Combination Therapy with Ciprofloxacin and Pentamidine against Multidrug-Resistant Pseudomonas aeruginosa: Assessment of In Vitro and In Vivo Efficacy and the Role of Resistance–Nodulation–Division (RND) Efflux Pumps
Abstract
1. Introduction
2. Results
2.1. Sensitivity of P. aeruginosa Strains to Ciprofloxacin and Pentamidine
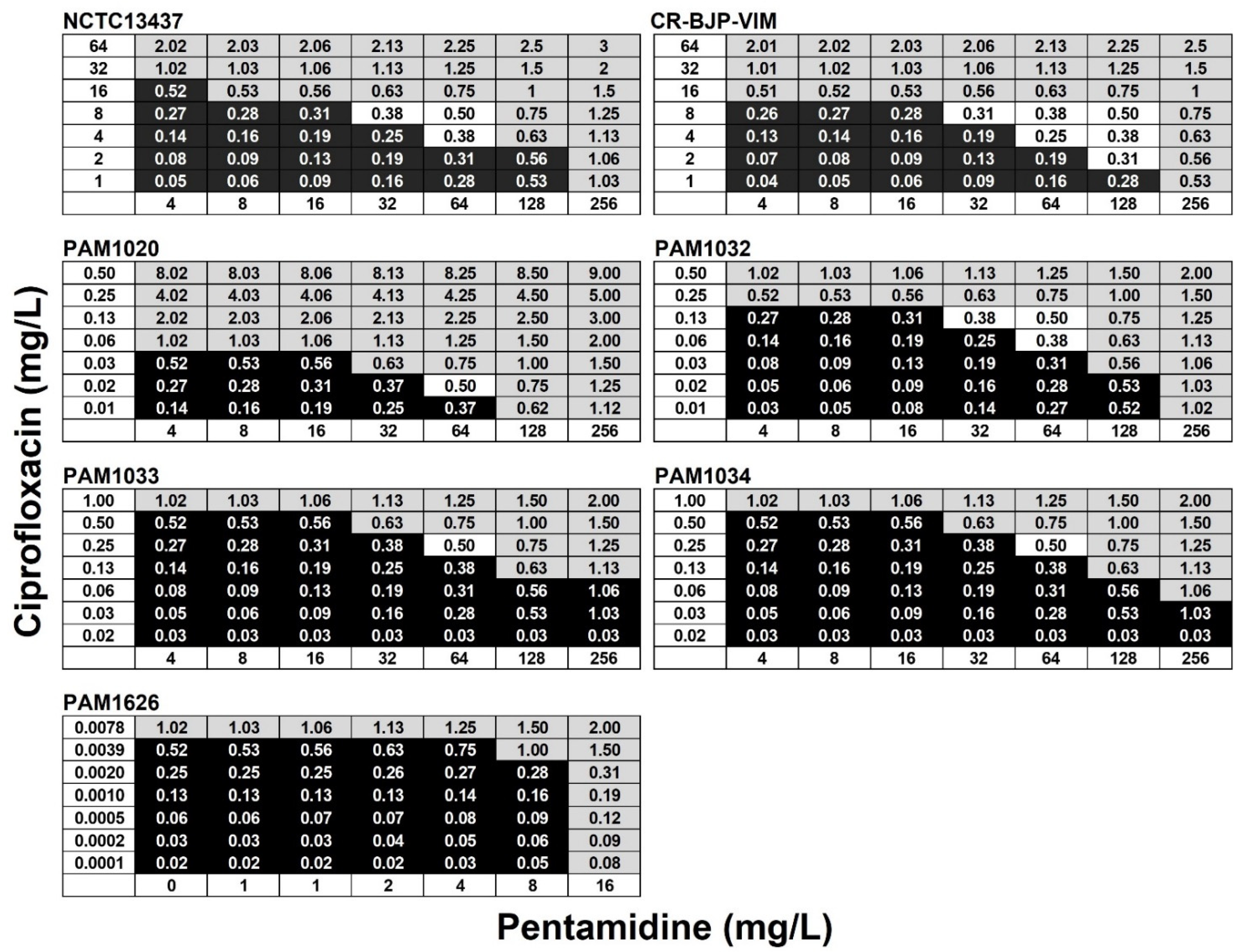
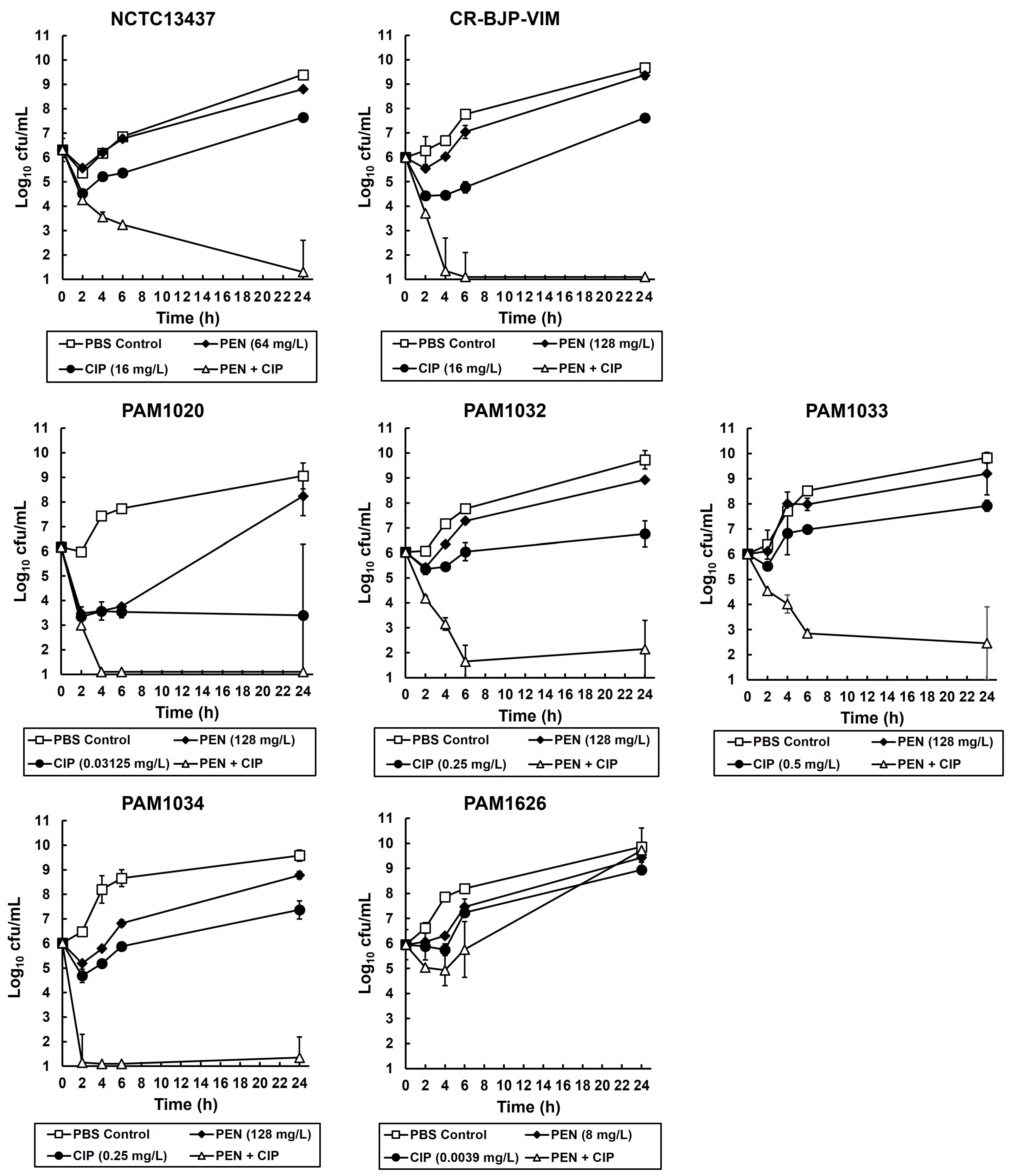
