Abstract
Expelling antibiotic molecules out of the cell wall through multiple efflux pumps is one of the potential mechanisms of developing resistance against a wide number of antibiotics in Staphylococcus aureus. The aim of this study was to investigate the association between the antibiotic susceptibility profile and the prevalence of different efflux pump genes i.e., norA, norB, norC, mepA, sepA, mdeA, qacA/B, and smr in the clinical isolates of S. aureus. Sixty clinical isolates were collected from a tertiary level hospital in Bangladesh. The disc diffusion method using ten antibiotics of different classes was used to discern the susceptibility profile. polymerase chain reaction (PCR) was employed to observe the resistance patterns and to detect the presence of plasmid and chromosomal encoded genes. Among the clinical isolates, 60% (36 out of 60) of the samples were Methicillin-resistant Staphylococcus aureus (MRSA), whereas 55% (33 out of 60) of the bacterial samples were found to be multi-drug resistant. The bacteria showed higher resistance to vancomycin (73.33%), followed by ciprofloxacin (60%), cefixime (53.33%), azithromycin (43.33%), and amoxicillin (31.67%). The prevalence of the chromosomally-encoded efflux genes norA (91.67%), norB (90%), norC (93.33%), mepA (93.33%), sepA (98.33%), and mdeA (93.33%) were extremely high with a minor portion of them carrying the plasmid-encoded genes qacA/B (20%) and smr (8.33%). Several genetic combinations of efflux pump genes were revealed, among which norA + norB + norC + mepA + sepA + mdeA was the most widely distributed combination among MRSA and MSSA bacteria that conferred resistance against ciprofloxacin and probably vancomycin. Based on the present study, it is evident that the presence of multiple efflux genes potentiated the drug extrusion activity and may play a pivotal role in the development of multidrug resistance in S. aureus.
1. Introduction
Antibiotic resistance is a major public health concern in the treatment of Staphylococcal infections, particularly the infections caused by methicillin-resistant S. aureus (MRSA). The widespread indiscriminate use of antimicrobial agents combined with the transmission of a significant portion of the organism through person-to-person contact generally develops antimicrobial resistance in the clinical isolates [1,2].
The most current S. aureus epidemiology is focused on the rise and dissemination of MRSA in clinical settings and in the population [3]. However, methicillin-susceptible S. aureus (MSSA) has been a major cause of outbreaks and the global expansion in healthcare settings over the last century, and it is still one of the most common bacteria found in hospital infections. A recent point-prevalence survey conducted in acute care hospitals in 33 countries and coordinated by the European Center for Disease Control and Prevention (ECDC) revealed that S. aureus is the second most commonly isolated microorganism after E. coli, and it remains the leading cause of surgical site infections, while MRSA proportion varies greatly by country [4]. The World Health Organization’s global report on antibiotic resistance surveillance provides a global picture of MRSA expansion [5,6]. Despite the fact that systematic antibiotic resistance data were only available in developed countries in Europe and also in America, and Australia, etc., MRSA was reported on all continents. The prevalence of MRSA in most nations exceeded 20%, and in some cases reached 80%.
The MDR Staphylococcal infection is also becoming a growing concern in Bangladesh. This increased resistance is caused by inadequate hospital administration, a lack of understanding, and cross resistance, which leads to major health consequences. One study in Chittagong, Bangladesh found that 21 of 30 S. aureus bacteria isolates from pus, two out of four from blood, and three out of four from miscellaneous materials were MDR [7]. The study also revealed that the majority of them were medication resistant, with respect to ciprofloxacin (38.06%), cefradine (32.27%) and ampicillin (5.26%) sensitivity. Another study reported that 85.11% of MRSA samples collected in Dhaka, Rajshahi, Mymensingh, and Chittagong over a 12-month period were resistant to penicillin and ampicillin [8].
Several mechanisms, ranging from drug target alteration to active efflux pump systems, are involved in the development of this resistance. Numerous studies have been conducted in recent years to determine the underlying reasons behind antibiotic resistance so that appropriate interventions can be taken. The MDR efflux pumps are either chromosomal or plasmid-encoded and are responsible for developing resistance to various antimicrobial drugs, as well as the selection of drug resistant strains, have received special attention. These pumps, alone or in combination with other pumps, may act as a first line of defense against antimicrobials, allowing the cells to survive and thereby become antibiotic resistant [9]. Due to their promiscuous substrate specificity, the overexpression of MDR efflux pumps can enhance resistance to numerous classes of antibiotics while also decreasing susceptibility to biocides shown to lead to low-level antimicrobial resistance and the formation of MDR phenotypes in clinical isolates [10]. For example, the overexpression of mepA and norB genes showed a pattern of resistance towards tetracyclines, fluoroquinolines, dyes and disinfectants [11]. A previous study identified the combinations of these genes as being in charge of ciprofloxacin resistance [12]. Aside from the risk of therapeutic failure, the emergence of these multidrug resistant phenotypes could lead to other issues, such as possible co-selection and cross-resistance between efflux mediated antibiotic and biocide resistance, which is especially concerning for drug-resistant strains such as multidrug-resistant S. aureus.
