Resistome and Genome Analysis of an Extensively Drug-Resistant Klebsiella michiganensis KMIB106: Characterization of a Novel KPC Plasmid pB106-1 and a Novel Cointegrate Plasmid pB106-IMP Harboring blaIMP-4 and blaSHV-12
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of the K. michiganensis KMIB106 Strain
2.2. Bacterial Identification and Antimicrobial Susceptibility Testing
2.3. Genome Sequencing and De Novo Assembly and Correction
2.4. Genome Annotation and Analysis
2.5. GenBank Accession Number
3. Results
3.1. Antibiotic Susceptibilities
3.2. Genomic Analysis of Bacterial Chromosome
3.3. Profile of Resistance Gene
3.4. Correlation between Antimicrobial Resistance Phenotype and Genes Profile
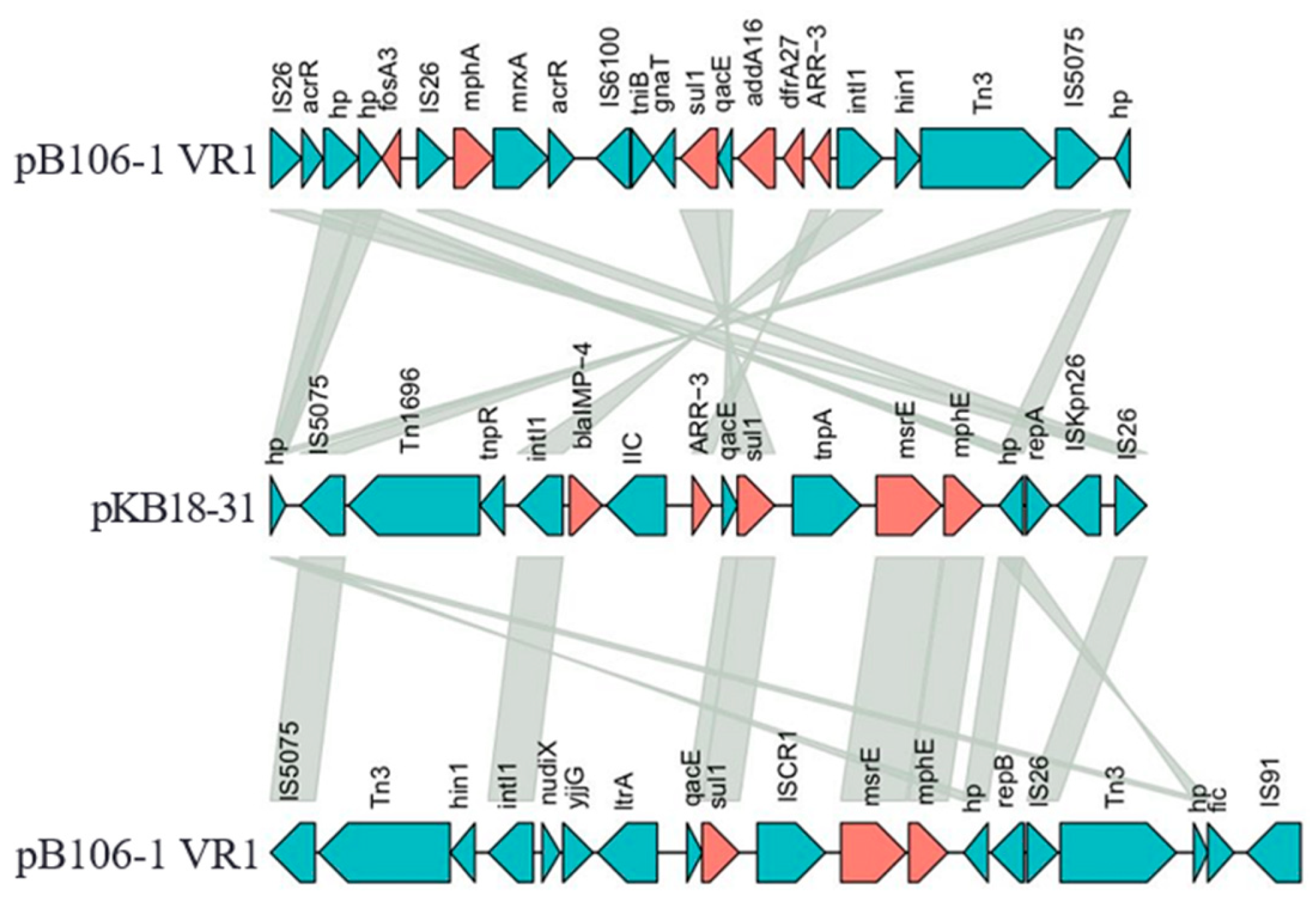
3.5. Genomic Analysis of the Novel KPC Plasmid pB106-1
3.6. Genomic Analysis of Plasmid pB106-2
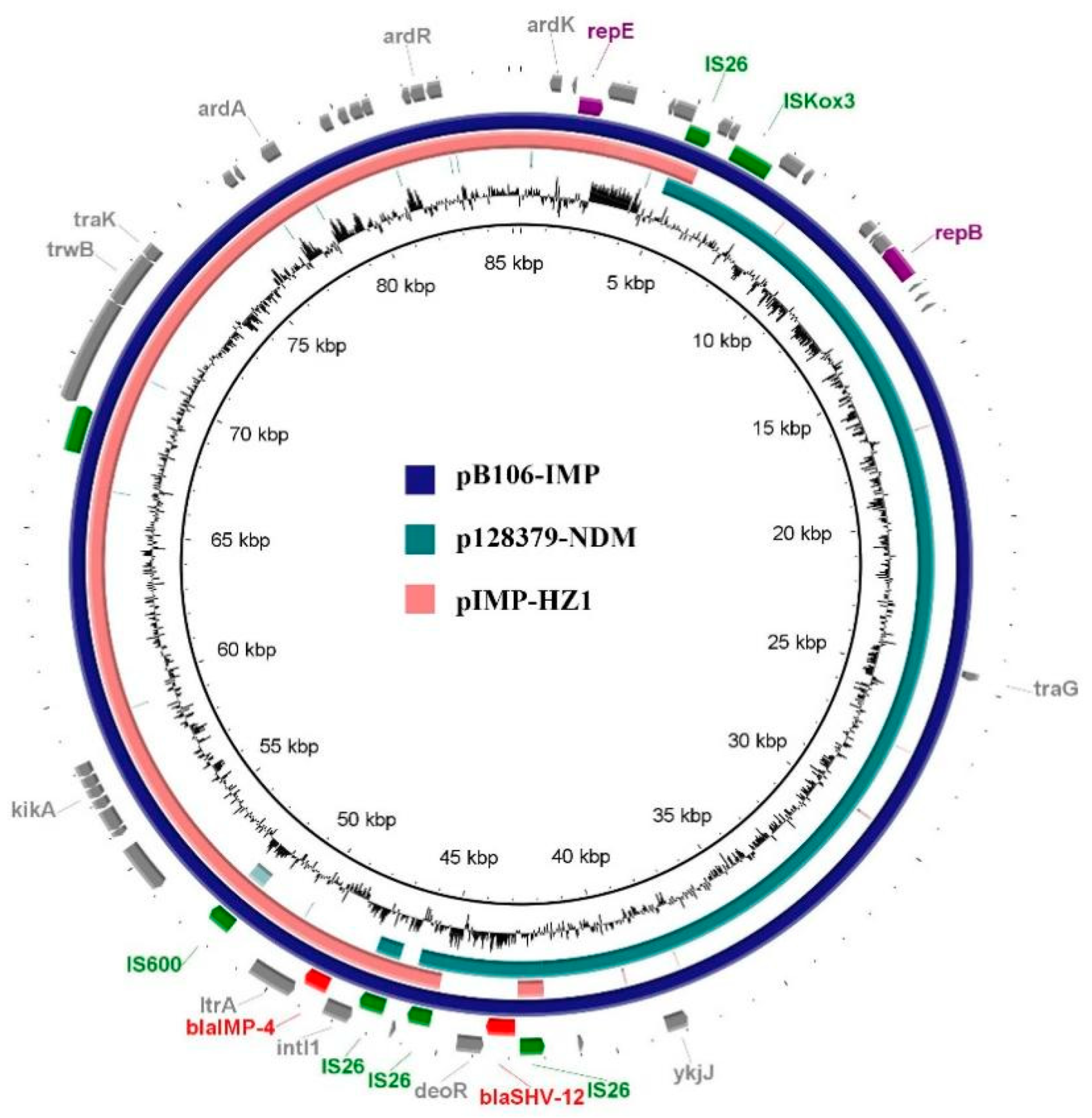
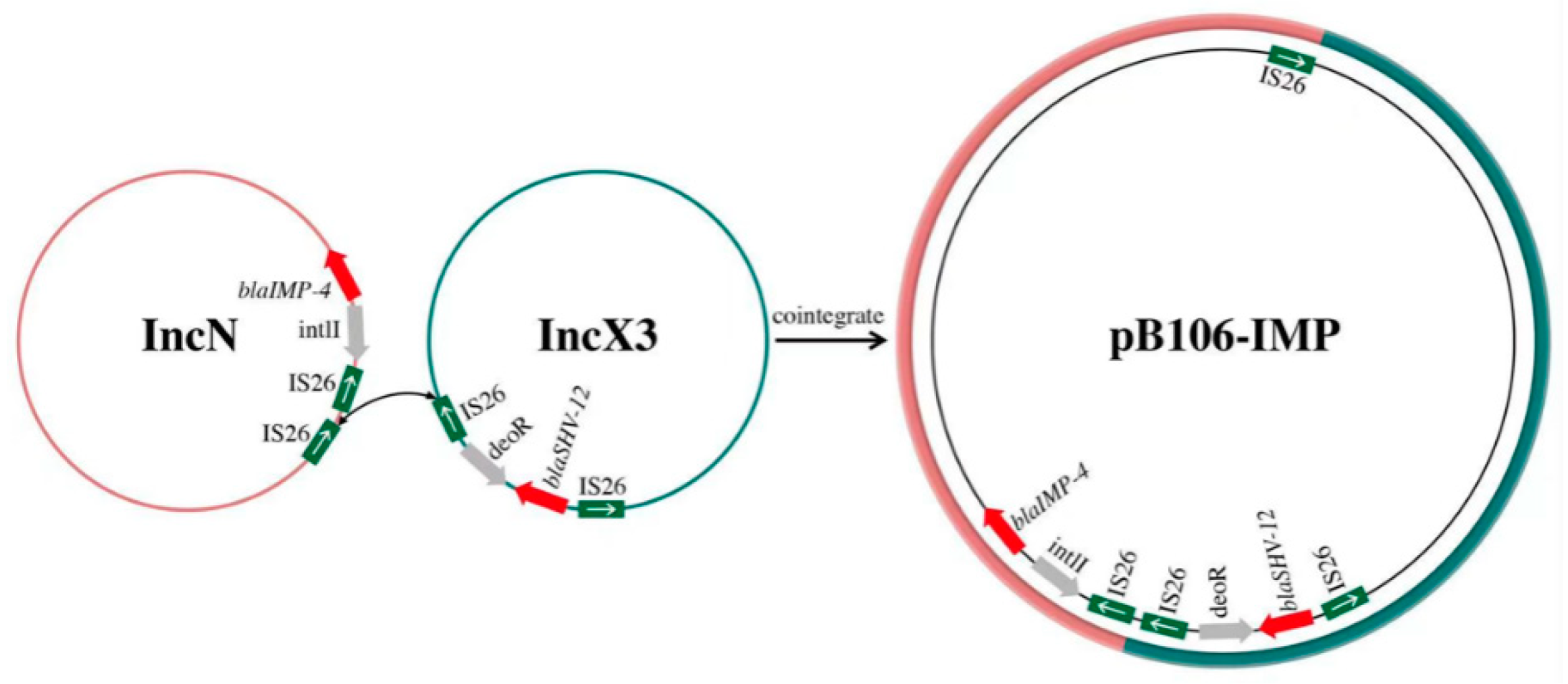
3.7. Genomic Analysis of Plasmid pB106-IMP
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saha, R.; Farrance, C.E.; Verghese, B.; Hong, S.; Donofrio, R.S. Klebsiella michiganensis sp. nov., a new bacterium isolated from a tooth brush holder. Urrent Microbiol. 2013, 66, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Seiffert, S.N.; Wüthrich, D.; Gerth, Y.; Egli, A.; Kohler, P.; Nolte, O. First clinical case of KPC-3-producing Klebsiella michiganensis in Europe. New Microbes New Infect. 2019, 29, 100516. [Google Scholar] [CrossRef] [PubMed]
- Campos-Madueno, E.I.; Sigrist, T.; Flückiger, U.M.; Risch, L.; Bodmer, T.; Endimiani, A. First report of a blaVIM-1 metallo-beta-lactamase-possessing Klebsiella michiganensis. J. Glob. Antimicrob. Resist. 2021, 25, 310–314. [Google Scholar] [CrossRef] [PubMed]
