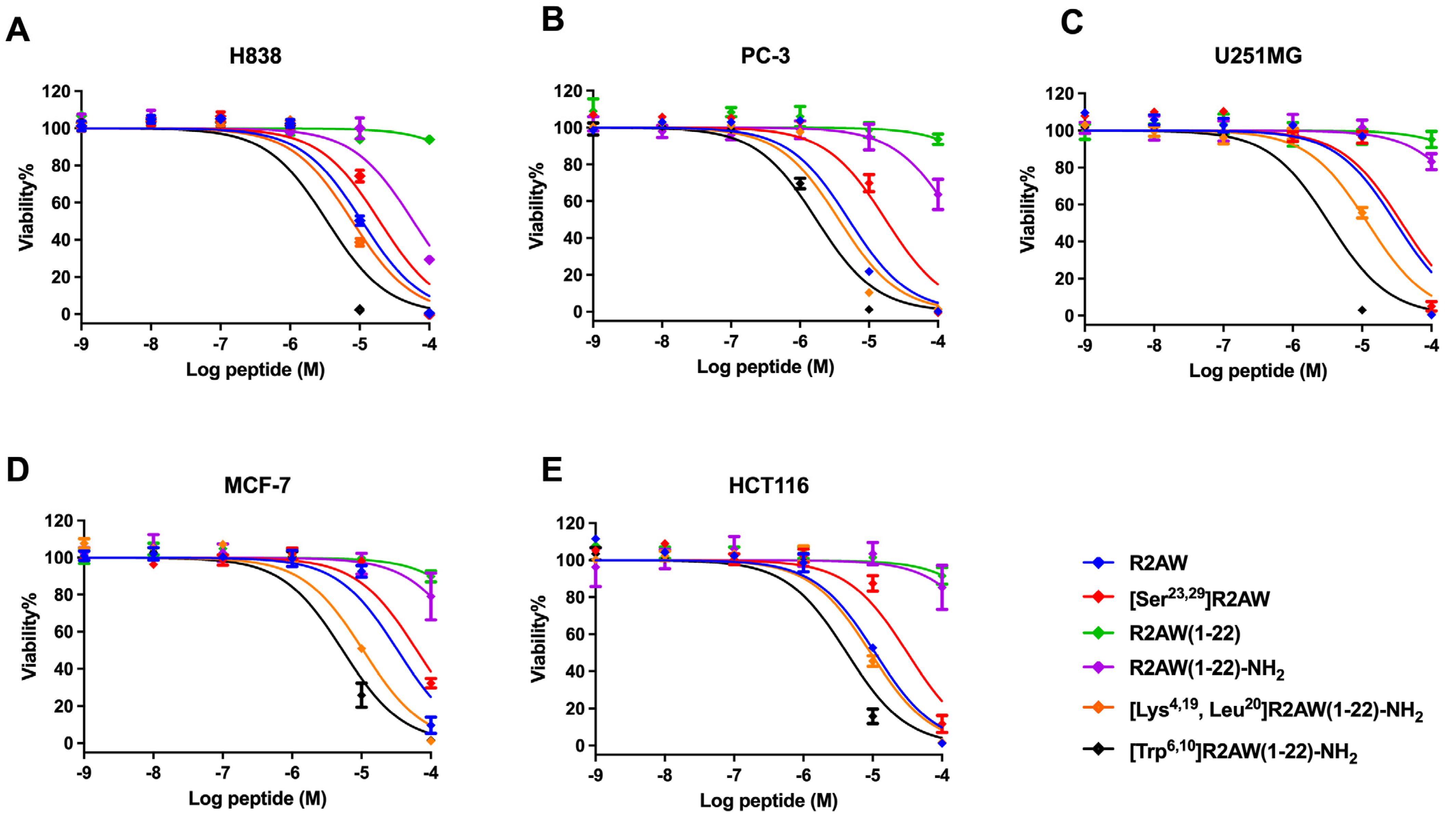
2.9. Antiproliferative Activity of Designed Analogues of R2AW against Cancer Cells
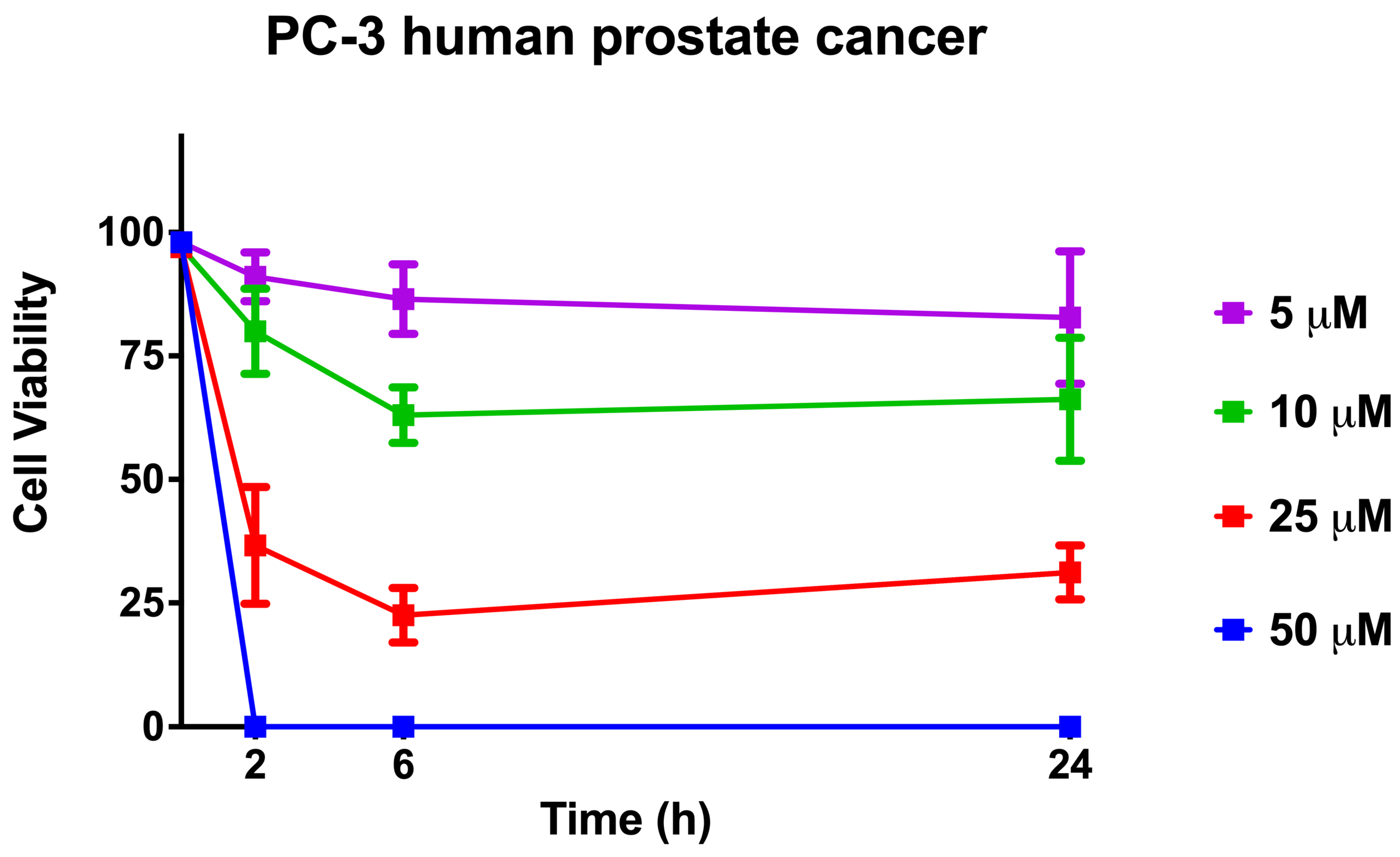
2.10. Cell Viability When Using [Lys4,19, Leu20]R2AW(1-22)-NH2 against Human Prostate Cancer Cells
2.11. Haemolytic Activities of R2AW and Its Designed Analogues
3. Discussion
4. Materials and Methods
4.1. Acquisition of Frog Skin Secretion
4.2. ‘Shotgun’ Cloning and Sequencing of Ranatuerin-2-AW Precursor-Encoding cDNA
4.3. Prediction of Secondary Structure
4.4. Synthesis of R2AW and Its Five Designed Analogues
4.5. Purification and Characterisation of R2AW and Its Designed Analogues
4.6. Circular Dichroism Spectra
4.7. Determination of MICs and MBCs
4.8. Antibiofilm Assays
4.9. Time–Killing Assays
4.10. Bacterial Cell Membrane Permeability Assays
4.11. Evaluation of Efficacy of [Lys4,19, Leu20]R2AW(1-22)-NH2 against MRSA in Insect Larvae
4.12. Anticancer MTT Assays
4.13. Trypan Blue Exclusion Assays
4.14. Haemolysis Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Munita, J.M.; Bayer, A.S.; Arias, C.A. Evolving resistance among Gram-positive pathogens. Clin. Infect. Dis. 2015, 61 (Suppl. 2), S48–S57. [Google Scholar] [CrossRef] [PubMed]
- Eichenberger, E.M.; Thaden, J.T. Epidemiology and mechanisms of resistance of extensively drug resistant gram-negative bacteria. Antibiotics 2019, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Castaneda, J.A.; Ruano-Ravina, A.; Barbosa-Lorenzo, R.; Paillier-Gonzalez, J.E.; Saldana-Campos, J.C.; Salinas, D.F.; Lemos-Luengas, E.V. Mortality due to KPC carbapenemase-producing Klebsiella pneumoniae infections: Systematic review and meta-analysis: Mortality due to KPC Klebsiella pneumoniae infections. J. Infect. 2018, 76, 438–448. [Google Scholar] [CrossRef]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Parsonnet, J. Bacterial infection as a cause of cancer. Environ. Health Perspect. 1995, 103, 263–268. [Google Scholar] [PubMed]
- Rolston, K.V. The spectrum of pulmonary infections in cancer patients. Curr. Opin. Oncol. 2001, 13, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Michaud, D.S. Role of bacterial infections in pancreatic cancer. Carcinogenesis 2013, 34, 2193–2197. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial peptides: Classification, design, application and research progress in multiple fields. Front. Microbiol. 2020, 11, 2559. [Google Scholar] [CrossRef]