Therefore, the present study was intended to find the association between the susceptibility profile of the clinical isolates and the prevalence of efflux pumps genes among them. The study will open up new avenues for future research on efflux pump encoding genes, efflux pumps, and their relationship with MDR. In addition, a new therapeutic concept based on the use of efflux pump inhibitors as adjuvants to conventional therapy could overcome the scarcity of therapeutic alternatives for multidrug resistance (MDR), as well as to help restore the activity of older and less expensive antibiotics.
2. Materials & Methods
2.1. Chemicals
Muller-Hinton agar and Muller-Hinton broth media were collected from TM media Ltd. of India. Amoxicillin (AMX 30 mg), ciprofloxacin (CIP 5 mg), cefixime (CFM 5 mg), tetracycline (TE 30 mg), chloramphenicol (C 30 mg), linezolid (LNZ 10 mg), azithromycin (AZM 15 mg), kanamycin (K 30 mg), vancomycin (VA 30 mg), and imipenem (IPM 10 mg) standards were generously donated by Advanced Chemical Industries Ltd., Dhaka, Bangladesh. The Promega DNA extraction kit was purchased from local suppliers. Specific primers and GoTaq® G2 Green Master Mix were purchased from Biotech Concern Ltd., Bangladesh.
2.2. Bacterial Strain Collection
All of the experiments were performed according to the Clinical Laboratory Standards Institute (CLSI) guidelines (M100, 30th Ed. 2020) and the European Committee on Antibiotic Susceptibility Testing (EUCAST) for Staphylococcus aureus V. 9.0. A total of sixty clinical isolates of Staphylococcus aureus were collected from the Department of Microbiology and Immunology Laboratory; Bangabandhu Sheikh Mujib Medical University, Bangladesh from June 2021 to September 2021.
2.3. Bacterial Identification
Clinical isolates of S. aureus were identified and authenticated by the standard protocol developed by the collection center. Molecular investigation was carried out for further confirmation of the S. aureus bacteria by PCR using specific primers (Forward primer—5′-AGAGTTTGATCCTGGCTCAG-3′ and Reverse primer—5′-TACGGTTACCTTGTTACGACTT-3′ constructed from the 16S rRNA region of S. aureus. MRSA strains were identified by molecular identification of the mecA gene by PCR.
2.4. Bacterial Cell Culture
Sterile tubes containing sterilized MHA were inoculated with a pure culture of an identified clinical isolate of S. aureus under a laminar airflow (LAF) bench and were incubated at 37 ± 2 °C for 2–3 h.
2.5. Antibacterial Susceptibility Test
An antibacterial susceptibility test was performed by using the Kirby-Bauer disc diffusion technique for ten antibiotics mentioned earlier.
MHA and MHB were prepared and sterilized using the appropriate guidelines. Clinical isolates cultured in the MHA media were again inoculated into the test tubes containing MHB and were placed into a shaker incubator for 2 h at a temperature of 37 °C. The bacterial growth was adjusted to 105 CFU/mL for 0.5 McFarland standard by adding phosphate buffer saline. Sterile MHA was poured into petri dishes and after the sterility of the plates was confirmed, bacteria were inoculated into the petri dishes with a cotton swab. The antibiotic discs were placed in the plates with sterilized forceps and then the plates were incubated upside down at 37 °C for 16–18 h. Post incubation, clear zones of inhibition (ZOI) were formed around the discs. The diameter of the zones was measured and interpreted in accordance with the Clinical and Laboratory Standards Institute (CLSI) antimicrobial susceptibility testing standards.
2.6. Amplification of Target Genes by Polymerase Chain Reaction
The extraction of total genomic DNA was done using a Wizard Genomic DNA Purification Kit. Specific primers for norA, norB, norC, mepA, sepA, mdeA, qacA/B, smr were used for the amplification (Table 1). An amount of 10 µL of PCR mixture was prepared by using 5 µL of GoTaq® G2 Green Master Mix, 0.5 µL of forward primer, 0.5 µL of reverse primer, and 3 µL of nuclease-free water with 1.0 µL of template DNA. A negative control without template DNA was also used.

