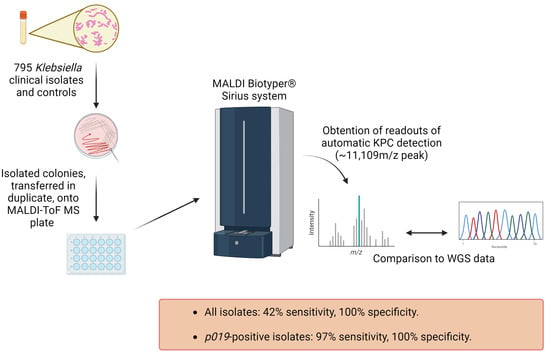
Poor Sensitivity of the MALDI Biotyper® MBT Subtyping Module for Detection of Klebsiella pneumoniae Carbapenemase (KPC) in Klebsiella Species
Abstract
1. Introduction
2. Results
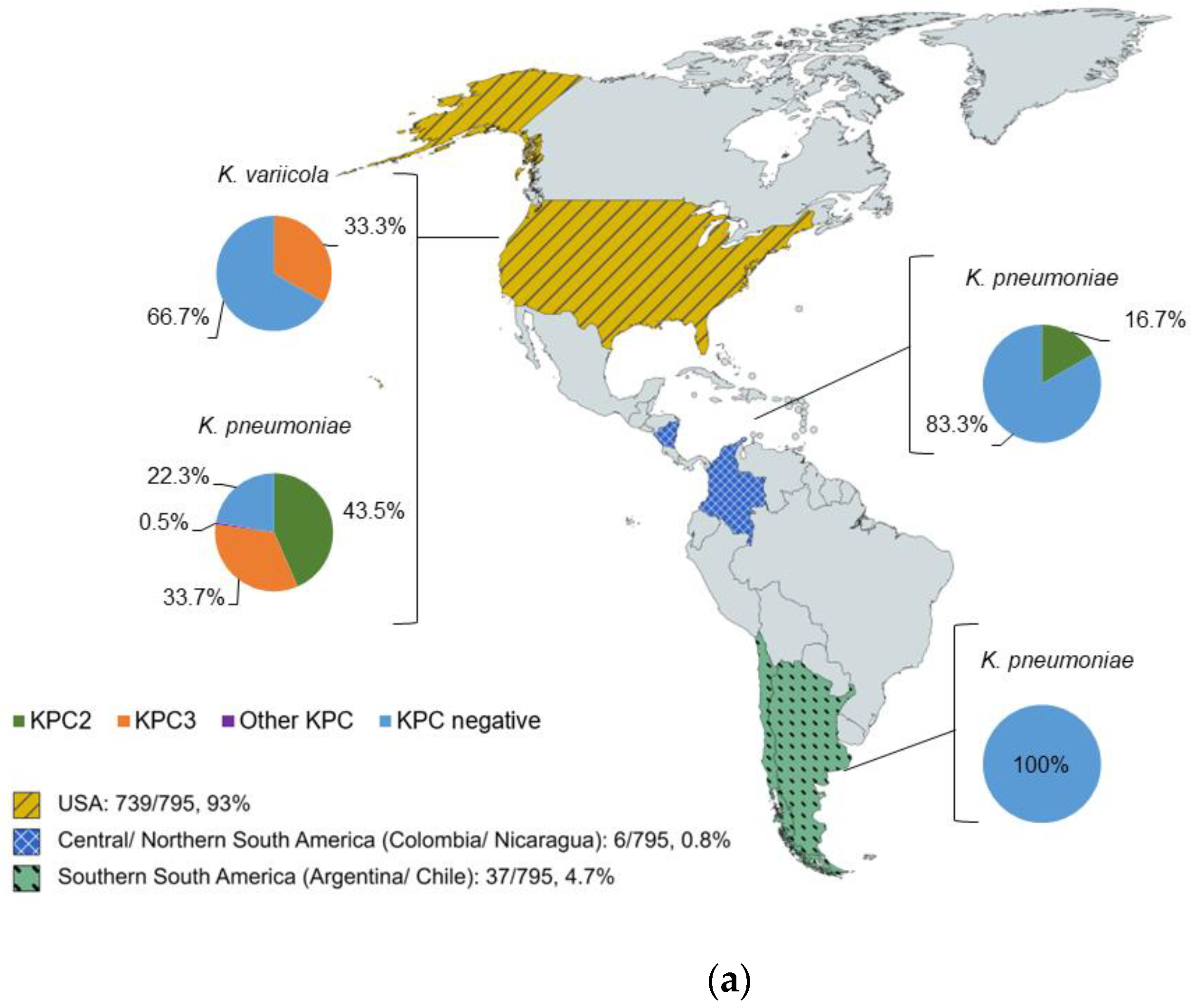
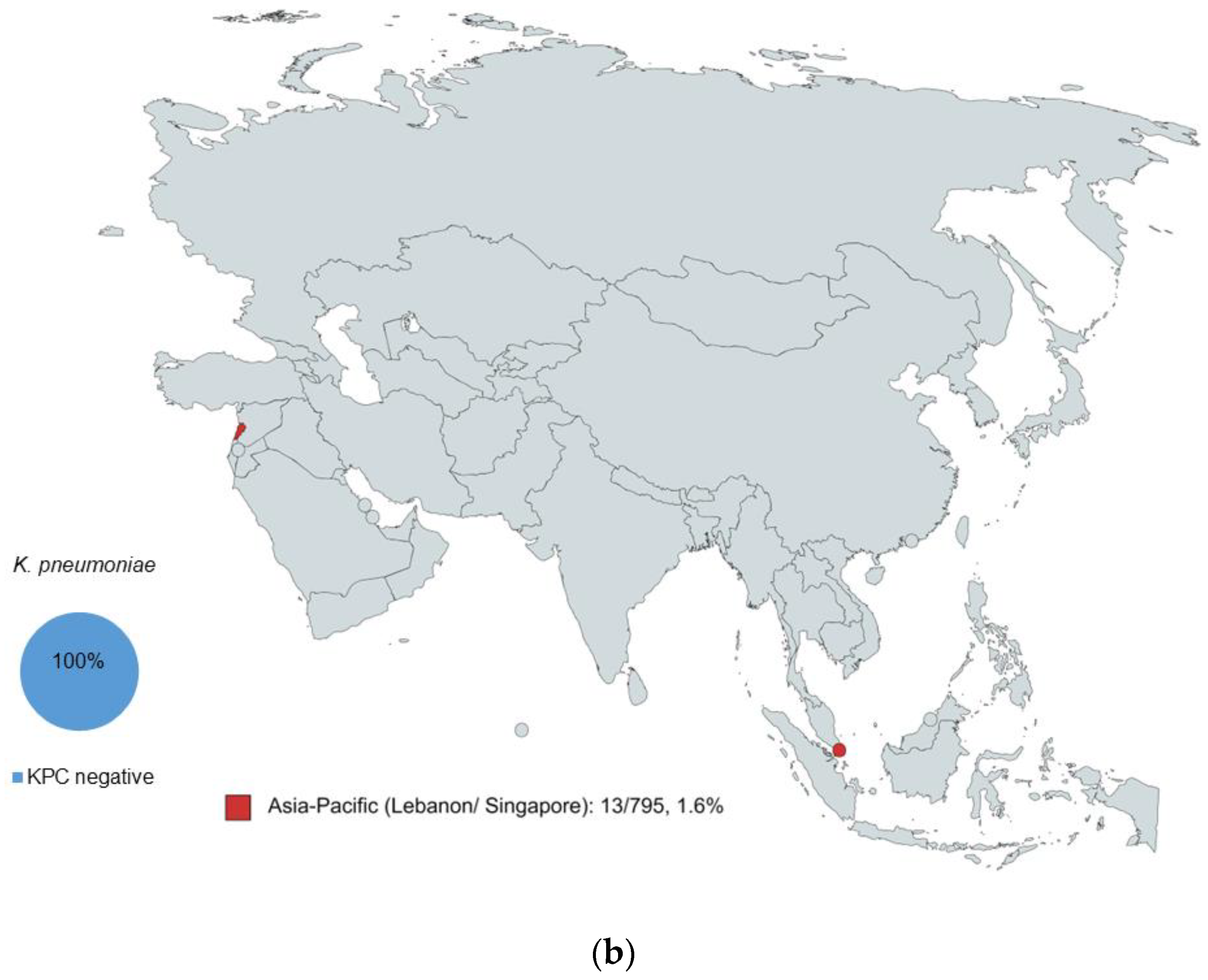
2.1. Characterization of Isolates by WGS
2.2. KPC Detection by WGS and MALDI-ToF MS
3. Discussion
4. Materials and Methods
4.1. Subculturing Isolates and Control Bacterial Strains
4.2. MALDI-ToF MS Plate Preparation
4.3. Obtaining MALDI-ToF MS Readouts
4.4. Statistical Analysis and Preparation of Figures
4.5. Whole Genome Sequencing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control. Antibiotic Resistance Threats in the United States; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2019.
- Armstrong, T.; Fenn, S.J.; Hardie, K.R. JMM Profile: Carbapenems: A broad-spectrum antibiotic. J. Med. Microbiol. 2021, 70, 001462. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report 2022; World Health Organization: Geneva, Switzerland, 2022.