- Conlon, J.M.; Mechkarska, M.; Lukic, M.L.; Flatt, P.R. Potential therapeutic applications of multifunctional host-defense peptides from frog skin as anti-cancer, anti-viral, immunomodulatory, and anti-diabetic agents. Peptides 2014, 57, 67–77. [Google Scholar] [CrossRef]
- Won, H.-S.; Kang, S.-J.; Lee, B.-J. Action mechanism and structural requirements of the antimicrobial peptides, gaegurins. Biochim. Biophys. Acta Biomembr. 2009, 1788, 1620–1629. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Kolodziejek, J.; Nowotny, N. Antimicrobial peptides from ranid frogs: Taxonomic and phylogenetic markers and a potential source of new therapeutic agents. Biochim. Biophys. Acta 2004, 1696, 1–14. [Google Scholar] [CrossRef]
- Goraya, J.; Knoop, F.C.; Conlon, J.M. Ranatuerins: Antimicrobial peptides isolated from the skin of the American bullfrog, Rana catesbeiana. Biochem. Biophys. Res. Commun. 1998, 250, 589–592. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Zhou, M.; Rao, P.; Walker, B.; Shaw, C. The Chinese bamboo leaf odorous frog (Rana (Odorrana) versabilis) and North American Rana frogs share the same families of skin antimicrobial peptides. Peptides 2006, 27, 1738–1744. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Kolodziejek, J.; Nowotny, N. Antimicrobial peptides from the skins of North American frogs. Biochim. Biophys. Acta Biomembr. 2009, 1788, 1556–1563. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, L.; Ma, C.; Zhang, Y.; Xi, X.; Wang, L.; Zhou, M.; Burrows, J.F.; Chen, T. A novel antimicrobial peptide, Ranatuerin-2PLx, showing therapeutic potential in inhibiting proliferation of cancer cells. Biosci. Rep. 2018, 38, BSR20180710. [Google Scholar] [CrossRef] [PubMed]
- Abraham, P.; Sundaram, A.; George, S.; Kumar, K.S. Structure-activity relationship and mode of action of a frog secreted antibacterial peptide B1CTcu5 using synthetically and modularly modified or deleted (SMMD) peptides. PLoS ONE 2015, 10, e0124210. [Google Scholar] [CrossRef] [PubMed]