Table 1.
List of primers with detailed information for the identification of different efflux pump genes.
The presence of chromosomally-encoded and plasmid-encoded efflux pump genes was examined in all of the isolated strains. For chromosomally-encoded efflux genes, the initial denaturation was set at 94 °C for 4 min, followed by 35 cycles of denaturation at 94 °C for 30 s. The annealing temperature was 57–60 °C (varied with genes) for 55 s, extension at 72 °C for 1 min, and a final extension of 72 °C for 5 min. For plasmid-encoded efflux genes, the initial denaturation was set at 95 °C for 1 min followed by 30 cycles of denaturation at 95 °C for 30 s, an annealing temperature of 56–58 °C for 45 s, extension at 72 °C for 1 min, and a final extension of 72 °C for 5 min. For the mecA gene, initial denaturation was set at 94 °C for 4 min followed by 35 cycles of final denaturation at 95 °C for 30 s. The annealing temperature was 53 °C for 30 s. An extension was conducted at 72 °C for 1 min with a final extension of 72 °C for 5 min.
The amplification products of the desired genes were visualized by resolving the PCR products in 0.6% agarose gel at 80 volts and 100 mA for 40 min. The gel was viewed using the Alpha Imager HP Gel documentation system (Cell Bioscience, Santa Clara, CA, 95051, USA), photographs were taken using a computer attached to the machine, and the bands were analyzed.
3. Result
3.1. Antibacterial Susceptibility Pattern in Clinical Isolates of S. aureus
In the present study, antibacterial sensitivity was tested using the Kirby-Bauer disc diffusion method to detect multidrug-resistant S. aureus among the collected clinical isolates. The clinical insolates of Staphylococcus aureus were evaluated for their susceptibility towards ten different antibiotics of different groups (Figure 1). A higher number of S. aureus showed resistance to vancomycin (73.33%), followed by ciprofloxacin (60%), cefixime (53.33%), azithromycin (43.33%), and amoxicillin (31.67%). However, the lower number of isolates showed resistance against chloramphenicol (3.33%), kanamycin (8.47%), linezolid (10%), and tetracycline (10%). Imipenem, on the other hand, turned out to be fully effective against the tested antibiotics. Among the isolates, 55% showed resistance to three or more classes of drugs, thereby meeting the classical definition of multidrug resistance (SM’s are enclosed herewith).
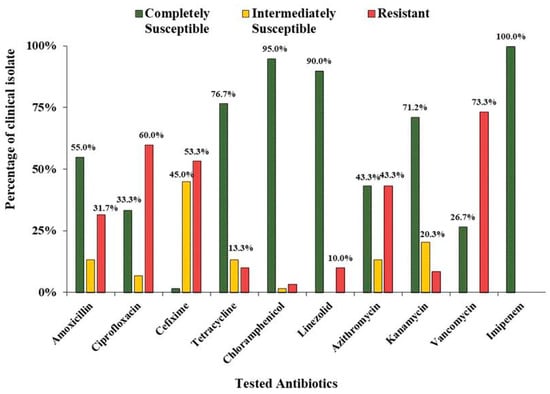
Figure 1.
Antibacterial susceptibility profile of the clinical isolates of S. aureus.
3.2. Characterizations of Different Types of S. aureus in Clinical Isolates
Different kinds of S. aureus have been found among the clinical isolates worldwide. Molecular characterization with PCR targeting the presence of the mecA gene, which is the hallmark of methicillin resistant S. aureus, was used to identify the presence of this bacteria among the samples. Of 60 isolates, 60% turned out to be MRSA, as they contained the mecA gene, which is responsible for methicillin resistance in S. aureus. If a clinical isolate showed resistance to at least three categories of different antibiotics, it is speculated to be multi-drug resistant. According to the susceptibility profile carried out with ten different antibiotics, a total of 33 clinical isolates of S. aureus showed resistance against at least three different antibiotics (Table 2). Among the MSSA (n = 24) clinical isolates, 15 samples were shown to have MDR characteristics, whereas 18 MRSA samples were found to be multi-drug resistant.

Table 2.
Characterization of different types of S. aureus in the clinical isolates (n = 60).
3.3. Prevalence of Different Efflux Pump Genes in the Clinical Isolates of S. aureus
Efflux pump genes were identified using polymerase chain reaction and were visualized by agarose gel electrophoresis. A sample representing the presence of norA gene in different clinical isolates has been shown below (Figure 2).