- Nordmann, P.; Naas, T.; Poirel, L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2011, 17, 1791–1798. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.; Komarow, L.; Chen, L.; Ge, L.; Hanson, B.M.; Cober, E.; Herc, E.; Alenazi, T.; Kaye, K.S.; Garcia-Diaz, J.; et al. Global epidemiology and clinical outcomes of carbapenem-resistant Pseudomonas aeruginosa and associated carbapenemases (POP): A prospective cohort study. Lancet Microbe 2023, 4, e159–e170. [Google Scholar] [CrossRef]
- Sawa, T.; Kooguchi, K.; Moriyama, K. Molecular diversity of extended-spectrum β-lactamases and carbapenemases, and antimicrobial resistance. J. Intensive Care 2020, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Queenan, A.M.; Bush, K. Carbapenemases: The versatile beta-lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef]
- Leavitt, A.; Chmelnitsky, I.; Carmeli, Y.; Navon-Venezia, S. Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob. Agents Chemother. 2010, 54, 4493–4496. [Google Scholar] [CrossRef]
- Hammoudi Halat, D.; Ayoub Moubareck, C. The current burden of carbapenemases: Review of significant properties and dissemination among Gram-negative bacteria. Antibiotics 2020, 9, 186. [Google Scholar] [CrossRef]
- Patel, R. MALDI-ToF MS for the diagnosis of infectious diseases. Clin. Chem. 2015, 61, 100–111. [Google Scholar] [CrossRef]
- Rychert, J. Benefits and limitations of MALDI-ToF mass spectrometry for the identification of microorganisms. J. Infect. 2019, 2, 1–5. [Google Scholar] [CrossRef]
- Leopold, J.; Popkova, Y.; Engel, K.M.; Schiller, J. Recent developments of useful MALDI matrices for the mass spectrometric characterization of lipids. Biomolecules 2018, 8, 173. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.F.; Wang, H.; Weingarten, R.A.; Drake, S.K.; Suffredini, A.F.; Garfield, M.K.; Chen, Y.; Gucek, M.; Youn, J.H.; Stock, F.; et al. A rapid matrix-assisted laser desorption ionization-time of flight mass spectrometry-based method for single-plasmid tracking in an outbreak of carbapenem-resistant Enterobacteriaceae. J. Clin. Microbiol. 2014, 52, 2804–2812. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R. Tn4401 carrying blaKPC is inserted within another insertion in pKpQIL and related plasmids. J. Clin. Microbiol. 2014, 52, 4448–4449. [Google Scholar] [CrossRef] [PubMed]
- Cordovana, M.; Kostrzewa, M.; Glandorf, J.; Bienia, M.; Ambretti, S.; Pranada, A.B. A full MALDI-based approach to detect plasmid-encoded KPC-producing Klebsiella pneumoniae. Front. Microbiol. 2018, 9, 2854. [Google Scholar] [CrossRef]
- Gaibani, P.; Galea, A.; Fagioni, M.; Ambretti, S.; Sambri, V.; Landini, M.P. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of KPC-producing Klebsiella pneumoniae. J. Clin. Microbiol. 2016, 54, 2609–2613. [Google Scholar] [CrossRef]
- Youn, J.H.; Drake, S.K.; Weingarten, R.A.; Frank, K.M.; Dekker, J.P.; Lau, A.F. Clinical performance of a matrix-assisted laser desorption ionization-time of flight mass spectrometry method for detection of certain blaKPC-containing plasmids. J. Clin. Microbiol. 2016, 54, 35–42. [Google Scholar] [CrossRef]
- Gato, E.; Constanso, I.P.; Rodino-Janeiro, B.K.; Guijarro-Sanchez, P.; Alioto, T.; Arroyo, M.J.; Mendez, G.; Mancera, L.; Gut, M.; Gut, I.; et al. Occurrence of the p019 gene in the blaKPC-harboring plasmids: Adverse clinical impact for direct tracking of KPC-producing Klebsiella pneumoniae by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2021, 59, e0023821. [Google Scholar] [CrossRef]
- Bruker Daltonics GmbH. MALDI Biotyper Subtyping Module; Bruker Daltonics GmbH: Bremen, Germany, 2019; Volume 4, p. 5. [Google Scholar]
- van Duin, D.; Arias, C.A.; Komarow, L.; Chen, L.; Hanson, B.M.; Weston, G.; Cober, E.; Garner, O.B.; Jacob, J.T.; Satlin, M.J.; et al. Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): A prospective cohort study. Lancet Infect. Dis. 2020, 20, 731–741. [Google Scholar] [CrossRef]
