The Klebsiella pneumoniae carbapenemase (KPC) β-Lactamase Has Evolved in Response to Ceftazidime Avibactam
Abstract
1. Introduction
2. Results
2.1. Alignment and Fisher’s Exact Test
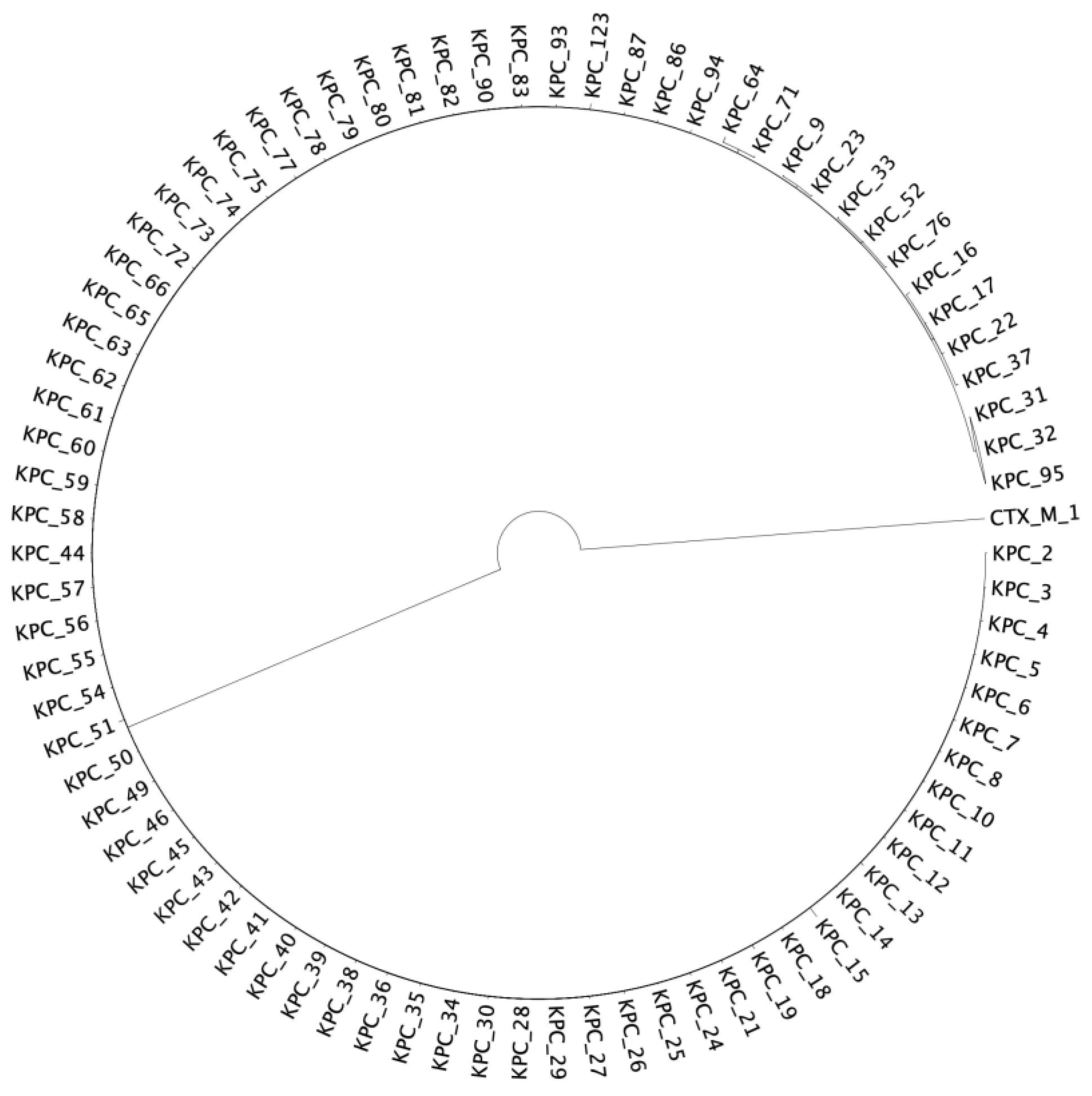
2.2. Phylogenetic Reconstruction
2.3. Analysis of Sites under Positive Selection
3. Discussion
4. Methods
4.1. Alignment
4.2. Test for Selection
4.3. β-Lactamase Numbering Scheme
4.4. Phylogenies
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nordmann, P.; Poirel, L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-Negative Bacteria. Clin. Infect. Dis. 2019, 69, S521–S528. [Google Scholar] [CrossRef] [PubMed]
- Plazak, M.E.; Tamma, P.D.; Heil, E.L. The Antibiotic Arms Race: Current and Emerging Therapy for Klebsiella Pneumoniae Carbapenemase (KPC)—Producing Bacteria. Expert. Opin. Pharmacother. 2018, 19, 2019–2031. [Google Scholar] [CrossRef] [PubMed]
- Tilahun, M.; kassa, Y.; Gedefie, A.; Ashagire, M. Emerging Carbapenem-Resistant Enterobacteriaceae Infection, Its Epidemiology and Novel Treatment Options: A Review. Infect. Drug Resist. 2021, 14, 4363–4374. [Google Scholar] [CrossRef] [PubMed]
- Ambler, R.P.; Coulson, A.F.; Frère, J.M.; Ghuysen, J.M.; Joris, B.; Forsman, M.; Levesque, R.C.; Tiraby, G.; Waley, S.G. A Standard Numbering Scheme for the Class A Beta-Lactamases. Biochem. J. 1991, 276 Pt 1, 269–270. [Google Scholar] [CrossRef] [PubMed]
- Chopra, T.; Rivard, C.; Awali, R.A.; Krishna, A.; Bonomo, R.A.; Perez, F.; Kaye, K.S. Epidemiology of Carbapenem-Resistant Enterobacteriaceae at a Long-Term Acute Care Hospital. Open Forum Infect. Dis. 2018, 5, ofy224. [Google Scholar] [CrossRef] [PubMed]
- Shields, R.K.; Chen, L.; Cheng, S.; Chavda, K.D.; Press, E.G.; Snyder, A.; Pandey, R.; Doi, Y.; Kreiswirth, B.N.; Nguyen, M.H.; et al. Emergence of Ceftazidime-Avibactam Resistance Due to Plasmid-Borne blaKPC-3 Mutations during Treatment of Carbapenem-Resistant Klebsiella Pneumoniae Infections. Antimicrob. Agents Chemother. 2017, 61, e02097-16. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Chen, J.; Shen, H.; Chen, Z.; Yang, Q.; Zhu, J.; Li, X.; Yang, Q.; Zhao, F.; Ji, J.; et al. Emergence and Rising of Ceftazidime-Avibactam Resistant KPC-Producing Pseudomonas Aeruginosa in China: A Molecular Epidemiology Study. medRxiv 2020. [Google Scholar] [CrossRef]
- Yigit, H.; Queenan, A.M.; Anderson, G.J.; Domenech-Sanchez, A.; Biddle, J.W.; Steward, C.D.; Alberti, S.; Bush, K.; Tenover, F.C. Novel Carbapenem-Hydrolyzing β-Lactamase, KPC-1, from a Carbapenem-Resistant Strain of Klebsiella Pneumoniae. Antimicrob. Agents Chemother. 2001, 45, 1151–1161. [Google Scholar] [CrossRef]
- Nordmann, P.; Naas, T.; Poirel, L. Global Spread of Carbapenemase-Producing Enterobacteriaceae. Emerg. Infect. Dis. 2011, 17, 1791–1798. [Google Scholar] [CrossRef]
- Aurilio, C.; Sansone, P.; Barbarisi, M.; Pota, V.; Giaccari, L.G.; Coppolino, F.; Barbarisi, A.; Passavanti, M.B.; Pace, M.C. Mechanisms of Action of Carbapenem Resistance. Antibiotics 2022, 11, 421. [Google Scholar] [CrossRef]