- Bao, K.; Yuan, W.; Ma, C.; Yu, X.; Wang, L.; Hong, M.; Xi, X.; Zhou, M.; Chen, T. Modification targeting the “rana box” motif of a novel nigrocin peptide from hylarana latouchii enhances and broadens its potency against multiple bacteria. Front. Microbiol. 2018, 9, 2846. [Google Scholar] [CrossRef]
- Robertson, L.S.; Fellers, G.M.; Marranca, J.M.; Kleeman, P.M. Expression analysis and identification of antimicrobial peptide transcripts from six North American frog species. Dis. Aquat. Organ. 2013, 104, 225–236. [Google Scholar] [CrossRef]
- Schibli, D.J.; Nguyen, L.T.; Kernaghan, S.D.; Rekdal, Ø.; Vogel, H.J. Structure-Function Analysis of Tritrpticin Analogs: Potential Relationships between Antimicrobial Activities, Model Membrane Interactions, and Their Micelle-Bound NMR Structures. Biophys. J. 2006, 91, 4413–4426. [Google Scholar] [CrossRef]
- Strandberg, E.; Tiltak, D.; Ieronimo, M.; Kanithasen, N.; Wadhwani, P.; Ulrich, A. Influence of C-terminal amidation on the antimicrobial and hemolytic activities of cationic α-helical peptides. Pure Appl. Chem. 2007, 79, 717–728. [Google Scholar] [CrossRef]
- Zhu, S.; Li, W.; O’Brien-Simpson, N.; Separovic, F.; Sani, M.A. C-terminus amidation influences biological activity and membrane interaction of maculatin 1.1. Amino Acids 2021, 53, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Jindal, M.; Le, C.; Mohd Yusof, M.; Sekaran, S. Net charge, hydrophobicity and specific amino acids contribute to the activity of antimicrobial peptides. J. Transl. Med. 2014, 17, 1–7. [Google Scholar]
- Andrushchenko, V.V.; Vogel, H.J.; Prenner, E.J. Solvent-dependent structure of two tryptophan-rich antimicrobial peptides and their analogs studied by FTIR and CD spectroscopy. Biochim. Biophys. Acta 2006, 1758, 1596–1608. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Greco, I.; Molchanova, N.; Holmedal, E.; Jenssen, H.; Hummel, B.D.; Watts, J.L.; Håkansson, J.; Hansen, P.R.; Svenson, J. Correlation between hemolytic activity, cytotoxicity and systemic in vivo toxicity of synthetic antimicrobial peptides. Sci. Rep. 2020, 10, 13206. [Google Scholar] [CrossRef]
- Ramachandran, G.N.; Ramakrishnan, C.; Sasisekharan, V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963, 7, 95–99. [Google Scholar] [CrossRef]