2.2. Exposure of P. aeruginosa Strains to Combinations of Ciprofloxacin and Pentamidine In Vitro Results in Synergistic Bactericidal Inhibition
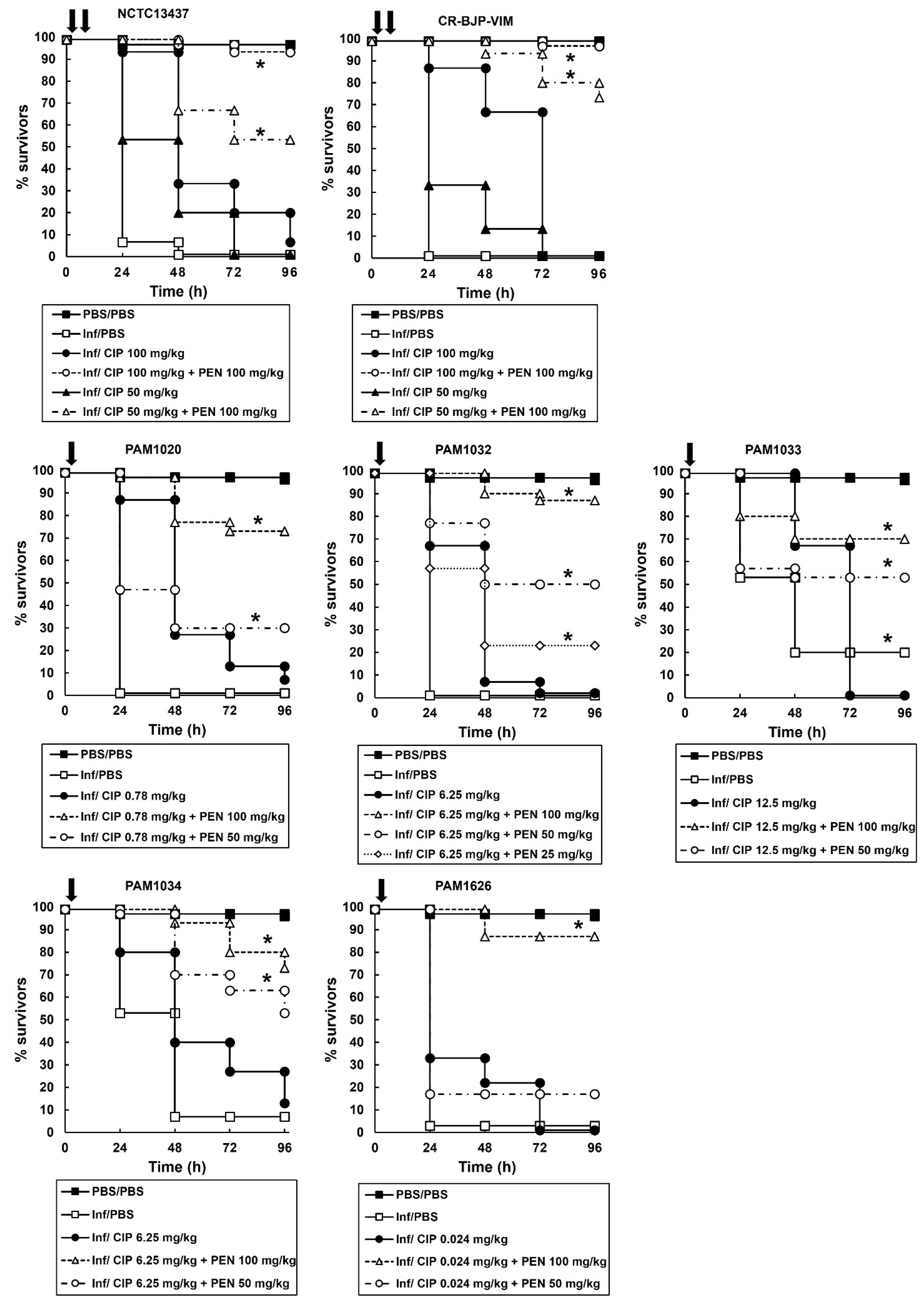
2.3. Survival of G. mellonella Larvae Infected with Different P. aeruginosa Strains and Treated with Ciprofloxacin or Pentamidine Alone Correlates with the In Vitro MIC Values