- Gupta, N.; Limbago, B.M.; Patel, J.B.; Kallen, A.J. Carbapenem-Resistant Enterobacteriaceae: Epidemiology and Prevention. Clin. Infect. Dis. 2011, 53, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Bratu, S.; Tolaney, P.; Karumudi, U.; Quale, J.; Mooty, M.; Nichani, S.; Landman, D. Carbapenemase-Producing Klebsiella Pneumoniae in Brooklyn, NY: Molecular Epidemiology and in Vitro Activity of Polymyxin B and Other Agents. J. Antimicrob. Chemother. 2005, 56, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Hobson, C.A.; Pierrat, G.; Tenaillon, O.; Bonacorsi, S.; Bercot, B.; Jaouen, E.; Jacquier, H.; Birgy, A. Klebsiella Pneumoniae Carbapenemase Variants Resistant to Ceftazidime-Avibactam: An Evolutionary Overview. Antimicrob. Agents Chemother. 2022, 66, e0044722. [Google Scholar] [CrossRef] [PubMed]
- Villegas, M.V.; Lolans, K.; Correa, A.; Kattan, J.N.; Lopez, J.A.; Quinn, J.P. Colombian Nosocomial Resistance Study Group First Identification of Pseudomonas Aeruginosa Isolates Producing a KPC-Type Carbapenem-Hydrolyzing Beta-Lactamase. Antimicrob. Agents Chemother. 2007, 51, 1553–1555. [Google Scholar] [CrossRef] [PubMed]
- Forero-Hurtado, D.; Corredor-Rozo, Z.L.; Ruiz-Castellanos, J.S.; Márquez-Ortiz, R.A.; Abril, D.; Vanegas, N.; Lafaurie, G.I.; Chambrone, L.; Escobar-Pérez, J. Worldwide Dissemination of blaKPC Gene by Novel Mobilization Platforms in Pseudomonas Aeruginosa: A Systematic Review. Antibiotics 2023, 12, 658. [Google Scholar] [CrossRef]
- Cuzon, G.; Naas, T.; Nordmann, P. Functional Characterization of Tn4401, a Tn3-Based Transposon Involved in blaKPC Gene Mobilization. Antimicrob. Agents Chemother. 2011, 55, 5370–5373. [Google Scholar] [CrossRef]
- Cuzon, G.; Naas, T.; Truong, H.; Villegas, M.-V.; Wisell, K.T.; Carmeli, Y.; Gales, A.C.; Navon-Venezia, S.; Quinn, J.P.; Nordmann, P. Worldwide Diversity of Klebsiella Pneumoniae That Produce β-Lactamase blaKPC-2 Gene. Emerg. Infect. Dis. 2010, 16, 1349–1356. [Google Scholar] [CrossRef]
- Chen, L.; Mathema, B.; Chavda, K.D.; DeLeo, F.R.; Bonomo, R.A.; Kreiswirth, B.N. Carbapenemase-Producing Klebsiella Pneumoniae: Molecular and Genetic Decoding. Trends Microbiol. 2014, 22, 686–696. [Google Scholar] [CrossRef]
- Jousset, A.B.; Bonnin, R.A.; Takissian, J.; Girlich, D.; Mihaila, L.; Cabanel, N.; Dortet, L.; Glaser, P.; Naas, T. Concomitant Carriage of KPC-Producing and Non-KPC-Producing Klebsiella Pneumoniae ST512 within a Single Patient. J. Antimicrob. Chemother. 2020, 75, 2087–2092. [Google Scholar] [CrossRef]
- Eilertson, B.; Chen, L.; Li, A.; Chavda, K.D.; Chavda, B.; Kreiswirth, B.N. CG258 Klebsiella Pneumoniae Isolates without β-Lactam Resistance at the Onset of the Carbapenem-Resistant Enterobacteriaceae Epidemic in New York City. J. Antimicrob. Chemother. 2019, 74, 17–21. [Google Scholar] [CrossRef]