Figure 2.
(A,B) Detection of norA gene in the clinical isolates using PCR and gel electrophoresis.
An extremely high prevalence of chromosomally encoded genes including norA (91.67%), norB (90%), norC (93.33%), mepA (93.33%), sepA (98.33%), and mdeA (93.33%) was observed among the clinical isolates (n = 60), while very few of them contained plasmid-encoded genes qacA/B (20%) and smr (8.33%) as well (Figure 3).
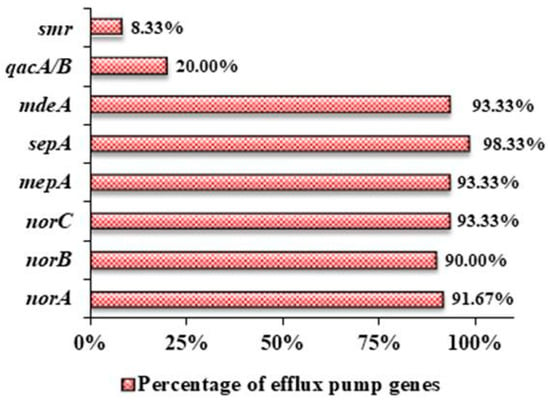
Figure 3.
Prevalence of different efflux pump genes in the clinical isolates of S. aureus.
Irrespective of their resistance status, the most effective combination of efflux pump genes was chromosomal encoded norA + norB + norC + mepA + sepA + mdeA among the clinical isolates including MRSA and MSSA, which provided a higher prevalence of resistance against the tested antibiotics (Figure 4). Six chromosomal genes were found to be 47.2% and 70.8% in methicillin-resistant S. aureus (MRSA) and methicillin-sensitive S. aureus (MSSA) clinical isolates, respectively. Two plasmid encoded genes such as qacA/B and smr along with chromosomal encoded genes were present in only MDR MRSA (11.1%). However, qacA/B along with six chromosomal encoded genes was found in both MDR MRSA (11.1%) and MDR MSSA (4.2%) isolates (Figure 4).
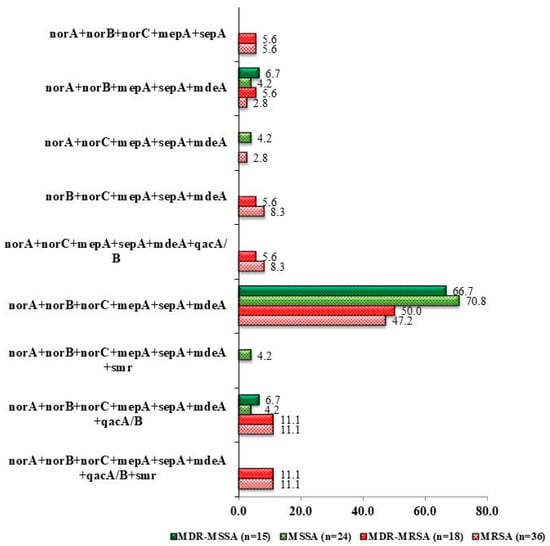
Figure 4.
Prevalence of different combinations of genes present in the collected clinical isolates of S. aureus.
3.4. Antibiotic Resistant Profile Associated with Efflux Pump Genes Combination
In the present study, the profiling of antibiotic resistant genes in the clinical isolates of S. aureus was investigated. The presence of at least three genes encoding efflux pumps in those clinical isolates of S. aureus that have largely showed resistance to several antibiotics was observed. More than six combinations of genes were found in the clinical isolates that showed resistance to five different antibiotics, namely vancomycin, amoxicillin, ciprofloxacin, cefixime, and azithromycin. The details of the different combinations of genes with respect to various antibiotic resistant isolates are shown in Table 3 and Table 4.

Table 3.
Cumulative frequency of different combinations of efflux pump genes in different clinical isolates.

Table 4.
Frequency of different combinations of efflux pump genes against different antibiotic resistant clinical isolates.
4. Discussion
This study was conducted on both methicillin resistant S. aureus (MRSA) and methicillin sensitive S. aureus (MSSA) collected from a tertiary hospital in Bangladesh. Based on our result, this is the first report in Bangladesh on the detection of several kinds of efflux pump genes in the clinical isolates of S. aureus. It enabled us to get a clear perception of the effectivity of different classes of antibiotics against S. aureus and to detect the prevalence of specific efflux protein encoding genes with a view to establishing a possible association between efflux pumps and multidrug resistance in S. aureus.
