Genotypic Characterization of Uropathogenic Escherichia coli from Companion Animals: Predominance of ST372 in Dogs and Human-Related ST73 in Cats
Abstract
1. Introduction
2. Results
2.1. Bacterial Isolates
2.2. Clonal Typing of E. coli Isolates
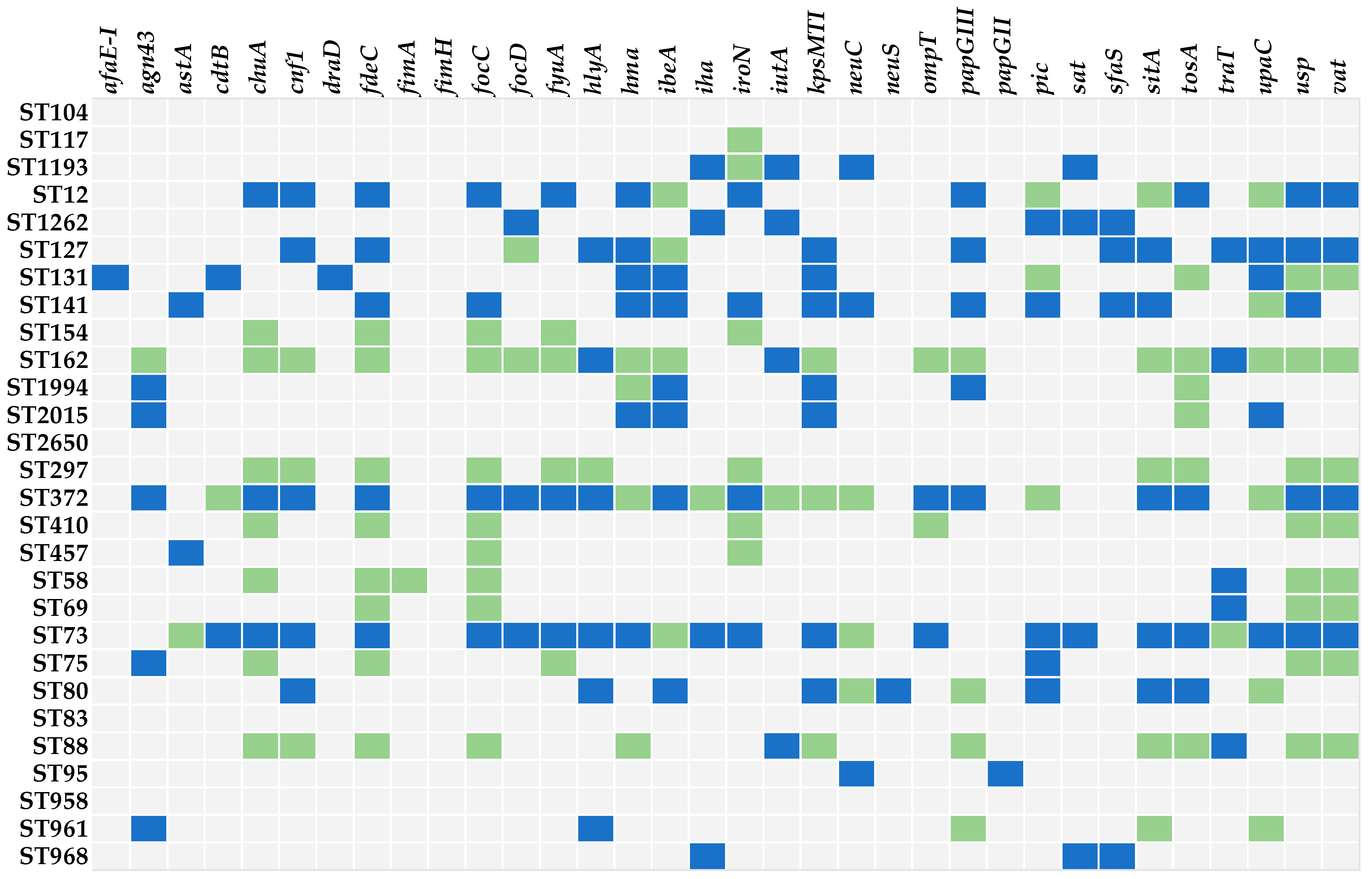
2.3. Distribution of Virulence-Associated Genes
2.4. Distribution of AMR Genes
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Sampling and Identification of Bacterial Isolates
5.2. Isolate Storage, DNA Preparation, and Whole Genome Sequencing
5.3. In Silico Methods
5.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stamm, W.E.; Norrby, S.R. Urinary tract infections: Disease panorama and challenges. J. Infect. Dis. 2001, 183 (Suppl. S1), S1–S4. [Google Scholar] [CrossRef] [PubMed]
- Aurich, S.; Prenger-Berninghoff, E.; Ewers, C. Prevalence and Antimicrobial Resistance of Bacterial Uropathogens Isolated from Dogs and Cats. Antibiotics 2022, 11, 1730. [Google Scholar] [CrossRef] [PubMed]
- Dorsch, R.; Remer, C.; Sauter-Louis, C.; Hartmann, K. Feline lower urinary tract disease in a German cat population. Tierarztl. Prax. 2014, 42, 231–239. [Google Scholar] [CrossRef]
- Thompson, M.F.; Litster, A.L.; Platell, J.L.; Trott, D.J. Canine bacterial urinary tract infections: New developments in old pathogens. Vet. J. 2011, 190, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Weese, J.S.; Blondeau, J.; Boothe, D.; Guardabassi, L.G.; Gumley, N.; Papich, M.; Jessen, L.R.; Lappin, M.; Rankin, S.; Westropp, J.L.; et al. International Society for Companion Animal Infectious Diseases (ISCAID) guidelines for the diagnosis and management of bacterial urinary tract infections in dogs and cats. Vet. J. 2019, 247, 8–25. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Z.; Zhang, Y.; Zhang, Z.; Lei, L.; Xia, Z. Increasing Prevalence of ESBL-Producing Multidrug Resistance Escherichia coli From Diseased Pets in Beijing, China From 2012 to 2017. Front. Microbiol. 2019, 10, 2852. [Google Scholar] [CrossRef] [PubMed]
- Marques, C.; Belas, A.; Franco, A.; Aboim, C.; Gama, L.T.; Pomba, C. Increase in antimicrobial resistance and emergence of major international high-risk clonal lineages in dogs and cats with urinary tract infection: 16 year retrospective study. J. Antimicrob. Chemother. 2018, 73, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Zogg, A.L.; Zurfluh, K.; Schmitt, S.; Nüesch-Inderbinen, M.; Stephan, R. Antimicrobial resistance, multilocus sequence types and virulence profiles of ESBL producing and non-ESBL producing uropathogenic Escherichia coli isolated from cats and dogs in Switzerland. Vet. Microbiol. 2018, 216, 79–84. [Google Scholar] [CrossRef]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum beta-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef]
- Jacoby, G.A. AmpC beta-lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef]
- Aldred, K.J.; Kerns, R.J.; Osheroff, N. Mechanism of quinolone action and resistance. Biochemistry 2014, 53, 1565–1574. [Google Scholar] [CrossRef] [PubMed]
- Weese, J.S.; Webb, J.; Ballance, D.; McKee, T.; Stull, J.W.; Bergman, P.J. Evaluation of antimicrobial prescriptions in dogs with suspected bacterial urinary tract disease. J. Vet. Intern. Med. 2021, 35, 2277–2286. [Google Scholar] [CrossRef] [PubMed]
- Weese, J.S.; Stull, J.W.; Evason, M.; Webb, J.; Ballance, D.; McKee, T.; Bergman, P.J. A multicenter study of antimicrobial prescriptions for cats diagnosed with bacterial urinary tract disease. J. Feline Med. Surg. 2022, 24, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Wiles, T.J.; Kulesus, R.R.; Mulvey, M.A. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp. Mol. Pathol. 2008, 85, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.A.; Stapleton, A.; Wenderoth, S.; Hooton, T.M.; Stamm, W.E. Chromosomal restriction fragment length polymorphism analysis of Escherichia coli strains causing recurrent urinary tract infections in young women. J. Infect. Dis. 1995, 172, 440–445. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Kaster, N.; Kuskowski, M.A.; Ling, G.V. Identification of urovirulence traits in Escherichia coli by comparison of urinary and rectal E. coli isolates from dogs with urinary tract infection. J. Clin. Microbiol. 2003, 41, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Foxman, B. The epidemiology of urinary tract infection. Nat. Rev. Urol. 2010, 7, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Mann, R.; Mediati, D.G.; Duggin, I.G.; Harry, E.J.; Bottomley, A.L. Metabolic Adaptations of Uropathogenic E. coli in the Urinary Tract. Front. Cell. Infect. Microbiol. 2017, 7, 241. [Google Scholar] [CrossRef]