- Li, D.; Li, P.; Peng, M.; Zhao, X.; Jiang, X.; Wang, D.; Yuan, Y.; Guo, Q.; Wang, M.; Xu, X.; et al. Transmission Barrier of the blaKPC Plasmid Mediated by Type I Restriction-Modification Systems in Escherichia Coli. J. Antimicrob. Chemother. 2022, 77, 952–956. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wu, Y.; Huang, L.; Zhang, Y.; Sun, Q.; Lu, J.; Zeng, Y.; Dong, N.; Cai, C.; Shen, Z.; et al. The Rapid Emergence of Ceftazidime-Avibactam Resistance Mediated by KPC Variants in Carbapenem-Resistant Klebsiella Pneumoniae in Zhejiang Province, China. Antibiotics 2022, 11, 731. [Google Scholar] [CrossRef] [PubMed]
- Campogiani, L.; Vitale, P.; Lodi, A.; Imeneo, A.; Fontana, C.; D’Agostini, C.; Compagno, M.; Coppola, L.; Spalliera, I.; Malagnino, V.; et al. Resistance to Ceftazidime/Avibactam in Klebsiella Pneumoniae KPC-Producing Isolates: A Real-Life Observational Study. Antibiotics 2023, 12, 820. [Google Scholar] [CrossRef] [PubMed]
- Ehmann, D.E.; Jahić, H.; Ross, P.L.; Gu, R.-F.; Hu, J.; Kern, G.; Walkup, G.K.; Fisher, S.L. Avibactam Is a Covalent, Reversible, Non–β-Lactam β-Lactamase Inhibitor. Proc. Natl. Acad. Sci. USA 2012, 109, 11663–11668. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, J.; Wang, R.; Cai, Y. Resistance to Ceftazidime-Avibactam and Underlying Mechanisms. J. Glob. Antimicrob. Resist. 2020, 22, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Barnes, M.D.; Winkler, M.L.; Taracila, M.A.; Page, M.G.; Desarbre, E.; Kreiswirth, B.N.; Shields, R.K.; Nguyen, M.-H.; Clancy, C.; Spellberg, B.; et al. Klebsiella Pneumoniae Carbapenemase-2 (KPC-2), Substitutions at Ambler Position Asp179, and Resistance to Ceftazidime-Avibactam: Unique Antibiotic-Resistant Phenotypes Emerge from β-Lactamase Protein Engineering. mBio 2017, 8, e00528-17. [Google Scholar] [CrossRef]
- Compain, F.; Dorchène, D.; Arthur, M. Combination of Amino Acid Substitutions Leading to CTX-M-15-Mediated Resistance to the Ceftazidime-Avibactam Combination. Antimicrob. Agents Chemother. 2018, 62, e00357-18. [Google Scholar] [CrossRef]
- Livermore, D.M.; Mushtaq, S.; Doumith, M.; Jamrozy, D.; Nichols, W.W.; Woodford, N. Selection of Mutants with Resistance or Diminished Susceptibility to Ceftazidime/Avibactam from ESBL- and AmpC-Producing Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 3336–3345. [Google Scholar] [CrossRef]
- Winkler, M.L.; Papp-Wallace, K.M.; Taracila, M.A.; Bonomo, R.A. Avibactam and Inhibitor-Resistant SHV β-Lactamases. Antimicrob. Agents Chemother. 2015, 59, 3700–3709. [Google Scholar] [CrossRef]
- Fröhlich, C.; Sørum, V.; Thomassen, A.M.; Johnsen, P.J.; Leiros, H.-K.S.; Samuelsen, Ø. OXA-48-Mediated Ceftazidime-Avibactam Resistance Is Associated with Evolutionary Trade-Offs. mSphere 2019, 4, e00024-19. [Google Scholar] [CrossRef]
- Venditti, C.; Butera, O.; Meledandri, M.; Balice, M.P.; Cocciolillo, G.C.; Fontana, C.; D’Arezzo, S.; De Giuli, C.; Antonini, M.; Capone, A.; et al. Molecular Analysis of Clinical Isolates of Ceftazidime-Avibactam-Resistant Klebsiella Pneumoniae. Clin. Microbiol. Infect. 2021, 27, 1040.e1–1040.e6. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Tamura, K.; Nei, M. MEGA: Molecular Evolutionary Genetics Analysis Software for Microcomputers. Comput. Appl. Biosci. 1994, 10, 189–191. [Google Scholar] [CrossRef] [PubMed]