- Hoenigl, M.; Valentin, T.; Zarfel, G.; Wuerstl, B.; Leitner, E.; Salzer, H.J.; Posch, J.; Krause, R.; Grisold, A.J. Nosocomial outbreak of Klebsiella pneumoniae carbapenemase-producing Klebsiella oxytoca in Austria. Antimicrob. Agents Chemother. 2012, 56, 2158–2161. [Google Scholar] [CrossRef]
- Ismail, A.; Founou, R.C.; Founou, L.L.; Allam, M.; Essack, S.Y. Genomic characterisation of Klebsiella michiganensis co-producing OXA-181 and NDM-1 carbapenemases isolated from a cancer patient in uMgungundlovu District, KwaZulu-Natal Province. South Afr. Med. J. 2018, 109, 7–8. [Google Scholar]
- Zheng, B.; Xu, H.; Yu, X.; Lv, T.; Jiang, X.; Cheng, H.; Zhang, J.; Chen, Y.; Huang, C.; Xiao, Y. Identification and genomic characterization of a KPC-2-, NDM-1- and NDM-5-producing Klebsiella michiganensis isolate. J. Antimicrob. Chemother. 2018, 73, 536–538. [Google Scholar] [CrossRef]
- Bonomo, R.A.; Burd, E.M.; Conly, J.; Limbago, B.M.; Poirel, L.; Segre, J.A.; Westblade, L.F. Carbapenemase-Producing Organisms: A Global Scourge. Clin. Infect. Dis. 2018, 66, 1290–1297. [Google Scholar] [CrossRef]
- Cheng, Z.; Bethel, C.R.; Thomas, P.W.; Shurina, B.A.; Alao, J.P.; Thomas, C.A.; Yang, K.; Marshall, S.H.; Zhang, H.; Sturgill, A.M.; et al. Carbapenem Use Is Driving the Evolution of Imipenemase 1 Variants. Antimicrob. Agents Chemother. 2021, 65, e01714-20. [Google Scholar] [CrossRef]
- Wang, Y.; Lo, W.U.; Lai, R.W.; Tse, C.W.; Lee, R.A.; Luk, W.K.; Cheng, V.C.; Que, T.L.; Chow, K.H.; Ho, P.L. IncN ST7 epidemic plasmid carrying blaIMP-4 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. J. Antimicrob. Chemother. 2017, 72, 99–103. [Google Scholar] [CrossRef]
- Chu, Y.W.; Afzal-Shah, M.; Houang, E.T.; Palepou, M.F.; Lyon, D.J.; Woodford, N.; Livermore, D.M. IMP-4, a novel metallo-beta-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob. Agents Chemother. 2001, 45, 710–714. [Google Scholar] [CrossRef]