2.4. Combination Therapy of Ciprofloxacin with Pentamidine of G. mellonella Larvae Infected with P. aeruginosa Results in Enhanced Efficacy Compared to Monotherapies
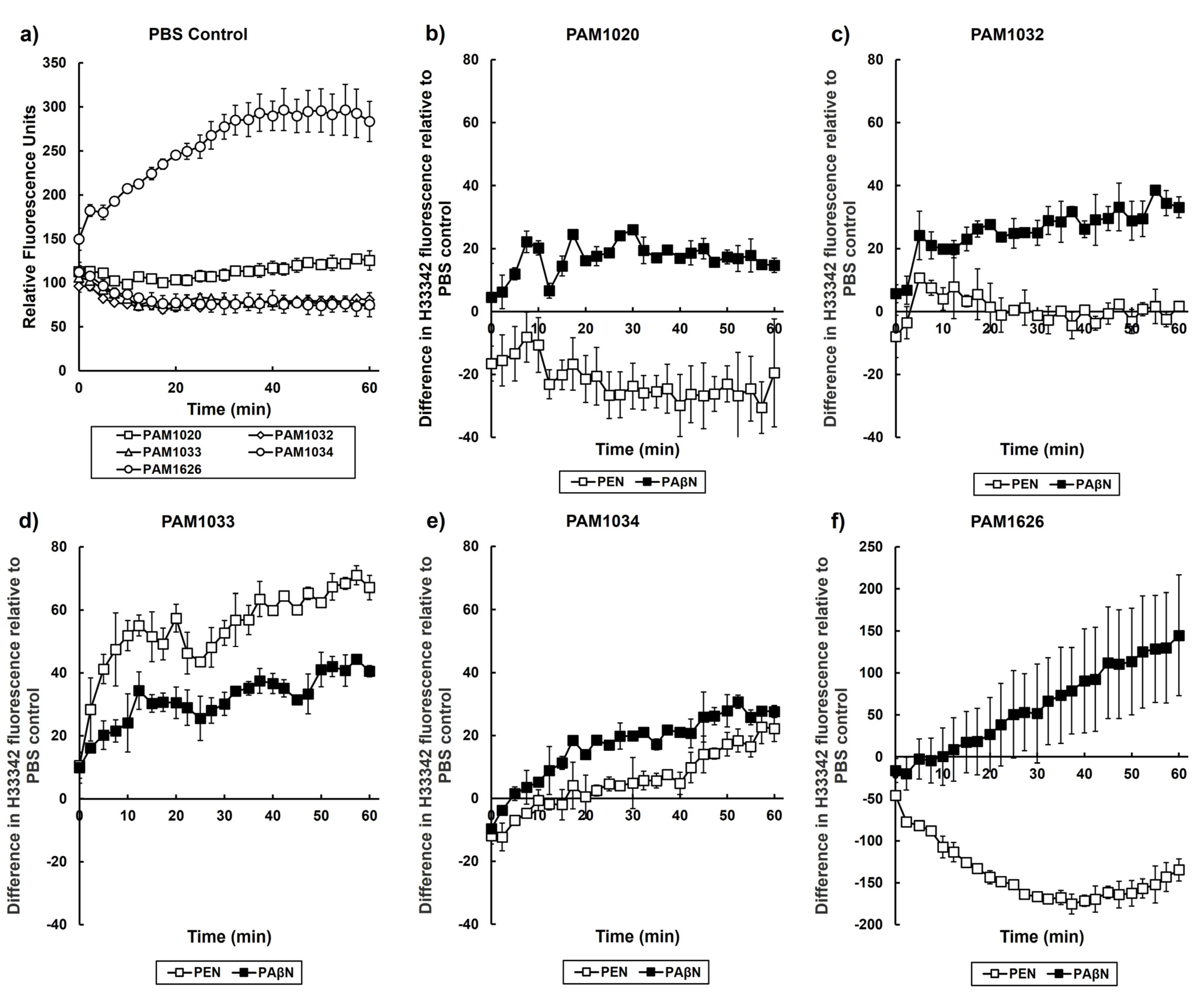
2.5. Exposure to Pentamidine Disrupts the Activity of Specific RND Efflux Pumps in P. aeruginosa
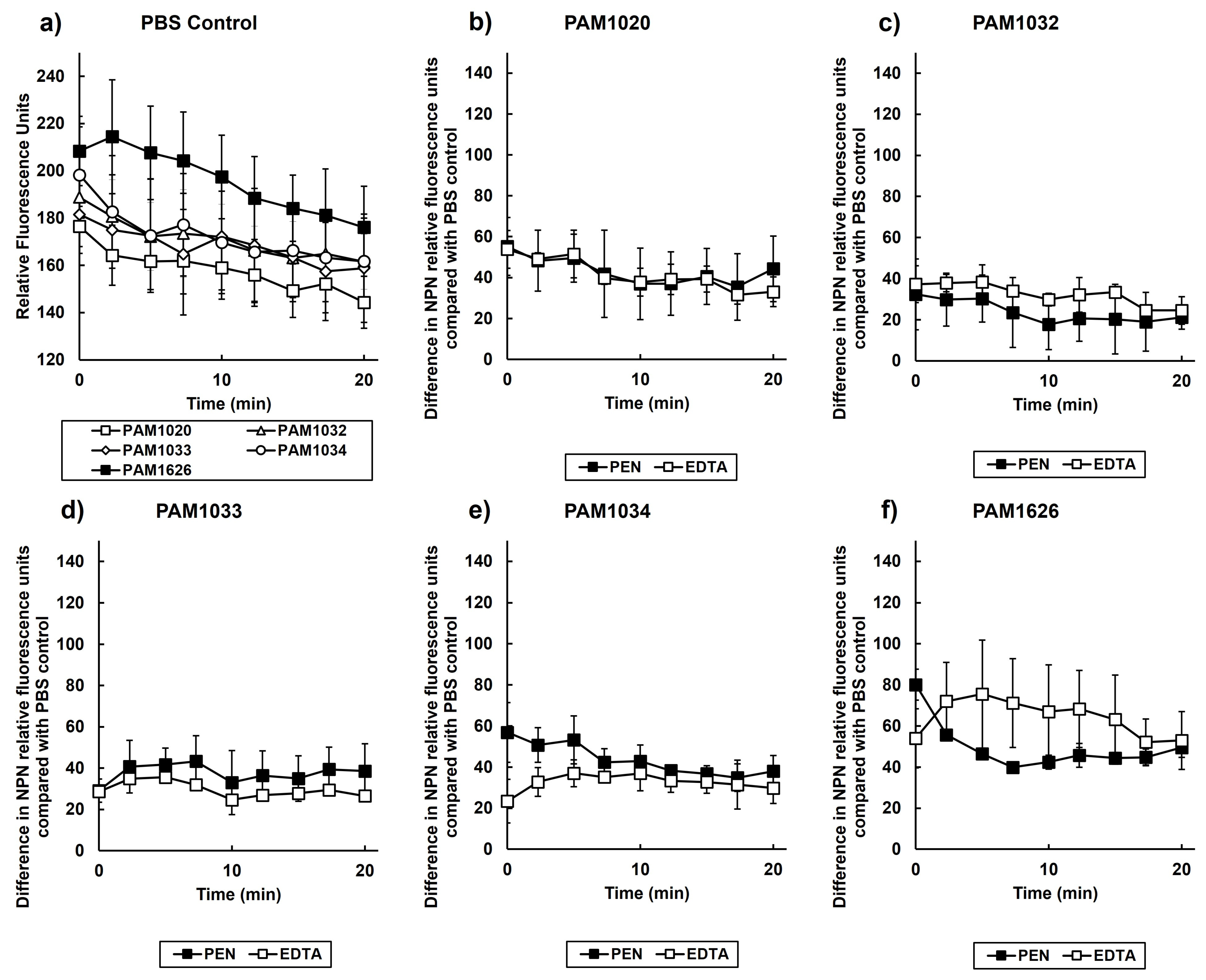
2.6. Exposure to Pentamidine Disrupts the Outer Membrane of P. aeruginosa
3. Discussion
4. Materials and Methods
4.1. Bacteria and Growth Media
4.2. Reagents and G. mellonella Larvae
4.3. Antibiotic Susceptibility and Checkerboard Assay
4.4. Time-Kill Assay
4.5. G. mellonella Infection Model
4.6. H33342 Uptake Assay Measuring Efflux Pump Inhibition
4.7. NPN Fluorescence Assay Measuring Membrane Permeabilization
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shrivastava, S.R.; Shrivastava, P.S.; Ramasamy, J. World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J. Med. Soc. 2018, 32, 76–77. [Google Scholar] [CrossRef]
- UKHSA. Thirty-Day All-Cause Mortality following MRSA, MSSA and Gram- Negative Bacteraemia and C. difficile Infections, 2020 to 2021. 2021. Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1039272/hcai-all-cause-fatality-report-2021.pdf (accessed on 29 June 2023).