- Barlow, M.; Fatollahi, J.; Salverda, M. Evidence for Recombination among the Alleles Encoding TEM and SHV Beta-Lactamases. J. Antimicrob. Chemother. 2009, 63, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Oueslati, S.; Tlili, L.; Exilie, C.; Bernabeu, S.; Iorga, B.; Bonnin, R.A.; Dortet, L.; Naas, T. Different Phenotypic Expression of KPC β-Lactamase Variants and Challenges in Their Detection. J. Antimicrob. Chemother. 2020, 75, 769–771. [Google Scholar] [CrossRef]
- Tooke, C.L.; Hinchliffe, P.; Bonomo, R.A.; Schofield, C.J.; Mulholland, A.J.; Spencer, J. Natural Variants Modify Klebsiella Pneumoniae Carbapenemase (KPC) Acyl-Enzyme Conformational Dynamics to Extend Antibiotic Resistance. J. Biol. Chem. 2021, 296, 100126. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.C.; Rice, K.; Palzkill, T. Natural Variants of the KPC-2 Carbapenemase Have Evolved Increased Catalytic Efficiency for Ceftazidime Hydrolysis at the Cost of Enzyme Stability. PLoS Pathog. 2015, 11, e1004949. [Google Scholar] [CrossRef] [PubMed]
- Shields, R.K.; Nguyen, M.H.; Press, E.G.; Chen, L.; Kreiswirth, B.N.; Clancy, C.J. In Vitro Selection of Meropenem Resistance among Ceftazidime-Avibactam-Resistant, Meropenem-Susceptible Klebsiella Pneumoniae Isolates with Variant KPC-3 Carbapenemases. Antimicrob. Agents Chemother. 2017, 61, e00079-17. [Google Scholar] [CrossRef]
- Shields, R.K.; Potoski, B.A.; Haidar, G.; Hao, B.; Doi, Y.; Chen, L.; Press, E.G.; Kreiswirth, B.N.; Clancy, C.J.; Nguyen, M.H. Clinical Outcomes, Drug Toxicity, and Emergence of Ceftazidime-Avibactam Resistance Among Patients Treated for Carbapenem-Resistant Enterobacteriaceae Infections. Clin. Infect. Dis. 2016, 63, 1615–1618. [Google Scholar] [CrossRef]
- Zhang, P.; Shi, Q.; Hu, H.; Hong, B.; Wu, X.; Du, X.; Akova, M.; Yu, Y. Emergence of Ceftazidime/Avibactam Resistance in Carbapenem-Resistant Klebsiella Pneumoniae in China. Clin. Microbiol. Infect. 2020, 26, 124.e1–124.e4. [Google Scholar] [CrossRef]
- Winkler, M.L.; Papp-Wallace, K.M.; Bonomo, R.A. Activity of Ceftazidime/Avibactam against Isogenic Strains of Escherichia Coli Containing KPC and SHV β-Lactamases with Single Amino Acid Substitutions in the Ω-Loop. J. Antimicrob. Chemother. 2015, 70, 2279–2286. [Google Scholar] [CrossRef]
- Parwana, D.; Gu, J.; Wang, Q.; Bethel, C.R.; Marshall, E.; Hujer, A.M.; Bonomo, R.A.; Haider, S. The Structural Role of N170 in Substrate-Assisted Deacylation in KPC-2 β-Lactamase. bioRxiv 2023. [Google Scholar] [CrossRef]
- Hemarajata, P.; Humphries, R.M. Ceftazidime/Avibactam Resistance Associated with L169P Mutation in the Omega Loop of KPC-2. J. Antimicrob. Chemother. 2019, 74, 1241–1243. [Google Scholar] [CrossRef] [PubMed]
- Levitt, P.S.; Papp-Wallace, K.M.; Taracila, M.A.; Hujer, A.M.; Winkler, M.L.; Smith, K.M.; Xu, Y.; Harris, M.E.; Bonomo, R.A. Exploring the Role of a Conserved Class A Residue in the Ω-Loop of KPC-2 β-Lactamase: A Mechanism for Ceftazidime Hydrolysis. J. Biol. Chem. 2012, 287, 31783–31793. [Google Scholar] [CrossRef] [PubMed]
