Site Mutation Improves the Expression and Antimicrobial Properties of Fungal Defense
Abstract
1. Introduction
2. Results
2.1. Sequence Design and Screening
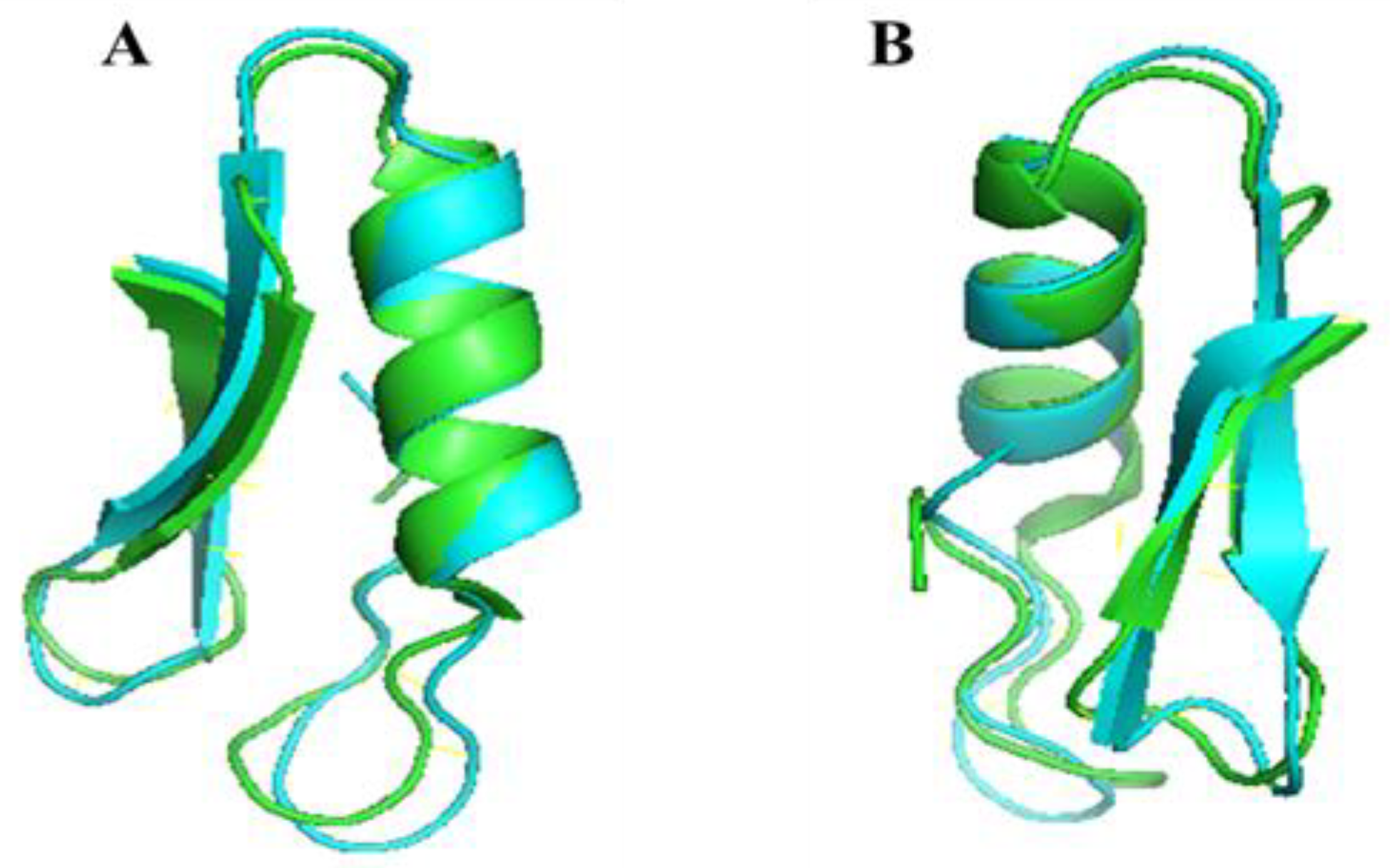
2.2. Structure Analysis of PN7
2.3. PN7 Had Potent Antimicrobial Activity
2.4. In Vitro and In Vivo Toxicity of PN7
2.5. Desirable Stability of PN7
2.6. In Vitro Antimicrobial Analysis of PN7
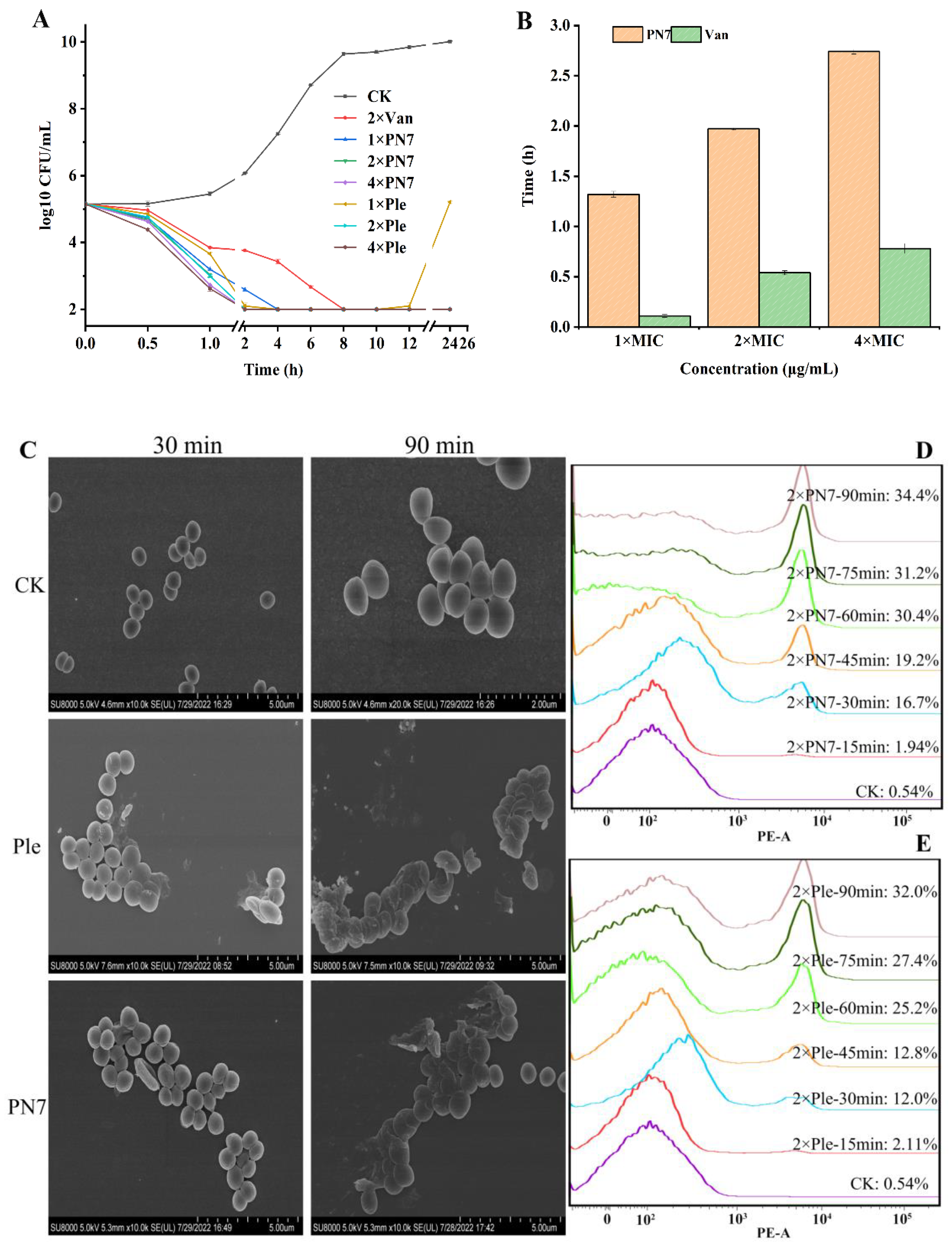
2.6.1. Time-Killing Curves
2.6.2. The Post Antibiotic Effect (PAE) of PN7 against S. aureus
2.6.3. Morphological Observation
2.6.4. Effect of PN7 on Membrane Penetrating
3. Discussion
4. Materials and Methods
4.1. Construction of the Library
4.2. Screening of Active Clones
4.3. Structure Analysis
4.4. Antibacterial Activity (MIC)
4.5. Safety Evaluation
4.6. Stability Analysis
4.6.1. Thermal and pH Stability
4.6.2. Salt Stability
4.6.3. Serum and Proteolytic Stability
4.7. In Vitro Bactericidal Kinetics
4.8. PAE of PN7 against S. aureus
4.9. SEM Observations
4.10. Effect of PN7 on Membrane Penetrating
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Browne, K.; Chakraborty, S.; Chen, R.; Willcox, M.D.; Black, D.S.; Walsh, W.R.; Kumar, N. A New Era of Antibiotics: The Clinical Potential of Antimicrobial Peptides. Int. J. Mol. Sci. 2020, 21, 7047. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.Y.; Yan, Z.B.; Meng, Y.M.; Hong, X.Y.; Shao, G.; Ma, J.J.; Cheng, X.R.; Liu, J.; Kang, J.; Fu, C.Y. Antimicrobial peptides: Mechanism of action, activity and clinical potential. Mil. Med. Res. 2021, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zietz, C.M.; Mudgapalli, A.; Wang, S.; Wang, Z. The evolution of the antimicrobial peptide database over 18 years: Milestones and new features. Protein Sci. 2022, 31, 92–106. [Google Scholar] [CrossRef]
- Schäfer, A.B.; Wenzel, M. A How-To Guide for Mode of Action Analysis of Antimicrobial Peptides. Front. Cell. Infect. Microbiol. 2020, 10, 540898. [Google Scholar] [CrossRef]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rojas, A.; Makarova, O.; Rolff, J. Antimicrobials, stress and mutagenesis. PLoS Pathog. 2014, 10, e1004445. [Google Scholar] [CrossRef]
- Fantner, G.E.; Barbero, R.J.; Gray, D.S.; Belcher, A.M. Kinetics of antimicrobial peptide activity measured on individual bacterial cells using high-speed atomic force microscopy. Nat. Nanotechnol. 2010, 5, 280–285. [Google Scholar] [CrossRef]
- Zhang, M.; Ouyang, J.; Fu, L.; Xu, C.; Ge, Y.; Sun, S.; Li, X.; Lai, S.; Ke, H.; Yuan, B.; et al. Hydrophobicity Determines the Bacterial Killing Rate of α-Helical Antimicrobial Peptides and Influences the Bacterial Resistance Development. J. Med. Chem. 2022, 65, 14701–14720. [Google Scholar] [CrossRef]