- Terlizzi, M.E.; Gribaudo, G.; Maffei, M.E. UroPathogenic Escherichia coli (UPEC) Infections: Virulence factors, bladder responses, antibiotic, and non-antibiotic antimicrobial strategies. Front. Microbiol. 2017, 8, 1566. [Google Scholar] [CrossRef]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef]
- Johnson, J.R.; Russo, T.A. Molecular epidemiology of extraintestinal pathogenic (uropathogenic) Escherichia coli. Int. J. Med. Microbiol. 2005, 295, 383–404. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Gajewski, A.; Lesse, A.J.; Russo, T.A. Extraintestinal pathogenic Escherichia coli as a cause of invasive nonurinary infections. J. Clin. Microbiol. 2003, 41, 5798–5802. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Scheutz, F.; Ulleryd, P.; Kuskowski, M.A.; O’Bryan, T.T.; Sandberg, T. Phylogenetic and pathotypic comparison of concurrent urine and rectal Escherichia coli isolates from men with febrile urinary tract infection. J. Clin. Microbiol. 2005, 43, 3895–3900. [Google Scholar] [CrossRef] [PubMed]
- Spurbeck, R.R.; Dinh, P.C.; Walk, S.T.; Stapleton, A.E.; Hooton, T.M.; Nolan, L.K.; Kim, K.S.; Johnson, J.R.; Mobley, H.L.T. Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect. Immun. 2012, 80, 4115–4122. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and Simple Determination of the Escherichia coli Phylogenetic Group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Dixit, O.V.A.; Vangchhia, B.; Condamine, B.; Dion, S.; Bridier-Nahmias, A.; Denamur, E.; Gordon, D. Characterization and rapid identification of phylogroup G in Escherichia coli, a lineage with high virulence and antibiotic resistance potential. Environ. Microbiol. 2019, 21, 3107–3117. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Russo, T.A. Extraintestinal pathogenic Escherichia coli: “the other bad E. coli”. J. Lab. Clin. Med. 2002, 139, 155–162. [Google Scholar] [CrossRef]
- Vangchhia, B.; Abraham, S.; Bell, J.M.; Collignon, P.; Gibson, J.S.; Ingram, P.R.; Johnson, J.R.; Kennedy, K.; Trott, D.J.; Turnidge, J.D.; et al. Phylogenetic diversity, antimicrobial susceptibility and virulence characteristics of phylogroup F Escherichia coli in Australia. Microbiology 2016, 162, 1904–1912. [Google Scholar] [CrossRef]
- LeCuyer, T.E.; Byrne, B.A.; Daniels, J.B.; Diaz-Campos, D.V.; Hammac, G.K.; Miller, C.B.; Besser, T.E.; Davis, M.A. Population structure and antimicrobial resistance of canine uropathogenic Escherichia coli. J. Clin. Microbiol. 2018, 56, 452–458. [Google Scholar] [CrossRef]
- Gilbertie, J.M.; Levent, G.; Norman, K.N.; Vinasco, J.; Scott, H.M.; Jacob, M.E. Comprehensive phenotypic and genotypic characterization and comparison of virulence, biofilm, and antimicrobial resistance in urinary Escherichia coli isolated from canines. Vet. Microbiol. 2020, 249, 108822. [Google Scholar] [CrossRef]
- Valat, C.; Drapeau, A.; Beurlet, S.; Bachy, V.; Boulouis, H.-J.; Pin, R.; Cazeau, G.; Madec, J.-Y.; Haenni, M. Pathogenic Escherichia coli in Dogs Reveals the Predominance of ST372 and the Human-Associated ST73 Extra-Intestinal Lineages. Front. Microbiol. 2020, 11, 580. [Google Scholar] [CrossRef] [PubMed]
- Fibke, C.D.; Croxen, M.A.; Geum, H.M.; Glass, M.; Wong, E.; Avery, B.P.; Daignault, D.; Mulvey, M.R.; Reid-Smith, R.J.; Parmley, E.J.; et al. Genomic Epidemiology of Major Extraintestinal Pathogenic Escherichia coli Lineages Causing Urinary Tract Infections in Young Women Across Canada. Open Forum Infect. Dis. 2019, 6, ofz431. [Google Scholar] [CrossRef] [PubMed]