The clinical isolates were classified as methicillin-sensitive S. aureus, methicillin-resistant S. aureus, MDR methicillin-sensitive S. aureus, and MDR methicillin-resistant S. aureus, where 60% of isolates turned out to be MRSA, which became a potential threat to the patients compared to MSSA [13,14]. Parvez et al. 2018 [15] reported a 72% prevalence of MRSA in the clinical isolates depending upon the presence of the mecA gene, which had a higher incidence rate than that indicated in another report from Bangladesh published in 2005 [8]. On the other hand, our findings are similar with a published report on the clinical samples of S. aureus collected from different hospitals of Chittagong, where they noted that 65.15% were of the MRSA strain [16]. Compared with a previous report from Western Europe where MRSA from the clinical isolates of S. aureus was found in a range between 5 to 54% [17], Bangladeshi isolates are more prone to be identified as MRSA.
In the present study, 98.33% of bacterial samples showed resistance against at least one class of drug. However, all of the strains showed complete susceptibility towards imipenem, a carbapenem derivative, which matched with the previous report from another hospital in Bangladesh [18]. Although the same report showed that all isolates were sensitive to vancomycin, we observed a significant difference here because 73.3% of isolates were resistant to it. It is evident that all of them can be designated as vancomycin resistant S. aureus strains (VRSA), which was first reported in the U.S. in 2002 [19]. Vancomycin resistance in the bacteria is generally conferred by the vanA operon encoded on Transposon Tn1546 in the Vancomycin resistant Enterococci (VRE), which may horizontally transfer to S. aureus isolates in our study isolates [20]. However, it is the first report of observing VRSA strains in Bangladesh which makes it a serious concern for clinicians. Surprisingly, a significant portion of the clinical isolates showed sensitivity towards amoxicillin compared to cefixime in this study (Figure 1), despite the latter antibiotic belonging to a more advanced class of beta-lactam antibiotics.
The extrusion of multiple substrates through efflux pump proteins being a predominant mechanism of antibacterial resistance, eight of the well-studied genes responsible for expressing different efflux proteins in S. aureus were taken into consideration and were used to find out whether there is any possible common genetic combination among the collected clinical isolates that may confer higher antibiotic resistance. It was found that irrespective of their resistance pattern, each bacterial sample contained at least four efflux protein-encoding genes in their DNA (Table 3). While the individual percentage of the prevalence of chromosomally-encoded genes exceeded an average of 90%, very few possessed plasmid-encoded efflux protein genes, with the smr gene being the least prevalent (8.33%). While several studies have been conducted individually on MFS transporter norA [21], norB [22], norC [23], mepA [24], sepA [25], and mdeA [26], as well as on their role in ciprofloxacin resistance, our findings are in accordance with these aforementioned works. A systematic review on the epidemiology of efflux pump genes on S. aureus also revealed similar findings, where norA (75%) was highly reported in Asia, and the least reported was qacA/B (20.8%) [27]. Table 5 shows a comparison of the prevalence of efflux pump genes in the clinical isolates of different countries.

Table 5.
Frequency of different efflux pump genes in S. aureus reported among different countries.
All 21 ciprofloxacin-resistant MRSA isolates had at least five chromosomal encoded genes. However, a combination of six genes such as norA + norB + norC + mepA + sepA + mdeA was found to be widely distributed among the clinical samples. All of these, except sepA, were found predominantly in the Iraqi patients as well [29]. Seven of the 21 ciprofloxacin-resistant MRSA contained at least one plasmid-encoded gene qacA/B and only two of them had smr gene along with six chromosomal encoded genes. A similar result was reported in African countries, where plasmid encoded qacA/B (40.5%) and smr (3.7%) genes were reported from the S. aureus isolates [34]. The presence of plasmid encoded qacA/B genes in the MRSA showed complete resistance towards ciprofloxacin and cefixime. This observation hinted toward a potentiated action of efflux pumps due to the presence of multiple efflux proteins in the same clinical isolate.
A similar distribution of genes was observed in the amoxicillin, cefixime, azithromycin, and vancomycin-resistant bacterial samples as well, since they possessed various genetic combinations, with the major portion containing at least six efflux genes in their DNA. However, whether these efflux pumps are the primary cause of developing resistance against other classes of drugs or not remains to be investigated. It is to be noted that efflux pumps are generally responsible for developing resistance against ciprofloxacin, tetracycline and azithromycin.