- Mathur, D.; Prakash, S.; Anand, P.; Kaur, H.; Agrawal, P.; Mehta, A.; Kumar, R.; Singh, S.; Raghava, G.P. PEPlife: A Repository of the Half-life of Peptides. Sci. Rep. 2016, 6, 36617. [Google Scholar] [CrossRef]
- Lazzaro, B.P.; Zasloff, M.; Rolff, J. Antimicrobial peptides: Application informed by evolution. Science 2020, 368, eaau5480. [Google Scholar] [CrossRef]
- Sampaio de Oliveira, K.B.; Leite, M.L.; Rodrigues, G.R.; Duque, H.M.; da Costa, R.A.; Cunha, V.A.; de Loiola Costa, L.S.; da Cunha, N.B.; Franco, O.L.; Dias, S.C. Strategies for recombinant production of antimicrobial peptides with pharmacological potential. Expert Rev. Clin. Pharmacol. 2020, 13, 367–390. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; Sharma, K.K.; Sharma, A.; Jain, R. Peptide-based drug discovery: Current status and recent advances. Drug Discov. Today 2023, 28, 103464. [Google Scholar] [CrossRef]
- Shi, G.; Kang, X.; Dong, F.; Liu, Y.; Zhu, N.; Hu, Y.; Xu, H.; Lao, X.; Zheng, H. DRAMP 3.0: An enhanced comprehensive data repository of antimicrobial peptides. Nucleic Acids Res. 2022, 50, D488–D496. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.; Teixeira, C.; Gomes, P.; Martins, M.C.L. Clinical Application of AMPs. Adv. Exp. Med. Biol. 2019, 1117, 281–298. [Google Scholar] [CrossRef] [PubMed]
- Usmani, S.S.; Bedi, G.; Samuel, J.S.; Singh, S.; Kalra, S.; Kumar, P.; Ahuja, A.A.; Sharma, M.; Gautam, A.; Raghava, G.P.S. THPdb: Database of FDA-approved peptide and protein therapeutics. PLoS ONE 2017, 12, e0181748. [Google Scholar] [CrossRef]
- Henninot, A.; Collins, J.C.; Nuss, J.M. The Current state of peptide drug discovery: Back to the future? J. Med. Chem. 2018, 61, 1382–1414. [Google Scholar] [CrossRef]
- Barreto-Santamaría, A.; Patarroyo, M.E.; Curtidor, H. Designing and optimizing new antimicrobial peptides: All targets are not the same. Crit. Rev. Clin. Lab. Sci. 2019, 56, 351–373. [Google Scholar] [CrossRef]
- Raventós, D.; Taboureau, O.; Mygind, P.H.; Nielsen, J.D.; Sonksen, C.P.; Kristensen, H.H. Improving on nature’s defenses: Optimization & high throughput screening of antimicrobial peptides. Comb. Chem. High Throughput Screen. 2005, 8, 219–233. [Google Scholar] [CrossRef]
- Blondelle, S.E.; Lohner, K. Optimization and high-throughput screening of antimicrobial peptides. Curr. Pharm. Des. 2010, 16, 3204–3211. [Google Scholar] [CrossRef]
- Ritter, S.C.; Yang, M.L.; Kaznessis, Y.N.; Hackel, B.J. Multispecies activity screening of microcin J25 mutants yields antimicrobials with increased specificity toward pathogenic Salmonella species relative to human commensal Escherichia coli. Biotechnol. Bioeng. 2018, 115, 2394–2404. [Google Scholar] [CrossRef]
- Tominaga, T.; Hatakeyama, Y. Development of innovative pediocin PA-1 by DNA shuffling among class IIa bacteriocins. Appl. Environ. Microbiol. 2007, 73, 5292–5299. [Google Scholar] [CrossRef][Green Version]