- Gopalakrishnan, K.V.; Saravanan, S.; Rangarajan, S.; Sekar, K. Rpms: Ramachandran plot for multiple structures. J. Appl. Crystallogr. 2008, 41, 219–221. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Kwon, M.-Y.; Hong, S.-Y.; Lee, K.-H. Structure-activity analysis of brevinin 1E amide, an antimicrobial peptide from Rana esculenta. Biochim. Biophys. Acta 1998, 1387, 239–248. [Google Scholar] [CrossRef]
- Sforça, M.L.; Oyama, S.; Canduri, F.; Lorenzi, C.C.; Pertinhez, T.A.; Konno, K.; Souza, B.M.; Palma, M.S.; Ruggiero Neto, J.; Azevedo, W.F. How C-terminal carboxyamidation alters the biological activity of peptides from the venom of the eumenine solitary wasp. Biochemistry 2004, 43, 5608–5617. [Google Scholar] [CrossRef]
- Dathe, M.; Wieprecht, T. Structural features of helical antimicrobial peptides: Their potential to modulate activity on model membranes and biological cells. Biochim. Biophys. Acta 1999, 1462, 71–87. [Google Scholar] [CrossRef]
- Tossi, A.; Sandri, L.; Giangaspero, A. Amphipathic, α-helical antimicrobial peptides. Biopolymers 2000, 55, 477–489. [Google Scholar] [CrossRef]
- da Silva, A.V.; De Souza, B.M.; Dos Santos Cabrera, M.P.; Dias, N.B.; Gomes, P.C.; Neto, J.R.; Stabeli, R.G.; Palma, M.S. The effects of the C-terminal amidation of mastoparans on their biological actions and interactions with membrane-mimetic systems. Biochim. Biophys. Acta 2014, 1838, 2357–2368. [Google Scholar] [CrossRef] [PubMed]
- Bechinger, B.; Gorr, S.-U. Antimicrobial peptides: Mechanisms of action and resistance. J. Dent. Res. 2017, 96, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.J.-Y.; Loh, J.M.S.; Proft, T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 2016, 7, 214–229. [Google Scholar] [CrossRef] [PubMed]
- Simmaco, M.; Mignogna, G.; Barra, D. Antimicrobial peptides from amphibian skin: What do they tell us? Biopolymers 1998, 47, 435–450. [Google Scholar] [CrossRef]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar]
- Auer, G.K.; Weibel, D.B. Bacterial cell mechanics. Biochemistry 2017, 56, 3710–3724. [Google Scholar] [CrossRef]
- Schweizer, F. Cationic amphiphilic peptides with cancer-selective toxicity. Eur. J. Pharmacol. 2009, 625, 190–194. [Google Scholar] [CrossRef]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef]