- Alsenani, T.A.; Viviani, S.L.; Kumar, V.; Taracila, M.A.; Bethel, C.R.; Barnes, M.D.; Papp-Wallace, K.M.; Shields, R.K.; Nguyen, M.H.; Clancy, C.J.; et al. Structural Characterization of the D179N and D179Y Variants of KPC-2 β-Lactamase: Ω-Loop Destabilization as a Mechanism of Resistance to Ceftazidime-Avibactam. Antimicrob. Agents Chemother. 2022, 66, e0241421. [Google Scholar] [CrossRef] [PubMed]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- Caspermeyer, J. MEGA Software Celebrates Silver Anniversary. Mol. Biol. Evol. 2018, 35, 1558–1560. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.G. Building Phylogenetic Trees from Molecular Data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef]
- PennState Eberly College of Science 4.5—Fisher’s Exact Test|STAT 504. Available online: https://online.stat.psu.edu/stat504/lesson/4/4.5 (accessed on 9 December 2023).
- Arakawa, Y.; Ohta, M.; Kido, N.; Fujii, Y.; Komatsu, T.; Kato, N. Close Evolutionary Relationship between the Chromosomally Encoded Beta-Lactamase Gene of Klebsiella Pneumoniae and the TEM Beta-Lactamase Gene Mediated by R Plasmids. FEBS Lett. 1986, 207, 69–74. [Google Scholar] [CrossRef]
- Barthélémy, M.; Peduzzi, J.; Labia, R. Complete Amino Acid Sequence of P453-Plasmid-Mediated PIT-2 Beta-Lactamase (SHV-1). Biochem. J. 1988, 251, 73–79. [Google Scholar] [CrossRef]
- Sutcliffe, J.G. Nucleotide Sequence of the Ampicillin Resistance Gene of Escherichia Coli Plasmid pBR322. Proc. Natl. Acad. Sci. USA 1978, 75, 3737–3741. [Google Scholar] [CrossRef]
- Boissinot, M.; Levesque, R.C. Nucleotide Sequence of the PSE-4 Carbenicillinase Gene and Correlations with the Staphylococcus Aureus PC1 Beta-Lactamase Crystal Structure. J. Biol. Chem. 1990, 265, 1225–1230. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.I.; Scahill, S.; Gibson, T.; Ambler, R.P. The Phototrophic Bacterium Rhodopseudomonas Capsulata Sp108 Encodes an Indigenous Class A Beta-Lactamase. Biochem. J. 1989, 260, 803–812. [Google Scholar] [CrossRef] [PubMed]
- Houba, S.; Willem, S.; Duez, C.; Molitor, C.; Dusart, J.; Frère, J.M.; Ghuysen, J.M. Nucleotide Sequence of the Gene Encoding the Active-Site Serine Beta-Lactamase from Actinomadura R39. FEMS Microbiol. Lett. 1989, 53, 241–246. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Madgwick, P.J.; Waley, S.G. Beta-Lactamase I from Bacillus Cereus. Structure and Site-Directed Mutagenesis. Biochem. J. 1987, 248, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Madonna, M.J.; Zhu, Y.F.; Lampen, J.O. Nucleotide Sequence of the Beta-Lactamase I Gene of Bacillus Cereus Strains 569/H and 5/B. Nucleic Acids Res. 1987, 15, 1877. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Pastor, F.I.; Lampen, J.O. Cloning and Sequencing of the blaZ Gene Encoding Beta-Lactamase III, a Lipoprotein of Bacillus Cereus 569/H. J. Bacteriol. 1987, 169, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Forsman, M.; Häggström, B.; Lindgren, L.; Jaurin, B. Molecular Analysis of Beta-Lactamases from Four Species of Streptomyces: Comparison of Amino Acid Sequences with Those of Other Beta-Lactamases. J. Gen. Microbiol. 1990, 136, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Lenzini, M.V.; Ishihara, H.; Dusart, J.; Ogawara, H.; Joris, B.; Van Beeumen, J.; Frère, J.-M.; Ghuysen, J.-M. Nucleotide Sequence of the Gene Encoding the Active-Site Serine β-Lactamase from Streptomyces Cacaoi. FEMS Microbiol. Lett. 1988, 49, 371–376. [Google Scholar] [CrossRef]