- Islam, M.R.; Shioya, K.; Nagao, J.; Nishie, M.; Jikuya, H.; Zendo, T.; Nakayama, J.; Sonomoto, K. Evaluation of essential and variable residues of nukacin ISK-1 by NNK scanning. Mol. Microbiol. 2009, 72, 1438–1447. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.C.; Kim, H.R.; Park, Y.S.; Park, S.M.; Kim, J.H. Design and screening of in vivo expressed antimicrobial peptide library. Biotechnol. Lett. 2002, 24, 251–256. [Google Scholar] [CrossRef]
- Guralp, S.A.; Murgha, Y.E.; Rouillard, J.M.; Gulari, E. From design to screening: A new antimicrobial peptide discovery pipeline. PLoS ONE 2013, 8, e59305. [Google Scholar] [CrossRef] [PubMed]
- Alecu, M.; Coman, G.; Mușetescu, A.; Coman, O.A. Antimicrobial peptides as an argument for the involvement of innate immunity in psoriasis (Review). Exp. Ther. Med. 2020, 20, 192. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Yang, N.; Teng, D.; Hao, Y.; Mao, R.; Wang, J. Molecular modification of Kex2 P1’ site enhances expression and druggability of fungal defensin. Antibiotics 2023, 12, 786. [Google Scholar] [CrossRef]
- Hara, S.; Mukae, H.; Sakamoto, N.; Ishimoto, H.; Amenomori, M.; Fujita, H.; Ishimatsu, Y.; Yanagihara, K.; Kohno, S. Plectasin has antibacterial activity and no affect on cell viability or IL-8 production. Biochem. Biophys. Res. Commun. 2008, 374, 709–713. [Google Scholar] [CrossRef]
- Yu, W.; Ma, J.; Chen, X.; Tan, Y.; Chen, P.; Zhu, X.; Liu, L. Expression and purification of recombinant Lactobacillus casei bacteriocin and analysis of its antibacterial activity. CyTA J. Food 2020, 18, 301–308. [Google Scholar] [CrossRef]
- Liu, Y.; Ding, S.; Shen, J.; Zhu, K. Nonribosomal antibacterial peptides that target multidrug-resistant bacteria. Nat Prod Rep. 2019, 36, 573–592. [Google Scholar] [CrossRef]
- Li, J.; Jaitzig, J.; Hillig, F.; Süssmuth, R.; Neubauer, P. Enhanced production of the nonribosomal peptide antibiotic valinomycin in Escherichia coli through small-scale high cell density fed-batch cultivation. Appl Microbiol Biotechnol. 2014, 98, 591–601. [Google Scholar] [CrossRef]
- Mygind, P.H.; Fischer, R.L.; Schnorr, K.M.; Hansen, M.T.; Sönksen, C.P.; Ludvigsen, S.; Raventós, D.; Buskov, S.; Christensen, B.; De Maria, L.; et al. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature 2005, 437, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Mandal, K.; Pentelute, B.L.; Tereshko, V.; Thammavongsa, V.; Schneewind, O.; Kossiakoff, A.A.; Kent, S.B. Racemic crystallography of synthetic protein enantiomers used to determine the X-ray structure of plectasin by direct methods. Protein Sci. 2009, 18, 1146–1154. [Google Scholar] [CrossRef]