- Gong, H.; Zhang, J.; Hu, X.; Li, Z.; Fa, K.; Liu, H.; Waigh, T.A.; McBain, A.; Lu, J.R. Hydrophobic control of the bioactivity and cytotoxicity of de novo-designed antimicrobial peptides. ACS Appl. Mater. 2019, 11, 34609–34620. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of peptide hydrophobicity in the mechanism of action of α-helical antimicrobial peptides. Antimicrob. Agents Chemother. 2007, 51, 1398–1406. [Google Scholar] [CrossRef] [PubMed]
- Yao, A.; Ma, Y.; Chen, X.; Zhou, M.; Xi, X.; Ma, C.; Ren, S.; Chen, T.; Shaw, C.; Wang, L. Modification Strategy of D-leucine Residue Addition on a Novel Peptide from Odorrana schmackeri, with Enhanced Bioactivity and In Vivo Efficacy. Toxins 2021, 13, 611. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yao, A.; Chen, X.; Wang, L.; Ma, C.; Xi, X.; Chen, T.; Shaw, C.; Zhou, M. Generation of truncated derivatives through in silico enzymatic digest of peptide GV30 target MRSA both in vitro and in vivo. Comput. Struct. Biotechnol. J. 2021, 19, 4984–4996. [Google Scholar] [CrossRef] [PubMed]
- Yao, A.; Ma, Y.; Sun, R.; Zou, W.; Chen, X.; Zhou, M.; Ma, C.; Chen, T.; Shaw, C.; Wang, L. A designed analog of an antimicrobial peptide, crabrolin, exhibits enhanced anti-proliferative and in vivo antimicrobial activity. Int. J. Mol. Sci. 2023, 24, 14472. [Google Scholar] [CrossRef] [PubMed]
- Desbois, A.P.; Coote, P.J. Wax moth larva (Galleria mellonella): An in vivo model for assessing the efficacy of antistaphylococcal agents. J. Antimicrob. Chemother. 2011, 66, 1785–1790. [Google Scholar] [CrossRef]
- Lin, Y.; Jiang, Y.; Zhao, Z.; Lu, Y.; Xi, X.; Ma, C.; Chen, X.; Zhou, M.; Chen, T.; Shaw, C.; et al. Discovery of a novel antimicrobial peptide, Temporin-pke, from the skin secretion of Pelophylax kl. esculentus, and evaluation of its structure-activity relationships. Biomolecules 2022, 12, 759. [Google Scholar] [CrossRef]
- Zou, W.; Zhang, Y.; Zhou, M.; Chen, X.; Ma, C.; Wang, T.; Jiang, Y.; Chen, T.; Shaw, C.; Wang, L. Exploring the active core of a novel antimicrobial peptide, palustrin-2LTb, from the Kuatun frog, Hylarana latouchii, using a bioinformatics-directed approach. Comput. Struct. Biotechnol. J. 2022, 20, 6192–6205. [Google Scholar] [CrossRef]




| Name | Sequence | Hydrophobicity | Hydrophobicity Moment | Net Charge |
|---|---|---|---|---|
| R2AW | GFMDTAKNVAKNVAATLLDKLKCKITGGC | 0.337 | 0.225 | +3 |