- Flament-Simon, S.-C.; Nicolas-Chanoine, M.-H.; García, V.; Duprilot, M.; Mayer, N.; Alonso, M.P.; García-Meniño, I.; Blanco, J.E.; Blanco, M.; Blanco, J. Clonal Structure, Virulence Factor-encoding Genes and Antibiotic Resistance of Escherichia coli, Causing Urinary Tract Infections and Other Extraintestinal Infections in Humans in Spain and France during 2016. Antibiotics 2020, 9, 161. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Thungrat, K.; Boothe, D.M. Multilocus Sequence Typing and Virulence Profiles in Uropathogenic Escherichia coli Isolated from Cats in the United States. PLoS ONE 2015, 10, e0143335. [Google Scholar] [CrossRef] [PubMed]
- Huber, H.; Zweifel, C.; Wittenbrink, M.M.; Stephan, R. ESBL-producing uropathogenic Escherichia coli isolated from dogs and cats in Switzerland. Vet. Microbiol. 2013, 162, 992–996. [Google Scholar] [CrossRef] [PubMed]
- Manges, A.R.; Johnson, J.R. Reservoirs of Extraintestinal Pathogenic Escherichia coli. Microbiol. Spectr. 2015, 3, 161–177. [Google Scholar] [CrossRef] [PubMed]
- Guardabassi, L.; Schwarz, S.; Lloyd, D.H. Pet animals as reservoirs of antimicrobial-resistant bacteria. J. Antimicrob. Chemother. 2004, 54, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Ewers, C.; Bethe, A.; Semmler, T.; Guenther, S.; Wieler, L.H. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: A global perspective. Clin. Microbiol. Infect. 2012, 18, 646–655. [Google Scholar] [CrossRef]
- Ksiezarek, M.; Novais, Â.; Felga, H.; Mendes, F.; Escobar, M.; Peixe, L. Phylogenomic analysis of a highly virulent Escherichia coli ST83 lineage with potential animal-human transmission. Microb. Pathog. 2021, 155, 104920. [Google Scholar] [CrossRef]
- Naziri, Z.; Derakhshandeh, A.; Soltani Borchaloee, A.; Poormaleknia, M.; Azimzadeh, N. Treatment Failure in Urinary Tract Infections: A Warning Witness for Virulent Multi-Drug Resistant ESBL- Producing Escherichia coli. Infect. Drug Resist. 2020, 13, 1839–1850. [Google Scholar] [CrossRef]
- Johnson, J.R.; Russo, T.A. Molecular Epidemiology of Extraintestinal Pathogenic Escherichia coli. EcoSal Plus 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Tracz, D.M.; Boyd, D.A.; Hizon, R.; Bryce, E.; McGeer, A.; Ofner-Agostini, M.; Simor, A.E.; Paton, S.; Mulvey, M.R. ampC gene expression in promoter mutants of cefoxitin-resistant Escherichia coli clinical isolates. FEMS Microbiol. Lett. 2007, 270, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Mammeri, H.; Poirel, L.; Fortineau, N.; Nordmann, P. Naturally occurring extended-spectrum cephalosporinases in Escherichia coli. Antimicrob. Agents Chemother. 2006, 50, 2573–2576. [Google Scholar] [CrossRef] [PubMed]
- Melo, L.C.; Haenni, M.; Saras, E.; Duprilot, M.; Nicolas-Chanoine, M.-H.; Madec, J.-Y. Emergence of the C1-M27 cluster in ST131 Escherichia coli from companion animals in France. J. Antimicrob. Chemother. 2019, 74, 3111–3113. [Google Scholar] [CrossRef] [PubMed]
- Price, L.B.; Johnson, J.R.; Aziz, M.; Clabots, C.; Johnston, B.; Tchesnokova, V.; Nordstrom, L.; Billig, M.; Chattopadhyay, S.; Stegger, M.; et al. The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio 2013, 4, e00377-13. [Google Scholar] [CrossRef] [PubMed]
- Boll, E.J.; Overballe-Petersen, S.; Hasman, H.; Roer, L.; Ng, K.; Scheutz, F.; Hammerum, A.M.; Dungu, A.; Hansen, F.; Johannesen, T.B.; et al. Emergence of Enteroaggregative Escherichia coli within the ST131 Lineage as a Cause of Extraintestinal Infections. mBio 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Hutton, T.A.; Innes, G.K.; Harel, J.; Garneau, P.; Cucchiara, A.; Schifferli, D.M.; Rankin, S.C. Phylogroup and virulence gene association with clinical characteristics of Escherichia coli urinary tract infections from dogs and cats. J. Vet. Diagn. Investig. 2018, 30, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Nüesch-Inderbinen, M.T.; Baschera, M.; Zurfluh, K.; Hächler, H.; Nüesch, H.; Stephan, R. Clonal Diversity, Virulence Potential and Antimicrobial Resistance of Escherichia coli Causing Community Acquired Urinary Tract Infection in Switzerland. Front. Microbiol. 2017, 8, 2334. [Google Scholar] [CrossRef]