- Schneider, T.; Kruse, T.; Wimmer, R.; Wiedemann, I.; Sass, V.; Pag, U.; Jansen, A.; Nielsen, A.K.; Mygind, P.H.; Raventós, D.S.; et al. Plectasin, a fungal defensin, targets the bacterial cell wall precursor Lipid II. Science 2010, 328, 1168–1172. [Google Scholar] [CrossRef] [PubMed]
- Eckert, R. Road to clinical efficacy: Challenges and novel strategies for antimicrobial peptide development. Future Microbiol. 2011, 6, 635–651. [Google Scholar] [CrossRef] [PubMed]
- Boman, H.G. Innate immunity and the normal microflora. Immunol. Rev. 2000, 173, 5–16. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, Y.; Teng, D.; Tian, Z.; Wang, S.; Wang, J. Expression of plectasin in Pichia pastoris and its characterization as a new antimicrobial peptide against Staphyloccocus and Streptococcus. Protein Expr. Purif. 2011, 78, 189–196. [Google Scholar] [CrossRef]
- Wang, G. Improved methods for classification, prediction, and design of antimicrobial peptides. Methods Mol. Biol. 2015, 1268, 43–66. [Google Scholar] [CrossRef]
- Zhang, K.; Teng, D.; Mao, R.; Yang, N.; Hao, Y.; Wang, J. Thinking on the Construction of Antimicrobial Peptide Databases: Powerful Tools for the Molecular Design and Screening. Int. J. Mol. Sci. 2023, 24, 3134. [Google Scholar] [CrossRef]
- Yang, N.; Teng, D.; Mao, R.; Hao, Y.; Wang, X.; Wang, Z.; Wang, X.; Wang, J. A recombinant fungal defensin-like peptide-P2 combats multidrug-resistant Staphylococcus aureus and biofilms. Appl. Microbiol. Biotechnol. 2019, 103, 5193–5213. [Google Scholar] [CrossRef]
- Tan, P.; Fu, H.; Ma, X. Design, optimization, and nanotechnology of antimicrobial peptides: From exploration to applications. Nano Today 2021, 39, 101229. [Google Scholar] [CrossRef]
- Higgs, R.; Lynn, D.J.; Cahalane, S.; Alaña, I.; Hewage, C.M.; James, T.; Lloyd, A.T.; O’Farrelly, C. Modification of chicken avian beta-defensin-8 at positively selected amino acid sites enhances specific antimicrobial activity. Immunogenetics 2007, 59, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Vasil, A.I.; Hale, J.; Hancock, R.E.; Vasil, M.L.; Hodges, R.S. Effects of net charge and the number of positively charged residues on the biological activity of amphipathic alpha-helical cationic antimicrobial peptides. Adv. Exp. Med. Biol. 2009, 611, 561–562. [Google Scholar] [CrossRef]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob. Agents Chemother. 2007, 51, 1398–1406. [Google Scholar] [CrossRef]
- Hao, Y.; Wang, J.; de la Fuente-Nunez, C.; Franco, O.L. Editorial: Antimicrobial Peptides: Molecular Design, Structure-Function Relationship, and Biosynthesis Optimization. Front. Microbiol. 2022, 13, 888540. [Google Scholar] [CrossRef]
- Schnorr, K.M.; Hansen, M.T.; Mygind, P.H.; Segura, D.R. Antimicrobial Polypeptides from Pseudoplectania nigrella. U.S. Patent 7972814B2, 5 July 2011. [Google Scholar]