- Manohar, P.; Leptihn, S.; Lopes, B.S.; Nachimuthu, R. Dissemination of carbapenem resistance and plasmids encoding carbapenemases in Gram-negative bacteria isolated in India. JAC-Antimicrob. Resist. 2021, 3, dlab015. [Google Scholar] [CrossRef] [PubMed]
- Djahmi, N.; Dunyach-Remy, C.; Pantel, A.; Dekhil, M.; Sotto, A.; Lavigne, J.P. Epidemiology of carbapenemase-producing Enterobacteriaceae and Acinetobacter baumannii in Mediterranean countries. Biomed Res. Int. 2014, 2014, 305784. [Google Scholar] [CrossRef] [PubMed]
- Bilal, H.; Zhang, G.; Rehman, T.; Han, J.; Khan, S.; Shafiq, M.; Yang, X.; Yan, Z.; Yang, X. First Report of blaNDM-1 Bearing IncX3 Plasmid in Clinically Isolated ST11 Klebsiella pneumoniae from Pakistan. Microorganisms 2021, 9, 951. [Google Scholar] [CrossRef]
- Xie, S.; Fu, S.; Li, M.; Guo, Z.; Zhu, X.; Ren, J.; Hu, F. Microbiological Characteristics of Carbapenem-Resistant Enterobacteriaceae Clinical Isolates Collected from County Hospitals. Infect. Drug Resist. 2020, 13, 1163–1169. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, B.; Cao, Q.; Huang, W.; Shen, L.; Qin, X. Two clinical strains of Klebsiella pneumoniae carrying plasmid-borne blaIMP-4, blaSHV-12, and armA isolated at a Pediatric Center in Shanghai, China. Antimicrob. Agents Chemother. 2009, 53, 1642–1644. [Google Scholar] [CrossRef]
- Guédon, G.; Libante, V.; Coluzzi, C.; Payot, S.; Leblond-Bourget, N. The Obscure World of Integrative and Mobilizable Elements, Highly Widespread Elements that Pirate Bacterial Conjugative Systems. Genes 2017, 8, 337. [Google Scholar] [CrossRef] [PubMed]