It has been documented that mecA gene is responsible for encoding the low-affinity penicillin-binding protein PBP 2A that confers methicillin resistance in S. aureus [35]. Among the amoxicillin-resistant strains, 57.89% of samples had the mecA gene, whereas the percentage was higher in cefixime-resistant strains (68.75%). The greater distribution of the mecA gene in cefixime compared to amoxicillin made us wonder about a shift in their evolutionary process, since bacterial cells stop undergoing mutational changes in the absence of antibiotic molecules, as doing otherwise adversely affects cellular homeostasis. Hence, the lower distribution of mecA in amoxicillin-resistant clinical strains and their improved susceptibility towards penicillin could be attributed to its lesser use in clinical practice compared to the other classes of antibiotics. The same inference can be made in the case of tetracycline, chloramphenicol and linezolid, because most of the isolates showed susceptibility towards these antibiotics in this study. If this is indeed the case, then it could make a significant contribution to the field of medicine in the field of antibiotic resistance. Nevertheless, the coexistence of the mecA gene with multiple efflux proteins in resistant bacterial strains made us ponder their synergistic effect. However, MSSA isolates possessing similar combinations of chromosomal genes exhibited a similar antibiotic resistance pattern when compared with the MRSA isolates. Notably, the combination of norA + norB + norC + mepA + sepA genes presenting in MSSA isolates were shown to be completely resistant to ciprofloxacin, azithromycin, and vancomycin, which shifted our focus to the expression of these genes and the specific conditions favoring it.
5. Conclusions
Antibiotic resistance is a worldwide phenomenon. With its constantly evolving molecular mechanism, Staphylococcus aureus has become a major catalyst behind this global health hazard. To combat it, we first need to identify the resistance pattern of this pathogen and the mechanisms responsible for its resistance. In this study, we found some common patterns of resistance and identified a number of clinically significant genes. However, our study had some limitations as well, since we have only observed the prevalence of specific genes but have not examined their expression in the clinical isolates. Hence, further study is needed to determine the association between efflux proteins and the waning sensitivity of S. aureus to different classes of drugs. However, based on the findings of our study, we could conclude that the resistance pattern of S. aureus in the Bangladeshi population is largely attributable to the prevalence of different efflux proteins.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics12020305/s1.
Author Contributions
T.A.S. and N.A. were both involved in sample collection, experimental design, data analysis, the interpretation of results, and manuscript writing. M.A.Z., S.C.M. and A.H. conducted the molecular works as well as helped in visualizing gel electrophoresis. S.Z.R. and A.H. characterized the bacterial sample as well. F.J.S. and R.K. were involved in D.N.A. extraction and susceptibility testing. A.H. and M.A.M. were involved in designing the study and in the supervision of the work. S.Z.R. and M.A.M. were involved in data interpretation and edited the manuscript. M.A.M. coordinated the experiment and supervised the team as well. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Centennial Research Grant from University of Dhaka to Md. Abdul Muhit (Grant letter no.Reg/Admin-3/47848) and the APC was partially funded by University of Dhaka administration.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of the Faculty of Biological Science, University of Dhaka (Ref. No. 161/Biol. Scs./2021).
Informed Consent Statement
Patient consent was waived due to collection of bacterial samples directly from the preserved agar media at microbiology laboratory of the Department of Microbiology and Immunology, Bangabandhu Sheikh Mujib Medical University, Bangladesh.
Data Availability Statement
The datasets originated in this study will be available upon request or in the Supplementary Files attached with this manuscript.
Acknowledgments
The authors would like to thank the Department of Microbiology and Immunology, Bangabandhu Sheikh Mujib Medical University, Bangladesh, for providing necessary support to collect the samples. In addition, the corresponding author, Md. Abdul Muhit, is grateful to the administration of the University of Dhaka for funding the project and APC.
Conflicts of Interest
The authors declare that they have no conflicts of interests.
References
- Okeke, I.N.; Lamikanra, A. Export of antimicrobial drugs by West African travelers. J. Travel Med. 2006, 10, 133–135. [Google Scholar] [CrossRef] [PubMed]
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.P.; Pearson, A.; Duckworth, G. Surveillance and epidemiology of MRSA bacteraemia in the UK. J. Antimicrob. Chemother. 2005, 56, 455–462. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Point Prevalence Survey of Healthcare-Associated Infections and Antimicrobial Use in European Acute Care Hospitals: 2011–2012; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2013. [Google Scholar]
- World Health Organization. WHO’s First Global Report on Antibiotic Resistance Reveals Serious, Worldwide Threat to Public Health. World Health Organization: Geneva, Switzerland. Available online: https://www.who.int/news/item/30-04-2014-who-s-first-global-report-on-antibiotic-resistance-reveals-serious-worldwide-threat-to-public-health (accessed on 5 August 2022).