- Jing, X.L.; Luo, X.G.; Tian, W.J.; Lv, L.H.; Jiang, Y.; Wang, N.; Zhang, T.C. High-level expression of the antimicrobial peptide plectasin in Escherichia coli. Curr. Microbiol. 2010, 61, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Zhang, Y.; Mao, R.; Teng, D.; Wang, X.; Wang, J. Design and recombination expression of a novel plectasin-derived peptide MP1106 and its properties against Staphylococcus aureus. Appl. Microbiol. Biotechnol. 2015, 99, 2649–2662. [Google Scholar] [CrossRef]
- Ramesh, S.; Govender, T.; Kruger, H.G.; de la Torre, B.G.; Albericio, F. Short AntiMicrobial Peptides (SAMPs) as a class of extraordinary promising therapeutic agents. J. Pept. Sci. 2016, 22, 438–451. [Google Scholar] [CrossRef]
- Ciarambino, T.; Giannico, O.V.; Campanile, A.; Tirelli, P.; Para, O.; Signoriello, G.; Giordano, M. Acute kidney injury and vancomycin/piperacillin/tazobactam in adult patients: A systematic review. Intern. Emerg. Med. 2020, 15, 327–331. [Google Scholar] [CrossRef]
- Ordooei Javan, A.; Shokouhi, S.; Sahraei, Z. A review on colistin nephrotoxicity. Eur. J. Clin. Pharmacol. 2015, 71, 801–810. [Google Scholar] [CrossRef]
- Gan, B.H.; Gaynord, J.; Rowe, S.M.; Deingruber, T.; Spring, D.R. The multifaceted nature of antimicrobial peptides: Current synthetic chemistry approaches and future directions. Chem. Soc. Rev. 2021, 50, 7820–7880. [Google Scholar] [CrossRef]
- Hancock, R.E.; Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Bowdish, D.M.; Davidson, D.J.; Hancock, R.E. A re-evaluation of the role of host defence peptides in mammalian immunity. Curr. Protein Pept. Sci. 2005, 6, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Xiao, Y.H. Pharmacokinetic/pharmacodynamic concepts of antimicrobial agents and their clinical significance. Natl. Med. J. China 2004, 88, 1914–1915. (In Chinese) [Google Scholar]
- Xiao, Y.H. Research and clinical application of antibiotic pharmacodynamics (Ⅰ). Chin. J. Clin. Pharmacol. 2002, 4, 310–316. (In Chinese) [Google Scholar]
- Gu, J.F. Research Progress on Post Antibiotic Effect and Its Clinical Significance. Anti Infect. Pharm. 2016, 13, 481–488. (In Chinese) [Google Scholar]
- Indrakumar, S.; Zalar, M.; Pohl, C.; Nørgaard, A.; Streicher, W.; Harris, P.; Golovanov, A.P.; Peters, G.H.J. Conformational Stability Study of a Therapeutic Peptide Plectasin Using Molecular Dynamics Simulations in Combination with NMR. J. Phys. Chem. B 2019, 123, 4867–4877. [Google Scholar] [CrossRef]
- Umerska, A.; Cassisa, V.; Bastiat, G.; Matougui, N.; Nehme, H.; Manero, F.; Eveillard, M.; Saulnier, P. Synergistic interactions between antimicrobial peptides derived from plectasin and lipid nanocapsules containing monolaurin as a cosurfactant against Staphylococcus aureus. Int. J. Nanomed. 2017, 12, 5687–5699. [Google Scholar] [CrossRef]