- Toval, F.; Köhler, C.-D.; Vogel, U.; Wagenlehner, F.; Mellmann, A.; Fruth, A.; Schmidt, M.A.; Karch, H.; Bielaszewska, M.; Dobrindt, U. Characterization of Escherichia coli isolates from hospital inpatients or outpatients with urinary tract infection. J. Clin. Microbiol. 2014, 52, 407–418. [Google Scholar] [CrossRef]
- Cepas, V.; Soto, S.M. Relationship between Virulence and Resistance among Gram-Negative Bacteria. Antibiotics 2020, 9, 719. [Google Scholar] [CrossRef]
- Soto, S.M.; Jimenez de Anta, M.T.; Vila, J. Quinolones induce partial or total loss of pathogenicity islands in uropathogenic Escherichia coli by SOS-dependent or -independent pathways, respectively. Antimicrob. Agents Chemother. 2006, 50, S649–S653. [Google Scholar] [CrossRef] [PubMed]
- Sabaté, M.; Moreno, E.; Pérez, T.; Andreu, A.; Prats, G. Pathogenicity island markers in commensal and uropathogenic Escherichia coli isolates. Clin. Microbiol. Infect. 2006, 12, 880–886. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.M.; Zaw, M.T.; Shamsudin, S.B.; Lin, Z. Polymerase chain reaction-restriction fragment length polymorphism method for differentiation of uropathogenic specific protein gene types. J. Microbiol. Immunol. Infect. 2016, 49, 591–594. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Kuskowski, M.A.; O’Bryan, T.T.; Maslow, J.N. Epidemiological correlates of virulence genotype and phylogenetic background among Escherichia coli blood isolates from adults with diverse-source bacteremia. J. Infect. Dis. 2002, 185, 1439–1447. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kanamaru, S.; Kurazono, H.; Nakano, M.; Terai, A.; Ogawa, O.; Yamamoto, S. Subtyping of uropathogenic Escherichia coli according to the pathogenicity island encoding uropathogenic-specific protein: Comparison with phylogenetic groups. Int. J. Urol. 2006, 13, 754–760. [Google Scholar] [CrossRef] [PubMed]
- Göttig, S.; Riedel-Christ, S.; Saleh, A.; Kempf, V.A.J.; Hamprecht, A. Impact of blaNDM-1 on fitness and pathogenicity of Escherichia coli and Klebsiella pneumoniae. Int. J. Antimicrob. Agents 2016, 47, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Pepin-Puget, L.; El Garch, F.; Bertrand, X.; Valot, B.; Hocquet, D. Genome analysis of enterobacteriaceae with non-wild type susceptibility to third-generation cephalosporins recovered from diseased dogs and cats in Europe. Vet. Microbiol. 2020, 242, 108601. [Google Scholar] [CrossRef]
- Shaheen, B.W.; Nayak, R.; Foley, S.L.; Kweon, O.; Deck, J.; Park, M.; Rafii, F.; Boothe, D.M. Molecular characterization of resistance to extended-spectrum cephalosporins in clinical Escherichia coli isolates from companion animals in the United States. Antimicrob. Agents Chemother. 2011, 55, 5666–5675. [Google Scholar] [CrossRef]
- Lo Piccolo, F.; Belas, A.; Foti, M.; Fisichella, V.; Marques, C.; Pomba, C. Detection of multidrug resistance and extended-spectrum/plasmid-mediated AmpC beta-lactamase genes in Enterobacteriaceae isolates from diseased cats in Italy. J. Feline Med. Surg. 2020, 22, 613–622. [Google Scholar] [CrossRef]
- Bortolami, A.; Zendri, F.; Maciuca, E.I.; Wattret, A.; Ellis, C.; Schmidt, V.; Pinchbeck, G.; Timofte, D. Diversity, Virulence, and Clinical Significance of Extended-Spectrum β-Lactamase- and pAmpC-Producing Escherichia coli From Companion Animals. Front. Microbiol. 2019, 10, 1260. [Google Scholar] [CrossRef]
- Pulss, S.; Stolle, I.; Stamm, I.; Leidner, U.; Heydel, C.; Semmler, T.; Prenger-Berninghoff, E.; Ewers, C. Multispecies and Clonal Dissemination of OXA-48 Carbapenemase in Enterobacteriaceae From Companion Animals in Germany, 2009–2016. Front. Microbiol. 2018, 9, 1265. [Google Scholar] [CrossRef] [PubMed]
- Feßler, A.T.; Scholtzek, A.D.; Schug, A.R.; Kohn, B.; Weingart, C.; Hanke, D.; Schink, A.-K.; Bethe, A.; Lübke-Becker, A.; Schwarz, S. Antimicrobial and biocide resistance among canine and feline Enterococcus faecalis, Enterococcus faecium, Escherichia coli, Pseudomonas aeruginosa, and Acinetobacter baumannii isolates from diagnostic submissions. Antibiotics 2022, 11, 152. [Google Scholar] [CrossRef] [PubMed]