- Abdulgader, S.M.; Shittu, A.O.; Nicol, M.P.; Kaba, M. Molecular epidemiology of Methicillin-resistant Staphylococcus aureus in Africa: A systematic review. Front. Microbiol. 2015, 6, 348. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Mahmud, M.; Chowdhury, M.; Hakim, M. Prevalence of multidrug resistant Staphylococcus aureus isolates in clinical specimens collected from local patients of Chittagong, Bangladesh. Chittagong Univ. J. Sci. 2013, 6, 175–185. [Google Scholar] [CrossRef]
- Haq, J.A.; Rahman, M.M.; Asna, S.M.Z.H.; Hossain, M.A.; Ahmed, I.; Haq, T.; Morshed, M.A.H.G. Methicillin-resistant Staphylococcus aureus in Bangladesh—A multicentre study. Int. J. Antimicrob. Agents 2005, 25, 276–277. [Google Scholar] [CrossRef]
- Costa, S.S.; Viveiros, M.; Amaral, L.; Couto, I. Multidrug efflux pumps in Staphylococcus aureus: An Update. Open Microbio. J. 2013, 7, 59–71. [Google Scholar] [CrossRef]
- Piddock, L.J.V. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 2006, 19, 382–402. [Google Scholar] [CrossRef]
- Vali, L.; Davies, S.E.; Lai, L.L.G.; Dave, J.; Amyes, S.G.B. Frequency of biocide resistance genes, antibiotic resistance and the effect of chlorhexidine exposure on clinical methicillin-resistant Staphylococcus aureus isolates. J. Antimicrob. Chemother. 2008, 61, 524–532. [Google Scholar] [CrossRef]
- Hassanzadeh, S.; Mashhadi, R.; Yousefi, M.; Askari, E.; Saniei, M.; Pourmand, M.R. Frequency of efflux pump genes mediating ciprofloxacin and antiseptic resistance in methicillin-resistant Staphylococcus aureus isolates. Microb. Pathog. 2017, 111, 71–74. [Google Scholar] [CrossRef]
- Watkins, R.R.; David, M.Z.; Salata, R.A. Current concepts on the virulence mechanisms of meticillin-resistant Staphylococcus aureus. J. Med. Microbiol. 2012, 61, 1179–1193. [Google Scholar] [CrossRef] [PubMed]
- Uhlemann, A.C.; Otto, M.; Lowy, F.D.; DeLeo, F.R. Evolution of community- and healthcare-associated methicillin-resistant Staphylococcus aureus. Infect. Genet. Evol. 2014, 21, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Parvez MA, K.; Ferdous, R.N.; Rahman, M.S.; Islam, S. Healthcare-associated (HA) and community-associated (CA) methicillin resistant Staphylococcus aureus (MRSA) in Bangladesh—Source, diagnosis and treatment. J. Genet. Eng. Biotechnol. 2018, 16, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Islam, T.; Kubra, K.; Chowdhury, M.M.H. Prevalence of Methicillin-Resistant Staphylococcus aureus in hospitals in Chittagong, Bangladesh: A threat of nosocomial infection. J. Microsc. Ultrastruct. 2018, 6, 188–191. [Google Scholar]
- Dulon, M.; Haamann, F.; Peters, C.; Schablon, A.; Nienhaus, A. MRSA prevalence in European healthcare settings: A review. BMC Infect. Dis. 2011, 11, 138. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.Z.; Naher, A.; Hasan, P.; Mozazfia, K.T.; Tasnim, H.; Ferdush, Z.; Towhid, K.M.; Simran, M.A.A. Prevalent bacteria and their sensitivity and resistance pattern to antibiotics: A study in Dhaka Medical College Hospital. J. Dhaka Med. Coll. 2017, 26, 52–64. [Google Scholar] [CrossRef]
- McGuinness, W.A.; Malachowa, N.; Deleo, F.R. Vancomycin resistance in Staphylococcus aureus. Yale J. Biol. Med. 2017, 90, 269–281. [Google Scholar] [PubMed]
- George, S.K.; Suseela, M.R.; Safi, S.E.; Elnagi, E.A.; Al-naam, Y.A.; Adam, A.A.M.; Jacob, A.M.; Al-Maqati, T.; KS, H.K. Molecular determination of van genes among clinical isolates of enterococci at a hospital setting. Saudi J. Biol. Sci. 2021, 28, 2895–2899. [Google Scholar] [CrossRef]
- Kaatz, G.W.; Seo, S.M. Inducible NorA-mediated multidrug resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1995, 39, 2650–2655. [Google Scholar] [CrossRef]