- Liu, H.; Yang, N.; Mao, R.; Teng, D.; Hao, Y.; Wang, X.; Wang, J. A new high-yielding antimicrobial peptide NZX and its antibacterial activity against Staphylococcus hyicus in vitro/vivo. Appl. Microbiol. Biotechnol. 2020, 104, 1555–1568. [Google Scholar] [CrossRef]
- Liu, H.; Yang, N.; Teng, D.; Mao, R.; Hao, Y.; Ma, X.; Wang, J. Design and pharmacodynamics of recombinant fungus defensin NZL with improved activity against Staphylococcus hyicus in vitro and in vivo. Int. J. Mol. Sci. 2021, 22, 5435. [Google Scholar] [CrossRef]
- Zhang, Y.; Teng, D.; Wang, X.; Mao, R.; Cao, X.; Hu, X.; Zong, L.; Wang, J. In vitro and in vivo characterization of a new recombinant antimicrobial peptide, MP1102, against methicillin-resistant Staphylococcus aureus. Appl. Microbiol. Biotechnol. 2015, 99, 6255–6266. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Teng, D.; Mao, R.; Wang, X.; Xi, D.; Hu, X.; Wang, J. High expression of a plectasin-derived peptide NZ2114 in Pichia pastoris and its pharmacodynamics, postantibiotic and synergy against Staphylococcus aureus. Appl. Microbiol. Biotechnol. 2014, 98, 681–694. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Mao, R.; Teng, D.; Wang, X.; Hao, Y.; Feng, X.; Wang, J. Design and pharmacodynamics of recombinant NZ2114 histidine mutants with improved activity against methicillin-resistant Staphylococcus aureus. AMB Express 2017, 7, 46. [Google Scholar] [CrossRef] [PubMed]
- Lakshmaiah Narayana, J.; Mishra, B.; Lushnikova, T.; Wu, Q.; Chhonker, Y.S.; Zhang, Y.; Zarena, D.; Salnikov, E.S.; Dang, X.; Wang, F.; et al. Two distinct amphipathic peptide antibiotics with systemic efficacy. Proc. Natl. Acad. Sci. USA 2020, 117, 19446–19454. [Google Scholar] [CrossRef]
- Li, B.; Yang, N.; Shan, Y.; Wang, X.; Hao, Y.; Mao, R.; Teng, D.; Fan, H.; Wang, J. Therapeutic potential of a designed CSαβ peptide ID13 in Staphylococcus aureus-induced endometritis of mice. Appl. Microbiol. Biotechnol. 2020, 104, 6693–6705. [Google Scholar] [CrossRef]
- Flamm, R.K.; Rhomberg, P.R.; Lindley, J.M.; Sweeney, K.; Ellis-Grosse, E.J.; Shortridge, D. Evaluation of the bactericidal activity of fosfomycin in combination with selected antimicrobial comparison agents tested against Gram-negative bacterial strains by using time-kill curves. Antimicrob. Agents Chemother. 2019, 63, e02549-18. [Google Scholar] [CrossRef]
- Eckert, R.; Qi, F.; Yarbrough, D.K.; He, J.; Anderson, M.H.; Shi, W. Adding selectivity to antimicrobial peptides: Rational design of a multidomain peptide against Pseudomonas spp. Antimicrob. Agents Chemother. 2006, 50, 1480–1488. [Google Scholar] [CrossRef]
- Giguère, S.; Lee, E.A.; Guldbech, K.M.; Berghaus, L.J. In vitro synergy, pharmacodynamics, and postantibiotic effect of 11 antimicrobial agents against Rhodococcus equi. Vet. Microbiol. 2012, 160, 207–213. [Google Scholar] [CrossRef]