- Bayot, M.L.; Bragg, B.N. Antimicrobial Susceptibility Testing; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Chin, C.S.; Peluso, P.; Sedlazeck, F.J.; Nattestad, M.; Concepcion, G.T.; Clum, A.; Dunn, C.; O’Malley, R.; Figueroa-Balderas, R.; Morales-Cruz, A.; et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 2016, 13, 1050–1054. [Google Scholar] [CrossRef]
- Lin, Y.; Yuan, J.; Kolmogorov, M.; Shen, M.W.; Chaisson, M.; Pevzner, P.A. Assembly of long error-prone reads using de Bruijn graphs. Proc. Natl. Acad. Sci. USA 2016, 113, e8396–e8405. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Berinson, B.; Olearo, F.; Both, A.; Brossmann, N.; Christner, M.; Aepfelbacher, M.; Rohde, H. EUCAST rapid antimicrobial susceptibility testing (RAST): Analytical performance and impact on patient management. J. Antimicrob. Chemother. 2021, 76, 1332–1338. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, W52–W57. [Google Scholar] [CrossRef] [PubMed]
- Bharatham, N.; Bhowmik, P.; Aoki, M.; Okada, U.; Sharma, S.; Yamashita, E.; Shanbhag, A.P.; Rajagopal, S.; Thomas, T.; Sarma, M.; et al. Structure and function relationship of OqxB efflux pump from Klebsiella pneumoniae. Nat. Commun. 2021, 12, 5400. [Google Scholar] [CrossRef] [PubMed]
- Lo, W.U.; Cheung, Y.Y.; Lai, E.; Lung, D.; Que, T.L.; Ho, P.L. Complete sequence of an IncN plasmid, pIMP-HZ1, carrying blaIMP-4 in a Klebsiella pneumoniae strain associated with medical travel to China. Antimicrob. Agents Chemother. 2013, 57, 1561–1562. [Google Scholar] [CrossRef]
- Feng, W.; Zhou, D.; Wang, Q.; Luo, W.; Zhang, D.; Sun, Q.; Tong, Y.; Chen, W.; Sun, F.; Xia, P. Dissemination of IMP-4-encoding pIMP-HZ1-related plasmids among Klebsiella pneumoniae and Pseudomonas aeruginosa in a Chinese teaching hospital. Sci. Rep. 2016, 6, 33419. [Google Scholar] [CrossRef]