- Stolle, I.; Prenger-Berninghoff, E.; Stamm, I.; Scheufen, S.; Hassdenteufel, E.; Guenther, S.; Bethe, A.; Pfeifer, Y.; Ewers, C. Emergence of OXA-48 carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in dogs. J. Antimicrob. Chemother. 2013, 68, 2802–2808. [Google Scholar] [CrossRef] [PubMed]
- Irrgang, A.; Pauly, N.; Tenhagen, B.-A.; Grobbel, M.; Kaesbohrer, A.; Hammerl, A.J.A. Spill-Over from Public Health? First Detection of an OXA-48-Producing Escherichia coli in a German Pig Farm. Microorganisms 2020, 8, 855. [Google Scholar] [CrossRef] [PubMed]
- Nation, R.L.; Li, J. Colistin in the 21st century. Curr. Opin. Infect. Dis. 2009, 22, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Luppi, A.; Gibellini, M.; Gin, T.; Vangroenweghe, F.; Vandenbroucke, V.; Bauerfeind, R.; Bonilauri, P.; Labarque, G.; Hidalgo, Á. Prevalence of virulence factors in enterotoxigenic Escherichia coli isolated from pigs with post-weaning diarrhoea in Europe. Porcine Health Manag. 2016, 2, 20. [Google Scholar] [CrossRef] [PubMed]
- Rhouma, M.; Fairbrother, J.M.; Beaudry, F.; Letellier, A. Post weaning diarrhea in pigs: Risk factors and non-colistin-based control strategies. Acta Vet. Scand. 2017, 59, 31. [Google Scholar] [CrossRef]
- Catry, B.; Cavaleri, M.; Baptiste, K.; Grave, K.; Grein, K.; Holm, A.; Jukes, H.; Liebana, E.; Lopez Navas, A.; Mackay, D.; et al. Use of colistin-containing products within the European Union and European Economic Area (EU/EEA): Development of resistance in animals and possible impact on human and animal health. Int. J. Antimicrob. Agents 2015, 46, 297–306. [Google Scholar] [CrossRef]
- Hamame, A.; Davoust, B.; Cherak, Z.; Rolain, J.-M.; Diene, S.M. Mobile Colistin Resistance (mcr) Genes in Cats and Dogs and Their Zoonotic Transmission Risks. Pathogens 2022, 11, 698. [Google Scholar] [CrossRef]
- Flament-Simon, S.-C.; de Toro, M.; García, V.; Blanco, J.E.; Blanco, M.; Alonso, M.P.; Goicoa, A.; Díaz-González, J.; Nicolas-Chanoine, M.-H.; Blanco, J. Molecular Characteristics of Extraintestinal Pathogenic E. coli (ExPEC), Uropathogenic E. coli (UPEC), and Multidrug Resistant E. coli Isolated from Healthy Dogs in Spain. Whole Genome Sequencing of Canine ST372 Isolates and Comparison with Human Isolates Causing Extraintestinal Infections. Microorganisms 2020, 8, 1712. [Google Scholar] [CrossRef]
- Elankumuran, P.; Browning, G.F.; Marenda, M.S.; Kidsley, A.; Osman, M.; Haenni, M.; Johnson, J.R.; Trott, D.J.; Reid, C.J.; Djordjevic, S.P. Identification of genes influencing the evolution of Escherichia coli ST372 in dogs and humans. Microb. Genom. 2023, 9, mgen.0.000930. [Google Scholar] [CrossRef] [PubMed]
- Ewers, C.; Grobbel, M.; Stamm, I.; Kopp, P.A.; Diehl, I.; Semmler, T.; Fruth, A.; Beutlich, J.; Guerra, B.; Wieler, L.H.; et al. Emergence of human pandemic O25:H4-ST131 CTX-M-15 extended-spectrum-beta-lactamase-producing Escherichia coli among companion animals. J. Antimicrob. Chemother. 2010, 65, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Elankumaran, P.; Cummins, M.L.; Browning, G.F.; Marenda, M.S.; Reid, C.J.; Djordjevic, S.P. Genomic and Temporal Trends in Canine ExPEC Reflect Those of Human ExPEC. Microbiology Spectrum 2022, 10, e0129122. [Google Scholar] [CrossRef] [PubMed]
- Schaufler, K.; Semmler, T.; Wieler, L.H.; Wöhrmann, M.; Baddam, R.; Ahmed, N.; Müller, K.; Kola, A.; Fruth, A.; Ewers, C.; et al. Clonal spread and interspecies transmission of clinically relevant ESBL-producing Escherichia coli of ST410--another successful pandemic clone? FEMS Microbiol. Ecol. 2016, 92, fiv155. [Google Scholar] [CrossRef] [PubMed]
- Kidsley, A.K.; White, R.T.; Beatson, S.A.; Saputra, S.; Schembri, M.A.; Gordon, D.; Johnson, J.R.; O’Dea, M.; Mollinger, J.L.; Abraham, S.; et al. Companion Animals Are Spillover Hosts of the Multidrug-Resistant Human Extraintestinal Escherichia coli Pandemic Clones ST131 and ST1193. Front. Microbiol. 2020, 11, 1968. [Google Scholar] [CrossRef]
- Tchesnokova, V.L.; Rechkina, E.; Larson, L.; Ferrier, K.; Weaver, J.L.; Schroeder, D.W.; She, R.; Butler-Wu, S.M.; Aguero-Rosenfeld, M.E.; Zerr, D.; et al. Rapid and Extensive Expansion in the United States of a New Multidrug-resistant Escherichia coli Clonal Group, Sequence Type 1193. Clin. Infect. Dis. 2019, 68, 334–337. [Google Scholar] [CrossRef]