- Shi, J.; Chen, C.; Wang, D.; Wang, Z.; Liu, Y. The antimicrobial peptide LI14 combats multidrug-resistant bacterial infections. Commun. Biol. 2022, 5, 926. [Google Scholar] [CrossRef]
- Li, L.; Shi, Y.; Cheserek, M.J.; Su, G.; Le, G. Antibacterial activity and dual mechanisms of peptide analog derived from cell-penetrating peptide against Salmonella typhimurium and Streptococcus pyogenes. Appl. Microbiol. Biotechnol. 2013, 97, 1711–1723. [Google Scholar] [CrossRef] [PubMed]


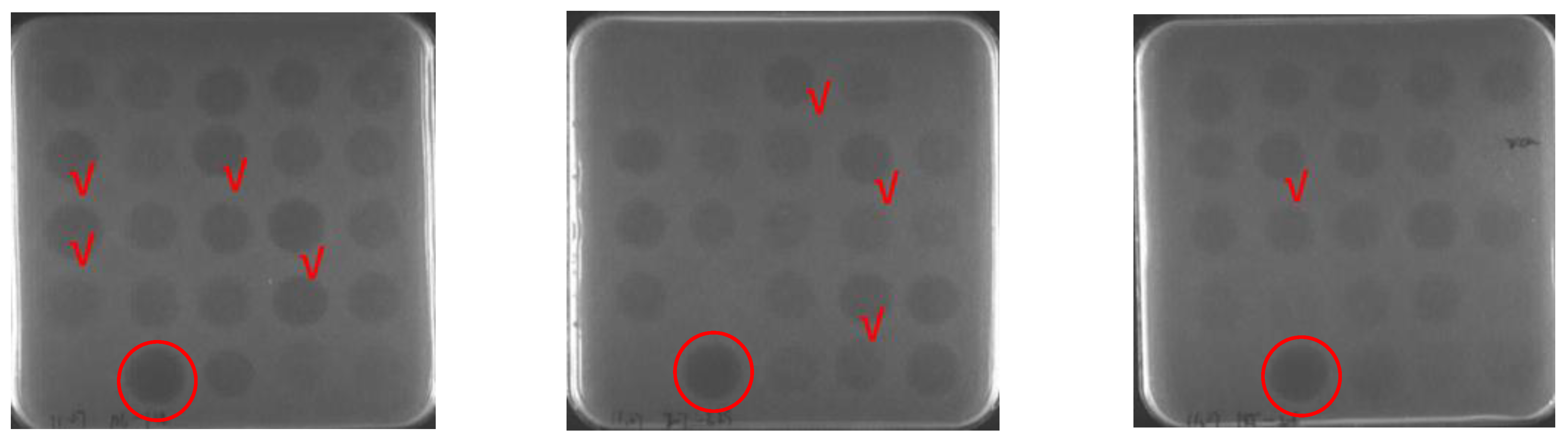
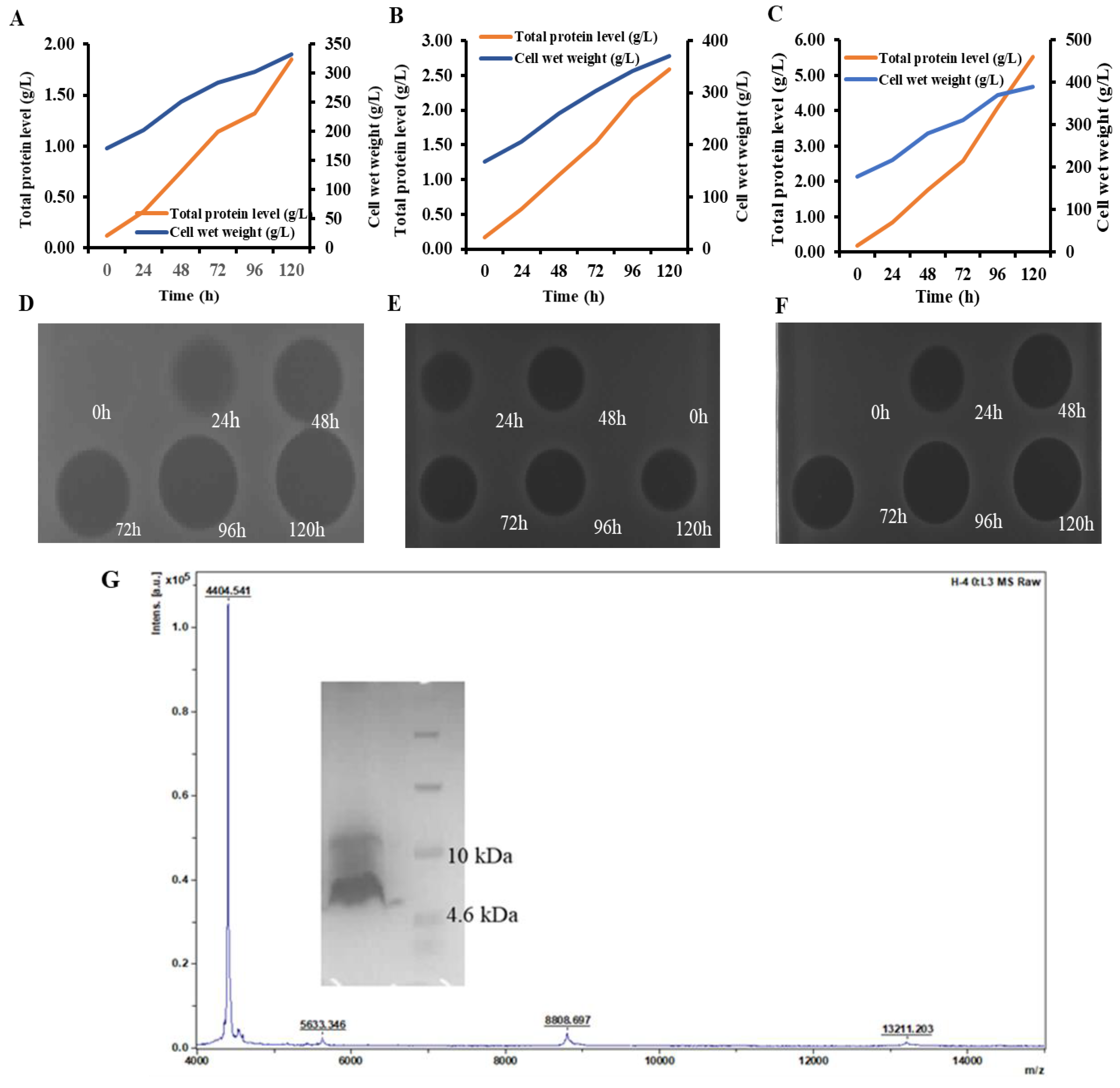
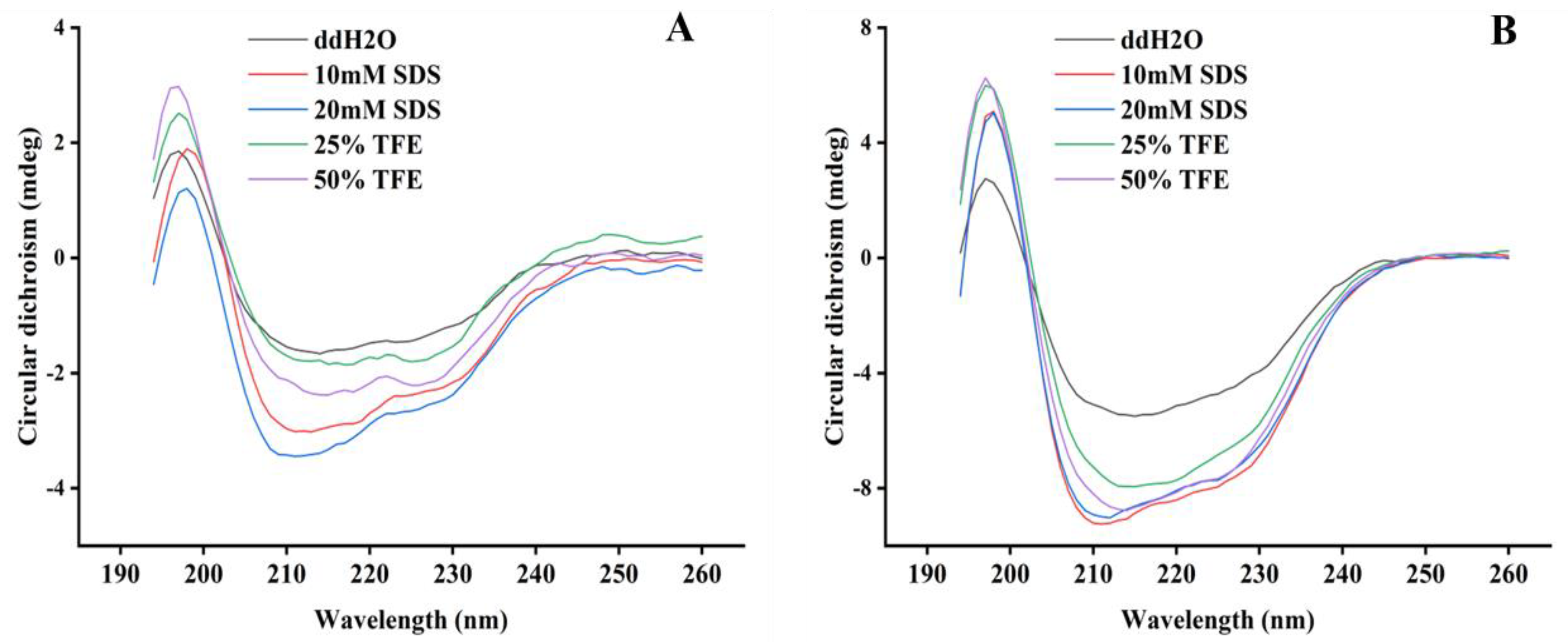

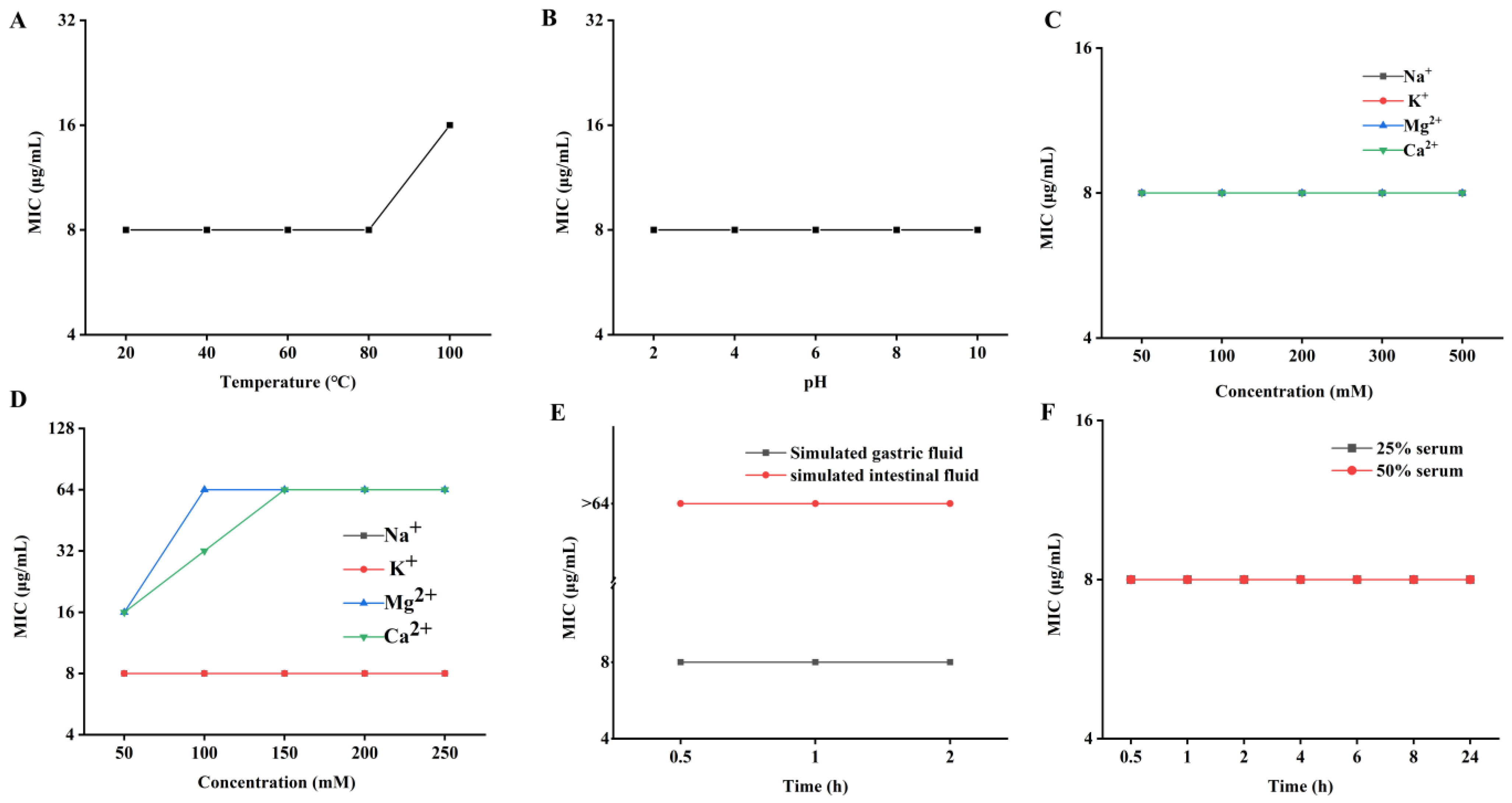

| ID | Sequence |
|---|---|
| PN-(3) | GGT TTT GGT TGT AAC GGT CCA TGG NNN GAA GAT GAT NNN NNN TGT CAT AAC CAT TGT AAG TCT ATT AAG GGT TAC AAG GGT GGT TAC TGT GCT AAG GGT GGT TTT GTT TGT AAG TGT TAC |
| PN-(5) | GGT TTT GGT TGT AAC GGT CCA TGG NNN GAA GAT GAT NNN NNN TGT CAT AAC CAT TGT AAG TCT ATT AAG GGT TAC AAG GGT GGT TAC TGT GCT NNN NNN GGT TTT GTT TGT AAG TGT TAC |
| PN-(7) | GGT TTT GGT TGT NNN GGT CCA TGG NNN GAA GAT GAT NNN NNN TGT CAT NNN CAT TGT AAG NNN ATT AAG GGT TAC AAG GGT GGT TAC TGT GCT AAG GGT GGT TTT NNN TGT AAG TGT TAC |
| Name | M.W. | PI | Charge | GRAVY | Instability Index | Sequences |
|---|---|---|---|---|---|---|
| Plec | 4407.99 | 7.77 | +1 | −0.695 | 13.82 | GFGCNGPWDEDDMQCHNHCKSIKGYKGGYCAKGGFVCKCY |
| PN1 | 4274.87 | 8.30 | +2 | −0.438 | 20.68 | GFGCNGPWLEDDAGCHNHCKSIKGYKGGYCAKGGFVCKCY |
| PN2 | 4347.92 | 8.62 | +3 | −0.708 | 11.71 | GFGCNGPWREDDTGCHNHCKSIKGYKGGYCAKGGFVCKCY |
| PN3 | 4447.06 | 8.86 | +4 | −0.810 | 15.71 | GFGCNGPWREDDRTCHNHCKSIKGYKGGYCAKGGFVCKCY |
| PN4 | 4318.92 | 8.30 | +2 | −0.428 | 28.58 | GFGCNGPWIEDDATCHNHCKSIKGYKGGYCAKGGFVCKCY |
| PN5 | 4347.71 | 8.30 | +2 | −0.568 | 26.75 | GFGCNGPWNEDDVTCHNHCKSIKGYKGGYCAKGGFVCKCY |
| PN6 | 4479.11 | 8.30 | +2 | −0.630 | 19.50 | GFGCNGPWWEDDMQCHNHCKSIKGYKGGYCAKGGFVCKCY |
| PN7 | 4409.99 | 8.61 | +3 | −0.722 | 19.50 | GFGCNGPWYEDDGRCHNHCKSIKGYKGGYCAKGGFVCKCY |
| PN8 | 4332.91 | 8.61 | +3 | −0.762 | 18.56 | GFGCNGPWNEDDGKCHNHCKSIKGYKGGYCAKGGFVCKCY |
| PN9 | 4333.89 | 8.62 | +3 | −0.710 | 19.50 | GFGCNGPWREDDGSCHNHCKSIKGYKGGYCAKGGFVCKCY |
| Peptide | MIC (μg/mL) | |||
|---|---|---|---|---|
| S. aureus 43300 | S. aureus 25923 | S. aureus E48 | S. aureus 546 | |
| PN1 | 8 | 16 | 4 | 8 |
| PN2 | 8 | 16 | 4 | 8 |
| PN3 | 8 | 16 | 8 | 8 |
| PN4 | 16 | 32 | 4 | 8 |
| PN5 | 4 | 16 | 4 | 4 |
| PN6 | 8 | 16 | 8 | 8 |
| PN7 | 4 | 16 | 4 | 4 |
| PN8 | 16 | >32 | 4 | 16 |
| PN9 | 16 | >32 | >16 | >16 |
| Secondary Structure | The Percentage of the Secondary Structure in Different Solvents (%) | ||||
|---|---|---|---|---|---|
| PN7-H2O | PN7-10 mM SDS | PN7-20 mM SDS | PN7-25% TFE | PN7-50% TFE | |