- Chen, C.M.; Yu, W.L.; Huang, M.; Liu, J.J.; Chen, I.C.; Chen, H.F.; Wu, L.T. Characterization of IS26-composite transposons and multidrug resistance in conjugative plasmids from Enterobacter cloacae. Microbiol. Immunol. 2015, 59, 516–525. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, W.; Schwarz, S.; Wang, C.; Liu, W.; Chen, F.; Luan, T.; Liu, S. Characterization of a blaIMP-4-carrying plasmid from Enterobacter cloacae of swine origin. J. Antimicrob. Chemother. 2019, 74, 1799–1806. [Google Scholar] [CrossRef]
- Kizny Gordon, A.; Phan, H.T.; Lipworth, S.I.; Cheong, E.; Gottlieb, T.; George, S.; Peto, T.E.; Mathers, A.J.; Walker, A.S.; Crook, D.W.; et al. Genomic dynamics of species and mobile genetic elements in a prolonged blaIMP-4-associated carbapenemase outbreak in an Australian hospital. J. Antimicrob. Chemother. 2020, 75, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Ginn, A.N.; Paulsen, I.T.; Iredell, J.R. pEl1573 Carrying blaIMP-4, from Sydney, Australia, is closely related to other IncL/M plasmids. Antimicrob. Agents Chemother. 2012, 56, 6029–6032. [Google Scholar] [CrossRef] [PubMed]
- Liakopoulos, A.; Mevius, D.; Ceccarelli, D. A Review of SHV Extended-Spectrum beta-Lactamases: Neglected Yet Ubiquitous. Front. Microbiol. 2016, 7, 1374. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef]
- Harmer, C.J.; Hall, R.M. IS26 cannot move alone. J. Antimicrob. Chemother. 2021, 76, 1428–1432. [Google Scholar] [CrossRef]
| Group | Antibiotics | Breakpoints (μg/mL) | MIC (μg/mL) | |
|---|---|---|---|---|
| S | R | |||
| Fluoroquinolones | Ciprofloxacin | ≤0.25 | ≥1 | ≤0.25 |
| Levofloxacin | ≤0.5 | ≥2 | ≤0.12 | |
| Beta-lactams | Piperacillin/tazobactam | ≤16/4 | ≥128/4 | ≥128/4 |
| Amoxicillin/clavulanic acid | ≤8/4 | ≥32/16 | ≥32/16 | |