- Bassetti, M.; Vena, A.; Croxatto, A.; Righi, E.; Guery, B. How to manage Pseudomonas aeruginosa infections. Drugs Context 2018, 7, 212527. [Google Scholar] [CrossRef] [PubMed]
- Langendonk, R.F.; Neill, D.R.; Fothergill, J.L. The building blocks of antimicrobial resistance in Pseudomonas aeruginosa: Implications for current resistance-breaking therapies. Front. Cell. Infect. Microbiol. 2021, 11, 665759. [Google Scholar] [CrossRef] [PubMed]
- Labovská, S. Pseudomonas aeruginosa as a cause of nosocomial infections. In Pseudomonas aeruginosa-Biofilm Formation, Infections and Treatments; Das, T., Ed.; IntechOpen: London, UK, 2021. [Google Scholar] [CrossRef]
- Weiner, L.M.; Webb, A.K.; Limbago, B.; Dudeck, M.A.; Patel, J.; Kallen, A.J.; Edwards, J.R.; Sievert, D.M. Antimicrobial-resistant pathogens associated with healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect. Control Hosp. Epidemiol. 2016, 37, 1288–1301. [Google Scholar] [CrossRef] [PubMed]
- Lund-Palau, H.; Turnbull, A.R.; Bush, A.; Bardin, E.; Cameron, L.; Soren, O.; Wierre-Gore, N.; Alton, E.W.F.W.; Bundy, J.G.; Connett, G.; et al. Pseudomonas aeruginosa infection in cystic fibrosis: Pathophysiological mechanisms and therapeutic approaches. Exp. Rev. Resp. Med. 2016, 10, 685–697. [Google Scholar] [CrossRef]
- Traugott, K.A.; Echevarria, K.; Maxwell, P.; Green, K.; Lewis, S.J. Monotherapy or combination therapy? The Pseudomonas aeruginosa conundrum. Pharmacotherapy 2011, 31, 598–608. [Google Scholar] [CrossRef]
- Lueangarun, S.; Leelarsamee, A. Impact of inappropriate empiric antimicrobial therapy on mortality of septic patients with bacteraemia: A retrospective study. Interdiscip. Perspect. Infect. Dis. 2012, 2012, 765205. [Google Scholar] [CrossRef]
- Babich, T.; Naucler, P.; Valik, J.K.; Giske, C.G.; Benito, N.; Cardona, R.; Rivera, A.; Pulcini, C.; Fattah, M.A.; Haquin, J.; et al. Combination versus monotherapy as definitive treatment for Pseudomonas aeruginosa bacteraemia: A multicentre retrospective observational cohort study. J. Antimicrob. Chemother. 2021, 76, 2172–2181. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-S.; Williamson, P.R.; Zheng, W. Improving therapy of severe infections through drug repurposing of synergistic combinations. Curr. Opin. Pharmacol. 2019, 48, 92–98. [Google Scholar] [CrossRef]
- Ejim, L.; Farha, M.A.; Falconer, S.B.; Wildenhain, J.; Coombes, B.K.; Tyers, M.; Brown, E.D.; Wright, G.D. Combinations of antibiotics and non-antibiotic drugs enhance antimicrobial efficacy. Nat. Chem. Biol. 2011, 7, 348–350. [Google Scholar] [CrossRef]
- Stokes, J.M.; MacNair, C.R.; Ilyas, B.; French, S.; Côté, J.-P.; Bouwman, C.; Farha, M.A.; Sieron, A.O.; Whitfield, C.; Coombes, B.K.; et al. Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat. Microbiol. 2017, 2, 17028. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Espejo, S.; Cebrero-Cangueiro, T.; Labrador-Herrera, G.; Pachón, J.; Pachón-Ibáńez, M.E.; Álvarez-Marín, R. In vitro activity of pentamidine alone and in combination with antibiotics against multidrug-resistant clinical Pseudomonas aeruginosa strains. Antibiotics 2020, 9, 885. [Google Scholar] [CrossRef] [PubMed]
- MacNair, C.R.; Farha, M.A.; Serrano-Wu, M.H.; Lee, K.K.; Hubbard, B.; Côté, J.P.; Carfrae, L.A.; Tu, M.M.; Gaulin, J.L.; Hunt, D.K.; et al. Preclinical development of pentamidine analogs identifies a potent and nontoxic antibiotic adjuvant. ACS Infect. Dis. 2022, 8, 768–777. [Google Scholar] [CrossRef]
- Venter, H.; Mowla, R.; Ohene-Agyei, T.; Ma, S. RND-type drug efflux pumps from Gram-negative bacteria: Molecular mechanism and inhibition. Front. Microbiol. 2015, 6, 377. [Google Scholar] [CrossRef] [PubMed]
- Poole, K. Pseudomonas aeruginosa: Resistance to the max. Front. Microbiol. 2011, 2, 65. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Nikaido, H.; Poole, K. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1995, 39, 1948–1953. [Google Scholar] [CrossRef] [PubMed]
- Lister, P.; Wolter, D.; Hanson, N. Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Komori, Y.; Mima, T.; Kuroda, T.; Mizushima, T.; Tsuchiya, T. Construction of a series of mutants lacking all of the four major mex operons for multidrug efflux pumps or possessing each one of the operons from Pseudomonas aeruginosa PAO1: MexCD-OprJ is an inducible pump. FEMS Microbiol. Lett. 2001, 202, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Yuan, M.; Livermore, D.M. Mechanisms of resistance to beta-lactam antibiotics amongst Pseudomonas aeruginosa isolates collected in the UK in 1993. J. Med. Microbiol. 1995, 43, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Muratani, T.; Iyobe, S.; Mitsuhashi, S. Mechanisms of high-level resistance to quinolones in urinary tract isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1994, 38, 1466–1469. [Google Scholar] [CrossRef]
- Fukuda, H.; Hosaka, M.; Iyobe, S.; Gotoh, N.; Nishino, T.; Hirai, K. nfxC-type quinolone resistance in a clinical isolate of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1995, 39, 790–792. [Google Scholar] [CrossRef] [PubMed]
- Lomovskaya, O.; Lee, A.; Hoshino, K.; Ishida, H.; Mistry, A.; Warren, M.S.; Boyer, E.; Chamberland, S.; Lee, V.J. Use of a genetic approach to evaluate the consequences of inhibition of efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1999, 43, 1340–1346. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 13.0. 2023. Available online: http://www.eucast.org (accessed on 3 May 2023).