- Valenza, G.; Werner, M.; Eisenberger, D.; Nickel, S.; Lehner-Reindl, V.; Höller, C.; Bogdan, C. First report of the new emerging global clone ST1193 among clinical isolates of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli from Germany. J. Glob. Antimicrob. Resist. 2019, 17, 305–308. [Google Scholar] [CrossRef]
- Roer, L.; Overballe-Petersen, S.; Hansen, F.; Schønning, K.; Wang, M.; Røder, B.L.; Hansen, D.S.; Justesen, U.S.; Andersen, L.P.; Fulgsang-Damgaard, D.; et al. Escherichia coli Sequence Type 410 Is Causing New International High-Risk Clones. mSphere 2018, 3. [Google Scholar] [CrossRef]
- Toombs-Ruane, L.J.; Benschop, J.; French, N.P.; Biggs, P.J.; Midwinter, A.C.; Marshall, J.C.; Chan, M.; Drinković, D.; Fayaz, A.; Baker, M.G.; et al. Carriage of Extended-Spectrum-Beta-Lactamase- and AmpC Beta-Lactamase-Producing Escherichia coli Strains from Humans and Pets in the Same Households. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef]
- Grönthal, T.; Österblad, M.; Eklund, M.; Jalava, J.; Nykäsenoja, S.; Pekkanen, K.; Rantala, M. Sharing more than friendship—transmission of NDM-5 ST167 and CTX-M-9 ST69 Escherichia coli between dogs and humans in a family, Finland, 2015. Euro Surveill. 2018, 23. [Google Scholar] [CrossRef]
- Johnson, J.R.; Davis, G.; Clabots, C.; Johnston, B.D.; Porter, S.; DebRoy, C.; Pomputius, W.; Ender, P.T.; Cooperstock, M.; Slater, B.S.; et al. Household Clustering of Escherichia coli Sequence Type 131 Clinical and Fecal Isolates According to Whole Genome Sequence Analysis. Open Forum Infect. Dis. 2016, 3, ofw129. [Google Scholar] [CrossRef] [PubMed]
- Ljungquist, O.; Ljungquist, D.; Myrenås, M.; Rydén, C.; Finn, M.; Bengtsson, B. Evidence of household transfer of ESBL-/pAmpC-producing Enterobacteriaceae between humans and dogs—a pilot study. Infect. Ecol. Epidemiol. 2016, 6, 31514. [Google Scholar] [CrossRef] [PubMed]
- Cozma, A.P.; Rimbu, C.M.; Zendri, F.; Maciuca, I.E.; Timofte, D. Clonal Dissemination of Extended-Spectrum Cephalosporin-Resistant Enterobacterales between Dogs and Humans in Households and Animal Shelters of Romania. Antibiotics 2022, 11, 1242. [Google Scholar] [CrossRef] [PubMed]
- Schubert, S.; Wieser, A. Einsatz der Matrix-Assisted Laser Desorption/ionisation-Time of Flight Massenspektrometrie (MALDI-TOF MS) in der mikrobiologischen Routinediagnostik/Matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS) in clinical microbiological routine diagnostics. LaboratoriumsMedizin 2011, 35, 195–203. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 2021, 7. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 2018, 34, 2490–2492. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- SAS® Institute Inc. Base SAS® 9.4 Procedures Guide: Statistical Procedures, 2nd ed.; Statistical Analysis System Institute Inc.: Cary, NC, USA, 2013. [Google Scholar]
- Kim, H.-Y. Statistical notes for clinical researchers: Chi-squared test and Fisher’s exact test. Restor. Dent. Endod. 2017, 42, 152–155. [Google Scholar] [CrossRef] [PubMed]
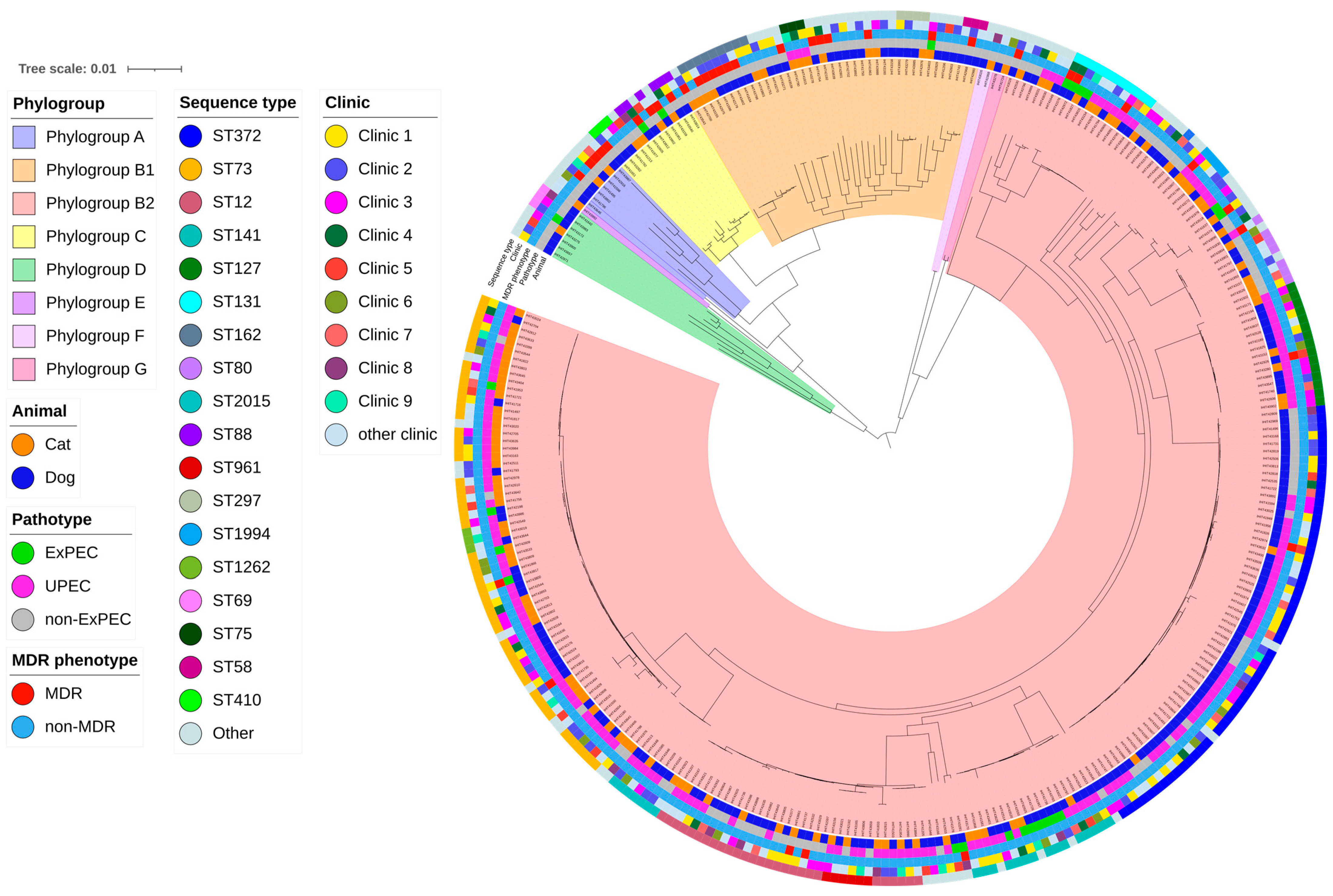
| Phylogenetic Group | Haemolytic Phenotype | Multilocus Sequence Type | Pathotype | Sero(geno)type | FimH Type | |
|---|---|---|---|---|---|---|
| (percentages of positive isolates are given in brackets) | ||||||
| Dog | A (3.0) B1 (12.1) B2 (76.8) C (4.0) D (3.0) E (0.0) F (0.5) G (0.5) | non-haemolytic (43.4) haemolytic (56.6) | ST372 (26.8) ST73 (9.1) ST12 (8.1) ST127 (5.1) ST141 (4.6) ST131 (3.0) ST162 (2.5) ST80 (1.52) ST2015 (2.53) Other (36.9) | ExPEC (8.6 *) UPEC (46.0) Other (45.5) | Ont (24.8) O4 (17.2) O2 (13.1) O6 (10.6) O25 (3.5) O8 (4.6) O83 (5.1) O15 (5.1) O25b (2.5) O9 (2.5) Other (11.1) | fimH9 (25.9) fimH5 (5.1) fimH32 (4.6) fimH2 (3.6) fimH102 (3.1) fimH13 (0.5) fimH27 (2.5) fimH14 (2.5) fimH197 (2.5) fimH1 (2.0) fimH39 (2.0) fimH31 (2.6) Other (44.2) |