| Ticacillin/clavulanic acid | ≤16/2 | ≥128/2 | ≥128/2 | |
| Ceftriaxone | ≤1 | ≥4 | ≥64 | |
| Ceftazidime | ≤4 | ≥16 | ≥64 | |
| Cefoxitin | ≤8 | ≥32 | ≥64 | |
| Cefuroxime axetil | ≤8 | ≥32 | ≥64 | |
| Cefuroxime sodium | ≤8 | ≥32 | ≥64 | |
| Aztreonam | ≤4 | ≥16 | ≤1 | |
| Cefepime | ≤2 | ≥16 | ≥32 | |
| Imipenem | ≤1 | ≥4 | ≥16 | |
| Ertapenem | ≤0.5 | ≥2 | ≥8 | |
| Tetracyclines | Tigecycline | ≤2 | ≥8 | ≤0.5 |
| Minocyline | ≤4 | ≥16 | ≥16 | |
| Doxycycline | ≤4 | ≥16 | ≥16 | |
| Aminoglycosides | Amikacin | ≤16 | ≥64 | ≤2 |
| Tobramycin | ≤4 | ≥16 | ≤1 | |
| Folate pathway inhibitors | Compound sulfamethoxazole | ≤2/38 | ≥4/76 | ≥320/80 |
| Lipopeptides | Colistin | / | ≥4 | ≤0.5 |
| Resistance Gene | Location | Identity | Query/Template Length | Position | Predicted Phenotype | Accession Number |
|---|---|---|---|---|---|---|
| oqxB | chromosome | 99.10 | 3131/3153 | 280,484..283,614 | FluoroquinoloneResistance | EU370913 |
| blaOXY-4-1 | chromosome | 99.88 | 869/873 | 5,016,109..5,016,977 | Beta-lactam resistance | AY077481 |
| tetD | pB106-1 | 100.0 | 1185/1185 | 213,924..215,108 | TetracyclineResistance | AF467077 |
| mphA | pB106-1 | 100.0 | 906/906 | 82,715..83,620 | Macrolide resistance | D16251 |
| msrE | pB106-1 | 100.0 | 1476/1476 | 188,852..190,327 | Macrolide resistance | FR751518 |
| mphE | pB106-1 | 100.0 | 885/885 | 190,383..191,267 | Macrolide resistance | DQ839391 |
| ARR-3 | pB106-1 | 100.0 | 453/453 | 90,760..91,212 | Rifamycin resistance | JF806499 |
| addA16 | pB106-1 | 99.65 | 846/846 | 89,128..89,973 | Aminoglycosides Resistance | EU675686 |
| fosA3 | pB106-1 | 100.0 | 417/417 | 81,087..81,503 | Fosfomycin resistance | AB522970 |
| sul1 | pB106-1 | 100.0 | 840/840 | 87,831..88,670 | Folate pathway antagonist | U12338 |