- Arakawa, Y.; Ohta, M.; Kido, N.; Mori, M.; Ito, H.; Komatsu, T.; Fujii, Y.; Kato, N. Chromosomal Beta-Lactamase of Klebsiella Oxytoca, a New Class A Enzyme That Hydrolyzes Broad-Spectrum Beta-Lactam Antibiotics. Antimicrob. Agents Chemother. 1989, 33, 63–70. [Google Scholar] [CrossRef]
- McLaughlin, J.R.; Murray, C.L.; Rabinowitz, J.C. Unique Features in the Ribosome Binding Site Sequence of the Gram-Positive Staphylococcus Aureus Beta-Lactamase Gene. J. Biol. Chem. 1981, 256, 11283–11291. [Google Scholar] [CrossRef]
- Dehottay, P.; Dusart, J.; De Meester, F.; Joris, B.; Van Beeumen, J.; Erpicum, T.; Frère, J.M.; Ghuysen, J.M. Nucleotide Sequence of the Gene Encoding the Streptomyces Albus G Beta-Lactamase Precursor. Eur. J. Biochem. 1987, 166, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Smith Moland, E.; Hanson, N.D.; Herrera, V.L.; Black, J.A.; Lockhart, T.J.; Hossain, A.; Johnson, J.A.; Goering, R.V.; Thomson, K.S. Plasmid-Mediated, Carbapenem-Hydrolysing Beta-Lactamase, KPC-2, in Klebsiella Pneumoniae Isolates. J. Antimicrob. Chemother. 2003, 51, 711–714. [Google Scholar] [CrossRef] [PubMed]
| Position | 6 | 8 | 13 | 18 | 34 | 49 | 62 | 89 | 92 | 93 | 104 | 105 | 120 | 147 | 163 | 164 | 165 | 169 | 170 | 171 | 172 | 179 | 180 | 191 | 202 | 207 | 240 | 241 | 243 | 254 | 264 | 270 | 274 | 292 | 293 | 294 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wildtype | R | V | L | A | A | M | A | G | D | T | P | W | A | G | D | R | W | L | N | S | A | D | T | Q | P | F | V | Y | T | T | Y | K | H | V | N | G | |
| KPC-2 | AY034847.1 | ||||||||||||||||||||||||||||||||||||
| KPC-3 | AF395881.1 | Y | |||||||||||||||||||||||||||||||||||
| KPC-4 | EU447304.1 | R | G | ||||||||||||||||||||||||||||||||||
| KPC-5 | EU400222.2 | R | |||||||||||||||||||||||||||||||||||
| KPC-6 | EU555534.1 | G | |||||||||||||||||||||||||||||||||||
| KPC-7 | EU729727.1 | I | Y | ||||||||||||||||||||||||||||||||||
| KPC-8 | FJ234412.1 | G | Y | ||||||||||||||||||||||||||||||||||
| KPC-9 | FJ624872.1 | A | Y | ||||||||||||||||||||||||||||||||||
| KPC-10 | GQ140348.1 | R | Y | ||||||||||||||||||||||||||||||||||
| KPC-11 | HM066995.1 | L | |||||||||||||||||||||||||||||||||||
| KPC-12 | HQ641421.1 | M | |||||||||||||||||||||||||||||||||||
| KPC-13 | HQ342889.1 | G | Y | ||||||||||||||||||||||||||||||||||
| KPC-14 | JX524191.1 | ||||||||||||||||||||||||||||||||||||
| KPC-15 | KC433553.1 | R | L | K | G | Y | |||||||||||||||||||||||||||||||
| KPC-16 | KC465199.1 | S | L | ||||||||||||||||||||||||||||||||||
| KPC-17 | KC465200.1 | L | |||||||||||||||||||||||||||||||||||
| KPC-18 | KP681699.1 | I | |||||||||||||||||||||||||||||||||||