| [Ser23,29]R2AW | GFMDTAKNVAKNVAATLLDKLKSKITGGS | 0.228 | 0.279 | +3 |
| R2AW(1-22) | GFMDTAKNVAKNVAATLLDKLK | 0.255 | 0.412 | +2 |
| R2AW(1-22)-NH2 | GFMDTAKNVAKNVAATLLDKLK-NH2 | 0.255 | 0.412 | +3 |
| [Lys4,19, Leu20]R2AW(1-22)-NH2 | GFMKTAKNVAKNVAATLLKLLK-NH2 | 0.358 | 0.516 | +6 |
| [Trp6,10]R2AW(1-22)-NH2 | GFMKTWKNVWKNVAATLLKLLK-NH2 | 0.534 | 0.674 | +6 |
| Peptides | MICs (mg/L and µM) | |||||
|---|---|---|---|---|---|---|
| S. aureus (NCTC 10788) | E. coli (ATCC 8739) | MRSA (NCTC 12493) | K. pneumoniae (ATCC 43816) | E. faecium (NTCC 12697) | P. aeruginosa (ATCC 9027) | |
| R2AW | 96.3 (32) | 96.3 (32) | 770 (256) | 385 (128) | 770 (256) | 385 (128) |
| [Ser23,29]R2AW | 190 (64) | 190 (64) | 763 (256) | 763 (256) | 763 (256) | 1526 (512) |
| R2AW(1-22) | >1203 (>512) | >1203 (>512) | >1203 (>512) | >1203 (>512) | >1203 (>512) | >1203 (>512) |
| R2AW(1-22)-NH2 | 150 (64) | 300 (128) | 601 (256) | 601 (256) | 601 (256) | 1202 (512) |
| [Lys4,19, Leu20]R2AW(1-22)-NH2 | 4.7 (2) | 4.7 (2) | 4.7 (2) | 9.4 (4) | 9.4 (4) | 18.9 (8) |
| [Trp6,10]R2AW(1-22)-NH2 | 10.4 (4) | 20.7 (8) | 10.4 (4) | 41.4 (16) | 20.7 (8) | 82.8 (32) |
| Vancomycin | 0.7 (0.5) | NA * | 0.7 (0.5) | NA * | 0.7 (0.5) | NA * |
| Colistin | NA * | 0.1 (0.125) | NA * | 18.5 (16) | NA * | 1.2 (1) |
| Melittin | 5.7 (2) | 11.4 (4) | 5.7 (2) | 91.1 (32) | 5.7 (2) | 45.5 (16) |
| MBCs (mg/L and µM) | ||||||
| R2AW | 193 (64) | 193 (64) | 1541 (512) | 771 (256) | 771 (256) | 1541 (512) |
| [Ser23,29]R2AW | 191 (64) | 381 (128) | 1526 (512) | 1526 (512) | 1526 (512) | >1526 (>512) |
| R2AW(1-22) | >1202 (>512) | >1202 (>512) | >1202 (>512) | >1202 (>512) | >1202 (>512) | >1202 (>512) |
| R2AW(1-22)-NH2 | 150 (64) | 301 (128) | 601 (256) | 601 (256) | 601 (256) | >1202 (>512) |
| [Lys4,19, Leu20]R2AW(1-22)-NH2 | 4.7 (2) | 9.4 (4) | 4.7 (2) | 18.9 (8) | 18.9 (8) | 37.7 (16) |
| [Trp6,10]R2AW(1-22)-NH2 | 20.7 (8) | 41.4 (16) | 20.7 (8) | 82.8 (32) | 41.4 (16) | 166 (64) |
| Vancomycin | 0.7 (0.5) | NA * | 0.7 (0.5) | NA * | >92.7 (>64) | NA * |
| Colistin | NA * | 0.3 (0.25) | NA * | 18.5 (16) | NA * | 2.3 (2) |
| Melittin | 5.7 (2) | 11.4 (4) | 5.7 (2) | 91.1 (32) | 5.7 (2) | 91.1 (32) |
| Peptides | MBIC/MBEC (µM) | |||||
|---|---|---|---|---|---|---|