| Cat | A (1.7) B1 (12.0) B2 (79.5) C (3.4) D (0.8) E (0.8) F (0.8) G (0.8) | non-haemolytic (29.9) haemolytic (70.1) | ST372 (1.7) ST73 (27.4) ST12 (9.4) ST127 (4.3) ST141 (5.1) ST131 (4.3) ST162 (3.4) ST80 (3.4) ST2015 (1.7) Other (39.3) | ExPEC (5.1 *) UPEC (59.8) Other (35.0) | Ont (26.5) O4 (11.1) O2 (17.1) O6 (8.6) O25 (12.0) O8 (5.1) O83 (1.7) O15 (0.0) O25b (3.4) O9 (3.4) Other (11.1) | fimH9 (11.1) fimH5 (5.1) fimH32 (5.1) fimH2 (6.0) fimH102 (3.4) fimH13 (6.0) fimH27 (2.6) fimH14 (1.7) fim197 (1.7) fimH1 (1.7) fimH39 (1.7) fimH31 (2.6) Other (51.3) |
| Category | Gene | Dog N (%) | Cat N (%) | Total N (%) | p-Value 1 |
|---|---|---|---|---|---|
| Adhesins | |||||
| Fimbrial | papGI | 1 (0.5) | 3 (2.6) | 4 (1.3) | 0.146 |
| papGII | 5 (2.5) | 2 (1.7) | 7 (2.2) | 0.483 | |
| papGIII | 92 (46.5) | 50 (42.7) | 142 (45.1) | 0.520 | |
| fimACDH | 194 (98.0) | 117 (100.0) | 311 (98.7) | 0.154 | |
| focAC/D | 70 (35.4) | 34 (29.1) | 104 (33.0) | 0.251 | |
| sfaSAF | 22 (11.1) | 17 (15.5) | 39 (12.4) | 0.373 | |
| Non-Fimbrial | csgABC/DEFG | 198 (100.0) | 117 (100.0) | 315 (100.0) | - |
| fdeC | 155 (78.3) | 96 (82.1) | 251 (79.7) | 0.422 | |
| iha | 10 (5.1) | 13 (11.1) | 23 (7.3) | 0.046 | |
| afaE-I | 2 (1.0) | - | 2 (0.6) | 0.394 | |
| draAD | 3 (1.5) | - | 3 (1.0) | 0.247 | |
| flgBCDEFGHIJ | 198 (100.0) | 117 (100.0) | 315 (100.0) | - | |
| Siderophore systems | ent/fep/fes | 197 (99.5) | 117 (100) | 314 (99.7) | 0.629 |
| irp1 + 2/fyuA | 168/167 (84.8/84.3) | 101/102 (86.3/87.2) | 269 (85.4) | 0.491 | |
| iroBCDEN | 144 (72.7) | 98 (83.8) | 242 (76.8) | 0.025 | |
| iuc/iutA | 28 (14.1) | 20 (17.1) | 48 (15.2) | 0.481 | |
| chuA/hma | 160/110 (80.8/55.6) | 97/84 (82.9/71.8) | 257/194 (81.6/61.6) | 0.643/0.004 | |
| sitABCD | 114 (54.0) | 62 (53.0) | 176 (55.9) | 0.429 | |
| Toxins | cnf1 | 107 (54.0) | 75 (64.1) | 182 (57.8) | 0.081 |
| vat | 144 (72.7) | 87 (74.4) | 231 (73.3) | 0.752 | |
| pic | 41 (20.7) | 60 (51.3) | 101 (32.1) | <0.001 | |
| astA | 25 (12.6) | 9 (7.7) | 31 (9.8) | 0.173 | |
| cdtB | 13 (6.6) | 15 (12.8) | 28 (8.9) | 0.059 | |
| hlyCABD | 97 (49.0) | 49 (41.9) | 146 (46.3) | 0.221 | |
| tosA | 102 (51.5) | 63 (53.8) | 165 (52.4) | 0.689 | |
| Autotransporter proteins | agn43 | 77 (38.9 | 37 (31.6) | 114 (36.2) | 0.195 |
| upaC | 69 (34.8) | 69 (59.0) | 138 (43.8) | <0.001 | |
| sat | 9 (4.5) | 8 (6.8) | 17 (5.4) | 0.384 | |
| Surface polysaccharides | kpsMTII | 80 (40.4) | 69 (59.0) | 149 (47.3) | <0.001 |
| neuC | 15 (7.6) | 7 (6.0) | 22 (7.0) | 0.592 | |
| neuS | 1 (0.5) | 1 (0.9) | 2 (0.6) | 0.606 | |
| Invasins | ompA | 198 (100.0) | 117 (100.0) | 315 (100.0) | - |
| ibeA | 84 (42.4) | 30 (25.6) | 114 (36.2) | 0.003 | |
| traT | 69 (34.8) | 41 (35.0) | 110 (34.9) | 0.972 | |
| Miscellaneous or unknown function | malX | 198 (100.0) | 117 (100.0) | 315 (100.0) | - |
| ompT | 174 (87.9) | 107 (91.5) | 281 (89.2) | 0.323 | |
| usp | 149 (75.3) | 90 (76.9) | 239 (75.9) | 0.738 |
| Antibiotic | Gene | Cat n (%) | Dog n (%) | p-Value 1 | B2 n (%) | non-B2 n (%) | p-Value 2 | All n (%) |
|---|---|---|---|---|---|---|---|---|
| Amino- glycoside | aac(3)-IId | - | 4 (2.0) | 0.301 | 3 (1.2) | 1 (1.4) | 1 | 4 (1.3) |
| aadA | 6 (5.1) | 22 (11.1) | 0.100 | 13 (5.3) | 15 (21.4) | <0.0001 | 28 (8.9) | |
| ant(2″)-Ia | 1 (0.9) | - | 0.371 | - | 1 (1.4) | 0.222 | 1 (0.3) | |
| aphAI-IAB | 32 (27.4) | 52 (26.3) | 0.895 | 50 (20.4) | 34 (48.6) | <0.0001 | 84 (26.7) | |
| Quinolone | aac(6′)-Ib-cr | 1 (0.9) | - | 0.371 | - | 1 (1.4) | 0.222 | 1 (0.3) |
| qnrB | - | 2 (1.0) | 0.532 | - | 2 (2.9) | 0.049 | 2 (0.6) | |
| qnrS | 1 (0.9) | 2 (1.0) | 1 | - | 3 (4.3) | 0.089 | 3 (1.0) | |
| Rifampicin | arr-3 | 1 (0.9) | - | 0.371 | - | 1 (1.4) | 0.222 | 1 (0.3) |
| β-Lactams | blaTEM | 23 (19.7) | 36 (18.2) | 0.766 | 36 (14.7) | 23 (32.9) | 0.002 | 59 (18.8) |
| blaOXA-1 | 2 (1.7) | 3 (1.5) | 1 | 1 (0.4) | 4 (5.7) | 0.009 | 5 (1.6) | |
| blaCTX-M-15 | - | 3 (1.5) | 0.297 | - | 3 (4.3) | 0.011 | 3 (1.0) | |
| blaCTX-M-27 | 1 (0.9) | - | 0.371 | - | 1 (1.4) | 0.222 | 1 (0.3) | |
| blaDHA-1 | - | 1 (0.5) | 1 | - | 1 (1.4) | 0.222 | 1 (0.3) | |
| blaCMY-2 | 1 (0.9) | - | 0.371 | 1 (0.4) | - | 1 | 1 (0.3) | |
| blaOXA-48 | 1 (0.9) | - | 0.371 | 1 (0.4) | - | 1 | 1 (0.3) | |