- Woodford, N.; Zhang, J.; Kaufmann, M.E.; Yarde, S.; del Mar Tomas, M.; Faris, C.; Vardhan, M.S.; Dawson, S.; Cotterill, S.L.; Livermore, D.M. Detection of Pseudomonas aeruginosa isolates producing VEB-type extended-spectrum β-lactamases in the United Kingdom. J. Antimicrob. Chemother. 2008, 62, 1265–1268. [Google Scholar] [CrossRef]
- Yasui, L.S.; Chen, K.; Wang, K.; Jones, T.P.; Caldwell, J.; Guse, D.; Kassis, A.I. Using Hoechst 33342 to target radioactivity to the cell nucleus. Radiat. Res. 2007, 167, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Siriyong, T.; Srimanote, P.; Chusri, S.; Yingyongnarongkul, B.-e.; Suaisom, C.; Tipmanee, V.; Voravuthikunchai, S.P. Conessine as a novel inhibitor of multidrug efflux pump systems in Pseudomonas aeruginosa. BMC Complement. Altern. Med. 2017, 17, 405. [Google Scholar] [CrossRef]
- Helander, I.M.; Mattila-Sandholm, T. Fluorometric assessment of Gram-negative bacterial permeabilization. J. Appl. Microbiol. 2000, 88, 213–219. [Google Scholar] [CrossRef]
- Träuble, H.; Overath, P. The structure of Escherichia coli membranes studied by fluorescence measurements of lipid phase transitions. Biochim. Biophys. Acta-Biomembr. 1973, 307, 491–512. [Google Scholar] [CrossRef]
- Prachayasittikul, V.; Isarankura-Na-Ayudhya, C.; Tantimongcolwat, T.; Nantasenamat, C.; Galla, H.-J. EDTA-induced membrane fluidization and destabilization: Biophysical studies on artificial lipid membranes. Acta Biochim. Biophys. Sin. 2007, 39, 901–913. [Google Scholar] [CrossRef]
- Cebrero-Cangueiro, T.; Álvarez-Marín, R.; Labrador-Herrera, G.; Smani, Y.; Cordero-Matίa, E.; Pachón, J.; Pachón-Ibáńez, M.E. In vitro activity of pentamidine alone and in combination with aminoglycosides, tigecycline, rifampicin, and doripenem against clinical strains of carbapenemase-producing and/or colistin-resistant Enterobacteriaceae. Front. Cell. Infect. Microbiol. 2018, 8, 363. [Google Scholar] [CrossRef] [PubMed]
- Sands, M.; Kron, M.A.; Brown, R.B. Pentamidine: A review. Rev. Infect. Dis. 1985, 7, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Patrick, W.M.; Lamont, I.L. Mechanisms of ciprofloxacin resistance in Pseudomonas aeruginosa: New approaches to an old problem. J. Med. Microbiol. 2019, 68, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hill, L.; Veli, N.; Coote, P.J. Evaluation of Galleria mellonella larvae for measuring the efficacy and pharmacokinetics of antibiotic therapies against Pseudomonas aeruginosa infection. Int. J. Antimicrob. Agents 2014, 43, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Eliopoulous, G.; Moellering, R. Antimicrobial combinations. In Antibiotics in Laboratory Medicine 3; Lorian, V., Ed.; Williams and Wilkins: Baltimore, MD, USA, 1996; pp. 330–396. [Google Scholar]
- Bland, J.M.; Altman, D.G. Survival probabilities (the Kaplan-Meier method). Br. Med. J. 1998, 317, 1572. [Google Scholar] [CrossRef] [PubMed]
- Bland, J.M. The logrank test. Br. Med. J. 2004, 328, 1073. [Google Scholar] [CrossRef] [PubMed]
- Krezdorn, J.; Adams, S.; Coote, P.J. A Galleria mellonella infection model reveals double and triple antibiotic combination therapies with enhanced efficacy versus a multidrug-resistant strain of Pseudomonas aeruginosa. J. Med. Microbiol. 2014, 63, 945–955. [Google Scholar] [CrossRef] [PubMed]