| S. aureus (NCTC 10788) | E. coli (ATCC 8739) | MRSA (NCTC 12493) | K. pneumoniae (ATCC 43816) | E. faecium (NTCC 12697) | P. aeruginosa (ATCC 9027) | |
| R2AW | 128/>512 | 128/512 | 256/>512 | 512/>512 | >512 | >512 |
| [Ser23,29]R2AW | 128/>512 | 128/512 | 256/>512 | >512 | >512 | >512 |
| R2AW(1-22) | >512 | >512 | >512 | >512 | >512 | >512 |
| R2AW(1-22)-NH2 | 128/>512 | 256/512 | 256/>256 | 512/>512 | >512 | >512 |
| [Lys4,19,Leu20]R2AW(1-22)-NH2 | 4/256 | 4/256 | 4/>512 | 8/256 | 8/>512 | 16/512 |
| [Trp6,10]R2AW(1-22)-NH2 | 8/512 | 4/256 | 8/>512 | 16/256 | 8/>512 | 32/512 |
| Peptides | IC50 (µM) | ||||
|---|---|---|---|---|---|
| H838 | PC3 | U251MG | MCF-7 | HCT116 | |
| R2AW | 10.78 | 5.093 | 30.78 | 33.20 | 10.82 |
| [Ser23,29]R2AW | 19.34 | 17.44 | 37.31 | 62.7 | 31.64 |
| R2AW(1-22) | 1423 | 1480 | 1818 | 894.5 | 1124 |
| R2AW(1-22)-NH2 | 58.91 | 194.3 | 515.8 | 373.6 | 609 |
| [Lys4,19, Leu20]R2AW(1-22)-NH2 | 7.828 | 3.671 | 12.40 | 10.58 | 9.284 |
| [Trp6,10]R2AW(1-22)-NH2 | 3.405 | 1.730 | 3.353 | 5.328 | 5.375 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, A.; Liu, T.; Cai, Y.; Zhou, S.; Chen, X.; Zhou, M.; Ma, C.; Chen, T.; Shaw, C.; Wang, L. Progressive Design of a Ranatuerin-2 Peptide from Amolops wuyiensis: Enhancement of Bioactivity and In Vivo Efficacy. Antibiotics 2024, 13, 5. https://doi.org/10.3390/antibiotics13010005
Yao A, Liu T, Cai Y, Zhou S, Chen X, Zhou M, Ma C, Chen T, Shaw C, Wang L. Progressive Design of a Ranatuerin-2 Peptide from Amolops wuyiensis: Enhancement of Bioactivity and In Vivo Efficacy. Antibiotics. 2024; 13(1):5. https://doi.org/10.3390/antibiotics13010005
Chicago/Turabian StyleYao, Aifang, Tianxing Liu, Yuhai Cai, Siqi Zhou, Xiaoling Chen, Mei Zhou, Chengbang Ma, Tianbao Chen, Chris Shaw, and Lei Wang. 2024. "Progressive Design of a Ranatuerin-2 Peptide from Amolops wuyiensis: Enhancement of Bioactivity and In Vivo Efficacy" Antibiotics 13, no. 1: 5. https://doi.org/10.3390/antibiotics13010005
APA StyleYao, A., Liu, T., Cai, Y., Zhou, S., Chen, X., Zhou, M., Ma, C., Chen, T., Shaw, C., & Wang, L. (2024). Progressive Design of a Ranatuerin-2 Peptide from Amolops wuyiensis: Enhancement of Bioactivity and In Vivo Efficacy. Antibiotics, 13(1), 5. https://doi.org/10.3390/antibiotics13010005