| Helix | 7.63 | 8.43 | 8.71 | 8.04 | 8.76 |
| Antiparallel | 41.39 | 35.91 | 33.03 | 40.10 | 39.17 |
| Parallel | 3.72 | 3.81 | 3.65 | 3.82 | 3.94 |
| β-turn | 17.22 | 18.25 | 19.45 | 17.35 | 17.52 |
| Random coli | 30.04 | 33.60 | 35.16 | 30.69 | 30.51 |
| Secondary Structure | The Percentage of the Secondary Structure in Different Solvents (%) | ||||
|---|---|---|---|---|---|
| Ple-H2O | Ple-10 mM SDS | Ple-20 mM SDS | Ple-25% TFE | Ple-50% TFE | |
| Helix | 15.4 | 17.6 | 17.2 | 19.3 | 19.9 |
| Antiparallel | 29.9 | 25.8 | 27.1 | 24.5 | 22.3 |
| Parallel | 4.7 | 5.1 | 5.0 | 5.5 | 5.6 |
| β-turn | 18.9 | 19.7 | 19.9 | 17.1 | 18.9 |
| Random coli | 31.0 | 32.0 | 31.5 | 32.9 | 32.8 |
| Strains | MIC (μg/mL) | Source | |
|---|---|---|---|
| PN7 | Plectasin | ||
| Gram-positive bacteria | |||
| Staphylococcus aureus ATCC 43300 | 8 | 4 a | ATCC |
| S. aureus ATCC 25923 | 16 | NT | ATCC |
| S. aureus CVCC 546 | 1 | 16 a | CVCC |
| S. aureus E48 | 1 | 4 a | Northwest A&F University |
| S. aureus CICC 10473 | 4 | NT | CICC |
| S. epidermidis ATCC 35984 | 16 | 16 | ATCC |
| S. hyicus 437-2 | 8 | NT | Tianjin Institute of Animal Sciences |
| S. hyicus NCTC 10350 | 4 | NT | NCTC |
| Streptococcus. suis CVCC 606 | 1 | 1 a | CVCC |
| S. agalactiae ATCC 13813 | 2 | 2 | ATCC |
| S. agalactiae CAU-FRI-4 | 1 | NT | Clinical strain (the laboratory) |
| Gram-negative bacteria | |||
| Escherichia coli CVCC 195 | >128 | >128 a | CVCC |
| E. coli CVCC 1515 | >128 | NT | CVCC |
| E. coli O157 | >128 | >128 a | CVCC |
| E. coli ATCC 25922 | >128 | NT | ATCC |
| Salmonella enterica ATCC 13076 | >128 | NT | ATCC |
| S. enteritidis CVCC 3377 | >128 | NT | CVCC |
| S. pullorum CVCC 1789 | >128 | >128 a | CVCC |
| Pseudomonas aeruginosa CICC 20625 | >128 | NT | CICC |
| P. aeruginosa CICC 21630 | >128 | >128 a | CICC |
| Fungus | |||
| Candida albicans CICC 98001 | >128 | >128 | CICC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, Y.; Teng, D.; Mao, R.; Yang, N.; Wang, J. Site Mutation Improves the Expression and Antimicrobial Properties of Fungal Defense. Antibiotics 2023, 12, 1283. https://doi.org/10.3390/antibiotics12081283
Hao Y, Teng D, Mao R, Yang N, Wang J. Site Mutation Improves the Expression and Antimicrobial Properties of Fungal Defense. Antibiotics. 2023; 12(8):1283. https://doi.org/10.3390/antibiotics12081283
Chicago/Turabian StyleHao, Ya, Da Teng, Ruoyu Mao, Na Yang, and Jianhua Wang. 2023. "Site Mutation Improves the Expression and Antimicrobial Properties of Fungal Defense" Antibiotics 12, no. 8: 1283. https://doi.org/10.3390/antibiotics12081283
APA StyleHao, Y., Teng, D., Mao, R., Yang, N., & Wang, J. (2023). Site Mutation Improves the Expression and Antimicrobial Properties of Fungal Defense. Antibiotics, 12(8), 1283. https://doi.org/10.3390/antibiotics12081283