| KPC-19 | KJ775801.1 | Y | T | ||||||||||||||||||||||||||||||||||
| KPC-21 | NG_049254.1 | R | |||||||||||||||||||||||||||||||||||
| KPC-22 | KM379100.1 | G | L | ||||||||||||||||||||||||||||||||||
| KPC-23 | MH450213.1 | A | Y | ||||||||||||||||||||||||||||||||||
| KPC-24 | KR052099.1 | P | |||||||||||||||||||||||||||||||||||
| KPC-25 | NG_051167.1 | ||||||||||||||||||||||||||||||||||||
| KPC-26 | KX619622.1 | S | |||||||||||||||||||||||||||||||||||
| KPC-27 | KX828722.1 | R | Y | ||||||||||||||||||||||||||||||||||
| KPC-28 | KY282958.1 | Y | |||||||||||||||||||||||||||||||||||
| KPC-29 | KY563764.1 | Y | |||||||||||||||||||||||||||||||||||
| KPC-30 | KY646302.1 | H | |||||||||||||||||||||||||||||||||||
| KPC-31 | MAPH01000113.1 | Y | Y | ||||||||||||||||||||||||||||||||||
| KPC-32 | MAPO01000050.1 | Y | M | Y | |||||||||||||||||||||||||||||||||
| KPC-33 | CP025144.1 | Y | |||||||||||||||||||||||||||||||||||
| KPC-34 | KU985429.1 | ||||||||||||||||||||||||||||||||||||
| KPC-35 | MH404098.1 | P | |||||||||||||||||||||||||||||||||||
| KPC-36 | MH593787.1 | E | Y | ||||||||||||||||||||||||||||||||||
| KPC-37 | MH718730.1 | R | L | ||||||||||||||||||||||||||||||||||
| KPC-38 | MK098861.1 | Y | A | ||||||||||||||||||||||||||||||||||
| KPC-39 | MK118771.1 | T | Y | ||||||||||||||||||||||||||||||||||
| KPC-40 | QRBR01000058.1 | S | Y | ||||||||||||||||||||||||||||||||||
| KPC-41 | MK497255.1 | ? | Y | ||||||||||||||||||||||||||||||||||
| KPC-42 | MK467612.1 | A | |||||||||||||||||||||||||||||||||||
| KPC-43 | MK628511.1 | R | |||||||||||||||||||||||||||||||||||
| KPC-44 | NG_065427.1 | ||||||||||||||||||||||||||||||||||||
| KPC-45 | MN104596.1 | K | |||||||||||||||||||||||||||||||||||
| KPC-46 | MN267701.1 | P | Y | ||||||||||||||||||||||||||||||||||
| KPC-49 | MN619655.1 | S | Y | ||||||||||||||||||||||||||||||||||
| KPC-50 | MN654342.1 | Y | |||||||||||||||||||||||||||||||||||
| KPC-51 | MN725731.1 | N | H | N | |||||||||||||||||||||||||||||||||
| KPC-52 | MN725732.1 | Y | |||||||||||||||||||||||||||||||||||
| KPC-54 | MN854706.1 | S | |||||||||||||||||||||||||||||||||||
| KPC-55 | MT028409.1 | N | |||||||||||||||||||||||||||||||||||
| KPC-56 | MT040751.1 | Y | W | ||||||||||||||||||||||||||||||||||
| KPC-57 | MT358626.1 | V | |||||||||||||||||||||||||||||||||||
| KPC-58 | NG_070177.1 | ||||||||||||||||||||||||||||||||||||
| KPC-59 | NG_070178.1 | D | |||||||||||||||||||||||||||||||||||
| KPC-60 | NG_070179.1 | T | |||||||||||||||||||||||||||||||||||
| KPC-61 | NG_070180.1 | P | Y | ||||||||||||||||||||||||||||||||||
| KPC-62 | NG_073465.1 | Q | Y | ||||||||||||||||||||||||||||||||||
| KPC-63 | NG_073466.1 | S | Y | ||||||||||||||||||||||||||||||||||
| KPC-64 | NG_073467.1 | S | A | H | Y | ||||||||||||||||||||||||||||||||
| KPC-65 | NG_073468.1 | Y | |||||||||||||||||||||||||||||||||||
| KPC-66 | NG_070739.1 | Y | |||||||||||||||||||||||||||||||||||
| KPC-71 | NG_070895.1 | S | |||||||||||||||||||||||||||||||||||
| KPC-72 | NG_070740.1 | D | |||||||||||||||||||||||||||||||||||
| KPC-73 | NG_070741.1 | ||||||||||||||||||||||||||||||||||||