- Truong-Bolduc, Q.C.; Dunman, P.M.; Strahilevitz, J.; Projan, S.J.; Hooper, D.C. MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J. Bacteriol. 2005, 187, 2395–2405. [Google Scholar] [CrossRef]
- Truong-Bolduc, Q.C.; Strahilevitz, J.; Hooper, D.C. NorC, a new efflux pump regulated by MgrA of Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 1104–1107. [Google Scholar] [CrossRef] [PubMed]
- Kaatz, G.W.; McAleese, F.; Seo, S.M. Multidrug Resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob. Agents Chemother. 2005, 49, 1857–1864. [Google Scholar] [CrossRef] [PubMed]
- Narui, K.; Noguchi, N.; Wakasugi, K.; Sasatsu, M. Cloning and characterization of a novel chromosomal drug efflux gene in Staphylococcus aureus. Biol. Pharm. Bull. 2002, 25, 1533–1536. [Google Scholar] [CrossRef]
- Huang, J.; O’Toole, P.W.; Shen, W.; Amrine-Madsen, H.; Jiang, X.; Lobo, N.; Palmer, L.M.; Voelker, L.; Fan, F.; Gwynn, M.N.; et al. Novel chromosomally encoded multidrug efflux transporter MdeA in Staphylococcus aureus. Antimicrob. Agents Chemother. 2004, 48, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Hassanzadeh, S.; Ganjloo, S.; Pourmand, M.R.; Mashhadi, R.; Ghazvini, K. Epidemiology of efflux pumps genes mediating resistance among Staphylococcus aureus: A systematic review. Microb. Pathog. 2020, 139, 103850. [Google Scholar] [CrossRef]
- Liu, Q.; Zhao, H.; Han, L.; Shu, W.; Wu, Q.; Ni, Y. Frequency of biocide-resistant genes and susceptibility to chlorhexidine in high-level mupirocin-resistant, methicillin-resistant Staphylococcus aureus (MuH MRSA). Diagn. Microbiol. Infect. Dis. 2015, 82, 278–283. [Google Scholar] [CrossRef]
- Shamkhi, G.J.; Saadedin, S.M.K.; Jassim, K.A. Detection the prevalence of some chromosomal efflux pump genes in Methicillin resistant Staphylococcus aureus isolated from Iraqi patients. Iraqi J. Biotechnol. 2019, 18, 33–42. [Google Scholar]
- Antiabong, J.F.; Kock, M.M.; Mbelle, N.M.; Ehlers, M.M. Diversity of multidrug efflux genes and phenotypic evaluation of the in vitro resistance dynamics of clinical Staphylococcus aureus isolates using methicillin; a model β-lactam. Open Microb. J. 2017, 11, 132–141. [Google Scholar] [CrossRef]
- Conceiçãoa, T.; Lencastrea, H.D.; Aires-de-Sousaa, M. Prevalence of biocide resistance genes and chlorhexidine and mupirocin non-susceptibility in Portuguese hospitals during a 31-year period (1985–2016). J. Glob. Antimicrob. Resist. 2021, 24, 169–174. [Google Scholar] [CrossRef]
- Shamsudin, M.N.; Alreshidi, M.A.; Hamat, R.A.; Alshrari, A.S.; Atshan, S.S.; Neela, V. High prevalence of qacA/B carriage among clinical isolates of meticillin-resistant Staphylococcus aureus in Malaysia. J. Hosp. Infect. 2012, 81, 206–208. [Google Scholar] [CrossRef]
- Longtin, J.; Seah, S.; Siebert, K.; McGeer, A.; Simor, A.; Longtin, Y.; Low, D.E.; Melano, R.G. Distribution of antiseptic resistance genes qacA, qacB, and smr in methicillin-resistant Staphylococcus aureus isolated in Toronto, Canada, from 2005 to 2009. Antimicrob. Agents Chemother. 2011, 55, 2999–3001. [Google Scholar] [CrossRef] [PubMed]
- Conceição, T.; Coelho, C.; Lencastre, H.D.; Aires-de-Sousa, M. High prevalence of biocide resistance determinants in Staphylococcus aureus isolates from three African countries. Antimicrob. Agents Chemother. 2016, 60, 678–681. [Google Scholar] [CrossRef] [PubMed]
- Beck, W.D.; Berger-Bächi, B.; Kayser, F.H. Additional DNA in methicillin-resistant Staphylococcus aureus and molecular cloning of mec-specific DNA. J. Bacteriol. 1986, 165, 373–378. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).