| dfrA27 | pB106-1 | 100.0 | 474/474 | 90,154..90,627 | Folate pathway antagonist | FJ459817 |
| blaSHV-12 | pB106-IMP | 98.88 | 861/861 | 42,832..43,692 | Beta-lactam resistance | KF976405 |
| blaIMP-4 | pB106-IMP | 100.0 | 741/741 | 48,488..49,228 | Beta-lactam resistance | AF244145 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Chen, H.; Liu, W.; Yang, L.; Xu, Z.; Chen, D. Resistome and Genome Analysis of an Extensively Drug-Resistant Klebsiella michiganensis KMIB106: Characterization of a Novel KPC Plasmid pB106-1 and a Novel Cointegrate Plasmid pB106-IMP Harboring blaIMP-4 and blaSHV-12. Antibiotics 2023, 12, 1463. https://doi.org/10.3390/antibiotics12091463
Wang L, Chen H, Liu W, Yang L, Xu Z, Chen D. Resistome and Genome Analysis of an Extensively Drug-Resistant Klebsiella michiganensis KMIB106: Characterization of a Novel KPC Plasmid pB106-1 and a Novel Cointegrate Plasmid pB106-IMP Harboring blaIMP-4 and blaSHV-12. Antibiotics. 2023; 12(9):1463. https://doi.org/10.3390/antibiotics12091463
Chicago/Turabian StyleWang, Linjing, Haijun Chen, Wanting Liu, Ling Yang, Zhenbo Xu, and Dingqiang Chen. 2023. "Resistome and Genome Analysis of an Extensively Drug-Resistant Klebsiella michiganensis KMIB106: Characterization of a Novel KPC Plasmid pB106-1 and a Novel Cointegrate Plasmid pB106-IMP Harboring blaIMP-4 and blaSHV-12" Antibiotics 12, no. 9: 1463. https://doi.org/10.3390/antibiotics12091463
APA StyleWang, L., Chen, H., Liu, W., Yang, L., Xu, Z., & Chen, D. (2023). Resistome and Genome Analysis of an Extensively Drug-Resistant Klebsiella michiganensis KMIB106: Characterization of a Novel KPC Plasmid pB106-1 and a Novel Cointegrate Plasmid pB106-IMP Harboring blaIMP-4 and blaSHV-12. Antibiotics, 12(9), 1463. https://doi.org/10.3390/antibiotics12091463