| KPC-74 | NG_070742.1 | ||||||||||||||||||||||||||||||||||||
| KPC-75 | NG_070743.1 | F | |||||||||||||||||||||||||||||||||||
| KPC-76 | NG_070896.1 | Y | |||||||||||||||||||||||||||||||||||
| KPC-77 | NG_070897.1 | P | |||||||||||||||||||||||||||||||||||
| KPC-78 | NG_071204.1 | A | |||||||||||||||||||||||||||||||||||
| KPC-79 | NG_071205.1 | ||||||||||||||||||||||||||||||||||||
| KPC-80 | NG_073469.1 | ||||||||||||||||||||||||||||||||||||
| KPC-81 | NG_073470.1 | ||||||||||||||||||||||||||||||||||||
| KPC-82 | NG_073471.1 | ||||||||||||||||||||||||||||||||||||
| KPC-83 | MW581775.1 | T | |||||||||||||||||||||||||||||||||||
| KPC-86 | MZ067229.1 | G | |||||||||||||||||||||||||||||||||||
| KPC-87 | MZ067230.1 | A | |||||||||||||||||||||||||||||||||||
| KPC-90 | MZ570431.1 | ||||||||||||||||||||||||||||||||||||
| KPC-93 | MZ569034.1 | ||||||||||||||||||||||||||||||||||||
| KPC-94 | MZ646140.1 | H | Y | ||||||||||||||||||||||||||||||||||
| KPC-95 | MZ646141.1 | T | Y | Y | |||||||||||||||||||||||||||||||||
| KPC-123 | ON012820.1 | A | |||||||||||||||||||||||||||||||||||
| MrBayes | 5 | 3 | 2 | 2 | 4 | 3 | 6 | 5 | 1 | 4 | 2 | 27 | |||||||||||||||||||||||||
| Parisomony1 | 3 | 3 | 2 | 2 | 4 | 3 | 7 | 4 | 2 | 4 | 2 | 10 | |||||||||||||||||||||||||
| Parisomony2 | 4 | 3 | 2 | 2 | 4 | 3 | 4 | 4 | 2 | 4 | 2 | 12 |
| Species/Gene | Accession | Citation |
|---|---|---|
| Klebsiella pneumoniae | NG049268.1 | [49] |
| PIT-2 (SHV-1) | P0AD63.1 | [50] |
| R-TEM | J01749.1 | [51] |
| PSE-4 | J05162 | [52] |
| Rhodopseudomas capulate (ampR gene) | X1579.1 | [53] |
| Actinomadura | NG_047541.1 | [54] |
| Bacillus cereus 569H | NG_047482.1 | [55] |
| Bacillus cereus 5/B | M12607.1 | [56] |
| Bacillus cereus III | M15195 | [57] |
| Bacillus licheniformis penicillinase | V00093.1 | [58] |
| Streptomyces badius | M34178.1 | [58] |
| Streptomyces cacaoi Ulg | BAA14224.1 | [59] |
| Klebsiella oxytoca | M27459.1 | [60] |
| Staphylococcus aureus | X04121 | [61] |
| Streptomyces albus | NG047481.1 | [62] |
| Streptomyces lavendulae | M34180.1 | [58] |
| Streptomyces fradiae | M34179.1 | [58] |
| KPC-2 | NG_049253 | [63] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garsevanyan, S.; Barlow, M. The Klebsiella pneumoniae carbapenemase (KPC) β-Lactamase Has Evolved in Response to Ceftazidime Avibactam. Antibiotics 2024, 13, 40. https://doi.org/10.3390/antibiotics13010040
Garsevanyan S, Barlow M. The Klebsiella pneumoniae carbapenemase (KPC) β-Lactamase Has Evolved in Response to Ceftazidime Avibactam. Antibiotics. 2024; 13(1):40. https://doi.org/10.3390/antibiotics13010040
Chicago/Turabian StyleGarsevanyan, Sona, and Miriam Barlow. 2024. "The Klebsiella pneumoniae carbapenemase (KPC) β-Lactamase Has Evolved in Response to Ceftazidime Avibactam" Antibiotics 13, no. 1: 40. https://doi.org/10.3390/antibiotics13010040
APA StyleGarsevanyan, S., & Barlow, M. (2024). The Klebsiella pneumoniae carbapenemase (KPC) β-Lactamase Has Evolved in Response to Ceftazidime Avibactam. Antibiotics, 13(1), 40. https://doi.org/10.3390/antibiotics13010040