- Wang, M.; Earley, M.; Chen, L.; Hanson, B.M.; Yu, Y.; Liu, Z.; Salcedo, S.; Cober, E.; Li, L.; Kanj, S.S.; et al. Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): A prospective, multicentre, cohort study. Lancet Infect. Dis. 2022, 22, 401–412. [Google Scholar] [CrossRef]
- Tacconelli, E.; Sifakis, F.; Harbarth, S.; Schrijver, R.; van Mourik, M.; Voss, A.; Sharland, M.; Rajendran, N.B.; Rodriguez-Bano, J.; Group, E.P.-N.C.-M. Surveillance for control of antimicrobial resistance. Lancet Infect. Dis. 2018, 18, e99–e106. [Google Scholar] [CrossRef]
- Angeletti, S.; Ciccozzi, M. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry in clinical microbiology: An updating review. Infect. Genet. Evol. 2019, 76, 104063. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.F.; Wang, H.; Weingarten, R.A.; Drake, S.K.; Suffredini, A.F.; Chen, Y.; Gucek, M.; Youn, J.H.; Stock, F.; Tso, H.; et al. Reply to “Tn4401 carrying blaKPC is inserted within another insertion in pKpQIL and related plasmids”. J. Clin. Microbiol. 2014, 52, 4450. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, Y.; Li, J.; Wang, Q.; Tang, K.; Li, C. Rapid detection of KPC-producing Klebsiella pneumoniae in China based on MALDI-ToF MS. J. Microbiol. Methods 2022, 192, 106385. [Google Scholar] [CrossRef] [PubMed]
- Gato, E.; Arroyo, M.J.; Mendez, G.; Candela, A.; Rodino-Janeiro, B.K.; Fernandez, J.; Rodriguez-Sanchez, B.; Mancera, L.; Arca-Suarez, J.; Beceiro, A.; et al. Direct detection of carbapenemase-producing Klebsiella pneumoniae by MALDI-ToF analysis of full spectra applying machine learning. J. Clin. Microbiol. 2023, 61, e0175122. [Google Scholar] [CrossRef] [PubMed]
| WGS | MALDI-ToF MS | ||
|---|---|---|---|
| Number | Number | ||
| blaKPC positive | 574 | KPC-positive | 239 |
| blaKPC2 | 321 | ||
| blaKPC3 | 249 | ||
| Other blaKPC | 4 | ||
| blaKPC negative | 221 | KPC negative | 556 |
| p019-Positive | p019-Negative | |||
|---|---|---|---|---|
| Number | Percent | Number | Percent | |
| blaKPC2 | 222 | 39 | 99 | 17 |
| blaKPC3 | 24 | 4 | 225 | 39 |
| Other blaKPC | 0 | 0 | 4 | 0.7 |
| Total | 246 | 43 | 328 | 57 |
| WGS | |||||
|---|---|---|---|---|---|
| p019-Positive | p019-Negative | ||||
| Number | Percent | Number | Percent | ||
| MALDI-ToF MS | KPC-positive | 239 | 30 | 0 | 0 |
| KPC-negative | 7 | 1 | 549 | 69 | |
| Total | 246 | 31 | 549 | 69 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuello, L.; Alvarez Otero, J.; Greenwood-Quaintance, K.E.; Chen, L.; Hanson, B.; Reyes, J.; Komarow, L.; Ge, L.; Lancaster, Z.D.; Gordy, G.G.; et al. Poor Sensitivity of the MALDI Biotyper® MBT Subtyping Module for Detection of Klebsiella pneumoniae Carbapenemase (KPC) in Klebsiella Species. Antibiotics 2023, 12, 1465. https://doi.org/10.3390/antibiotics12091465
Cuello L, Alvarez Otero J, Greenwood-Quaintance KE, Chen L, Hanson B, Reyes J, Komarow L, Ge L, Lancaster ZD, Gordy GG, et al. Poor Sensitivity of the MALDI Biotyper® MBT Subtyping Module for Detection of Klebsiella pneumoniae Carbapenemase (KPC) in Klebsiella Species. Antibiotics. 2023; 12(9):1465. https://doi.org/10.3390/antibiotics12091465
Chicago/Turabian StyleCuello, Luz, Judith Alvarez Otero, Kerryl E. Greenwood-Quaintance, Liang Chen, Blake Hanson, Jinnethe Reyes, Lauren Komarow, Lizhao Ge, Zane D. Lancaster, Garrett G. Gordy, and et al. 2023. "Poor Sensitivity of the MALDI Biotyper® MBT Subtyping Module for Detection of Klebsiella pneumoniae Carbapenemase (KPC) in Klebsiella Species" Antibiotics 12, no. 9: 1465. https://doi.org/10.3390/antibiotics12091465
APA StyleCuello, L., Alvarez Otero, J., Greenwood-Quaintance, K. E., Chen, L., Hanson, B., Reyes, J., Komarow, L., Ge, L., Lancaster, Z. D., Gordy, G. G., Schuetz, A. N., & Patel, R. (2023). Poor Sensitivity of the MALDI Biotyper® MBT Subtyping Module for Detection of Klebsiella pneumoniae Carbapenemase (KPC) in Klebsiella Species. Antibiotics, 12(9), 1465. https://doi.org/10.3390/antibiotics12091465