- Adamson, D.H.; Krikstopaityte, V.; Coote, P.J. Enhanced efficacy of putative efflux pump inhibitor/antibiotic combination treatments versus MDR strains of Pseudomonas aeruginosa in a Galleria mellonella in vivo infection model. J. Antimicrob. Chemother. 2015, 70, 2271–2278. [Google Scholar] [CrossRef]
- Lamers, R.P.; Cavallari, J.F.; Burrows, L.L. The efflux inhibitor phenylalanine-arginine beta-naphthylamide (PAβN) permeabilizes the outer membrane of gram-negative bacteria. PLoS ONE 2013, 8, e60666. [Google Scholar] [CrossRef]


| Strain | Genotype | Phenotype | Reference |
|---|---|---|---|
| NCTC13437 | Clinical isolate producing VEB-1; VIM-10 β-lactamases | Resistant to β-lactams and fluoroquinolones by an unknown mechanism | [26] |
| CR-BJP-VIM | Clinical isolate producing a VIM β-lactamase | Resistant to β-lactams, aminoglycosides, and fluoroquinolones | Clinical isolate |
| PAM1020 | PA01 prototroph | Wild-type parent strain | [24] |
| PAM1626 | ΔmexAB-oprM::Cm; ΔmexCD-oprJ::Gm; ΔmexEF-oprN::ΩHg | mexAB-oprM; mexCD-oprJ; and mexEF-oprN deleted | [24] |
| PAM1032 | nalB-type mutation | mexAB-oprM overexpressed | [24] |
| PAM1033 | nfxB-type mutation | mexCD-oprJ overexpressed | [24] |
| PAM1034 | nfxC-type mutation | mexEF-oprN overexpressed | [24] |
| MIC (mg/L) | |||
|---|---|---|---|
| Strain | Phenotype | PEN | CIP |
| NCTC13437 | Resistant to β-lactams and fluoroquinolones | 256 | 32 |
| CR-BJP-VIM | Resistant to β-lactams, aminoglycosides, and fluoroquinolones | 256–512 | 32 |
| PAM1020 | Isogenic parent strain of efflux pump mutants | 256 | 0.0625 |
| PAM1032 | Overexpression of MexAB-OprM | 256 | 0.5–1 |
| PAM1033 | Overexpression of MexCD-OprJ | 256–512 | 1 |
| PAM1034 | Overexpression of MexEF-OprN | 256–512 | 1 |
| PAM1626 | Triple deletion of MexAB-OprM, MexCD-OprJ, and MexEF-OprN | 16 | 0.0078 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fletcher, M.; McCormack, A.; Parcell, B.J.; Coote, P.J. Combination Therapy with Ciprofloxacin and Pentamidine against Multidrug-Resistant Pseudomonas aeruginosa: Assessment of In Vitro and In Vivo Efficacy and the Role of Resistance–Nodulation–Division (RND) Efflux Pumps. Antibiotics 2023, 12, 1236. https://doi.org/10.3390/antibiotics12081236
Fletcher M, McCormack A, Parcell BJ, Coote PJ. Combination Therapy with Ciprofloxacin and Pentamidine against Multidrug-Resistant Pseudomonas aeruginosa: Assessment of In Vitro and In Vivo Efficacy and the Role of Resistance–Nodulation–Division (RND) Efflux Pumps. Antibiotics. 2023; 12(8):1236. https://doi.org/10.3390/antibiotics12081236
Chicago/Turabian StyleFletcher, Megan, Alex McCormack, Benjamin J. Parcell, and Peter J. Coote. 2023. "Combination Therapy with Ciprofloxacin and Pentamidine against Multidrug-Resistant Pseudomonas aeruginosa: Assessment of In Vitro and In Vivo Efficacy and the Role of Resistance–Nodulation–Division (RND) Efflux Pumps" Antibiotics 12, no. 8: 1236. https://doi.org/10.3390/antibiotics12081236
APA StyleFletcher, M., McCormack, A., Parcell, B. J., & Coote, P. J. (2023). Combination Therapy with Ciprofloxacin and Pentamidine against Multidrug-Resistant Pseudomonas aeruginosa: Assessment of In Vitro and In Vivo Efficacy and the Role of Resistance–Nodulation–Division (RND) Efflux Pumps. Antibiotics, 12(8), 1236. https://doi.org/10.3390/antibiotics12081236