| blaSHV-1 | 3 (2.6) | 2 (1.0) | 0.364 | 5 (2.0) | - | 0.590 | 5 (1.6) | |
| Phenicols | catA1 | - | 6 (3.0) | 0.088 | 4 (1.6) | 2 (2.9) | 0.618 | 6 (1.9) |
| catB3 | 1 (0.9) | - | 0.371 | - | 1 (1.4) | 0.222 | 1 (0.3) | |
| cmlA1 | - | 2 (1.0) | 0.532 | 1 (0.4) | 1 (1.4) | 0.396 | 2 (0.6) | |
| floR | 3 (2.6) | 3 (1.5) | 0.674 | - | 6 (8.6) | <0.0001 | 6 (1.9) | |
| Folate pathway inhibitors | sul1 | 6 (5.1) | 14 (7.1) | 0.495 | 10 (4.1) | 10 (14.3) | 0.002 | 20 (6.3) |
| sul2 | 15 (12.8) | 22 (11.1) | 0.649 | 18 (7.3) | 19 (27.1) | <0.0001 | 37 (11.7) | |
| sul3 | - | 4 (2.0) | 0.301 | 1 (0.4) | 3 (4.3) | 0.036 | 4 (1.3) | |
| drfA | 12 (10.3) | 28 (14.1) | 0.383 | 14 (5.7) | 26 (37.1) | <0.0001 | 40 (12.7) | |
| Tetracycline | tet(A) | 8 (6.8) | 21 (10.6) | 0.264 | 17 (6.9) | 12 (17.1) | 0.009 | 29 (9.2) |
| tet(B) | 5 (4.3) | 7 (3.5) | 0.741 | 4 (1.6) | 8 (11.4) | <0.0001 | 12 (3.8) | |
| tet(D) | 1 (0.9) | - | 0.371 | 1 (0.4) | - | 1 | 1 (0.3) | |
| tet(M) | - | 1 (0.5) | 1 | 1 (0.4) | - | 1 | 1 (0.3) | |
| Macrolides | mph(A) | 1 (0.9) | 6 (3.0) | 0.265 | 4 (1.6) | 3 (4.3) | 0.186 | 7 (2.2) |
| mph(B) | - | 1 (0.5) | 1 | 1 (0.4) | - | 1 | 1 (0.3) | |
| mph(E) | - | 1 (0.5) | 1 | - | 1 (1.4) | 0.222 | 1 (0.3) | |
| msr(E) | - | 1 (0.5) | 1 | - | 1 (1.4) | 0.222 | 1 (0.3) | |
| Colistin | mcr-4.6 * | - | 1 (0.5) | 1 | 1 (0.4) | - | 1 | 1 (0.3) |
| Fosfomycin | fosA7 | 3 (2.6) | - | 0.050 | - | 3 (4.3) | 0.011 | 3 (1.0) |
| AMR genes total | 128 | 245 | 0.244 | 287 | 186 | <0.0001 | 373 | |
| Sample ID | Animal | Haemolysis | ST | Phylo-Group | CP/ESBL/ AmpC-Positive Isolate | AMR Genes | Mutations | |||
|---|---|---|---|---|---|---|---|---|---|---|
| ampC Promotor | gyrA | parC | parE | |||||||
| IHIT41651 | dog | 0 | 410 | C | ESBL | blaCTX-M-15, blaTEM-1B,aadA2, sul1, dfrA12, tet(A), mph(A) | - | S83L, D87N | S80I | S458A |
| IHIT41754 | dog | 0 | 1844 | B1 | AmpC | blaDHA-1, blaTEM-1B,sul1, dfrA17, tet(B), qnrB4, mph(A) | −1C > T, −18G > A | - | - | - |
| IHIT42192 | cat | 0 | 533 | B1 | ESBL | blaCTX-M-27,sul2, dfrA36, tet(B), sul2 | −1C > T −18G > A | D87N, S83L | S80I | |
| IHIT42968 | dog | 0 | 58 | B1 | AmpC | blaEC11-like, dfrA5 | −42C > T * −18G > A −1C > T | - | - | - |
| IHIT43081 | cat | 1 | 372 | B2 | CP | blaOXA-48 | - | - | - | - |
| IHIT43540 | dog | 0 | 88 | C | ESBL | blaCTX-M-15,aadA1, sul2, dfrA1, tet(B), qnrS1 | −18G > A −1C > T | S83L | - | - |
| IHIT43661 | cat | 1 | 12 | B2 | AmpC | blaCMY-2 | - | - | - | - |
| IHIT43802 | dog | 0 | 361 | A | ESBL | blaCTX-M-15, blaTEM-32,aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id, sul2, dfrA1 | - | D87N, S83L | S80I | S458A |
| IHIT44091 | cat | 1 | 131 | B2 | AmpC | blaTEM-1B,blaEC-6, aph(3″)-Ib, aph(6)-Id, sul2, dfrA8 | −28G > A | - | - | I529L |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aurich, S.; Wolf, S.A.; Prenger-Berninghoff, E.; Thrukonda, L.; Semmler, T.; Ewers, C. Genotypic Characterization of Uropathogenic Escherichia coli from Companion Animals: Predominance of ST372 in Dogs and Human-Related ST73 in Cats. Antibiotics 2024, 13, 38. https://doi.org/10.3390/antibiotics13010038
Aurich S, Wolf SA, Prenger-Berninghoff E, Thrukonda L, Semmler T, Ewers C. Genotypic Characterization of Uropathogenic Escherichia coli from Companion Animals: Predominance of ST372 in Dogs and Human-Related ST73 in Cats. Antibiotics. 2024; 13(1):38. https://doi.org/10.3390/antibiotics13010038
Chicago/Turabian StyleAurich, Sophie, Silver Anthony Wolf, Ellen Prenger-Berninghoff, Lakshmipriya Thrukonda, Torsten Semmler, and Christa Ewers. 2024. "Genotypic Characterization of Uropathogenic Escherichia coli from Companion Animals: Predominance of ST372 in Dogs and Human-Related ST73 in Cats" Antibiotics 13, no. 1: 38. https://doi.org/10.3390/antibiotics13010038
APA StyleAurich, S., Wolf, S. A., Prenger-Berninghoff, E., Thrukonda, L., Semmler, T., & Ewers, C. (2024). Genotypic Characterization of Uropathogenic Escherichia coli from Companion Animals: Predominance of ST372 in Dogs and Human-Related ST73 in Cats. Antibiotics, 13(1), 38. https://doi.org/10.3390/antibiotics13010038






