QUIRMIA—A Phenotype-Based Algorithm for the Inference of Quinolone Resistance Mechanisms in Escherichia coli
Abstract
1. Introduction
2. Results
2.1. Design of the Quinolone Resistance Mechanisms Inference Algorithm (QUIRMIA)
2.2. EUCAST-Based Classification of Resistance Phenotypes
2.3. WGS-Based Prediction of Resistance Phenotypes
3. Discussion
4. Materials and Methods
4.1. Clinical Isolates
4.2. Antimicrobial Susceptibility Testing
4.3. Whole Genome Sequencing
4.4. Detection of Resistance Genes
4.5. Software
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Drlica, K.; Malik, M. Fluoroquinolones: Action and resistance. Curr. Top. Med. Chem. 2003, 3, 249–282. [Google Scholar] [CrossRef] [PubMed]
- Aly, S.A.; Boothe, D.M.; Suh, S.-J. A novel alanine to serine substitution mutation in SoxS induces overexpression of efflux pumps and contributes to multidrug resistance in clinical Escherichia coli isolates. J. Antimicrob. Chemother. 2015, 70, 2228–2233. [Google Scholar] [CrossRef] [PubMed]
- Hansen, L.H.; Jensen, L.B.; Sørensen, H.I.; Sørensen, S.J. Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J. Antimicrob. Chemother. 2007, 60, 145–147. [Google Scholar] [CrossRef] [PubMed]
- Robicsek, A.; Strahilevitz, J.; Jacoby, G.A.; Macielag, M.; Abbanat, D.; Hye Park, C.; Bush, K.; Hooper, D.C. Fluoroquinolone-modifying enzyme: A new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 2006, 12, 83–88. [Google Scholar] [CrossRef]
- Briales, A.; Rodríguez-Martínez, J.M.; Velasco, C.; Díaz de Alba, P.; Domínguez-Herrera, J.; Pachón, J.; Pascual, A. In vitro effect of qnrA1, qnrB1, and qnrS1 genes on fluoroquinolone activi-ty against isogenic Escherichia coli isolates with mutations in gyrA and parC. Antimicrob. Agents Chemother. 2011, 55, 1266–1269. [Google Scholar] [CrossRef]
- Garoff, L.; Yadav, K.; Hughes, D. Increased expression of Qnr is sufficient to confer clinical resistance to ciprofloxacin in Escherichia coli. J. Antimicrob. Chemother. 2018, 73, 348–352. [Google Scholar] [CrossRef]
- Jiang, X.; Li, J.; Zhang, Y.; Yan, H.; Wang, Y.; Shi, L.; Zhou, L. Detection of plasmid-mediated quinolone resistance determinants and qnrS expression in Enterobacteriaceae clinical isolates. J. Infect. Dev. Ctries 2014, 8, 1625–1629. [Google Scholar] [CrossRef][Green Version]
- Dellgren, L.; Claesson, C.; Högdahl, M.; Forsberg, J.; Hanberger, H.; Nilsson, L.E.; Hällgren, A. Phenotypic screening for quinolone resistance in Escherichia coli. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1765–1771. [Google Scholar] [CrossRef]
- Marcusson, L.L.; Frimodt-Møller, N.; Hughes, D. Interplay in the Selection of Fluoroquinolone Resistance and Bacterial Fitness. PLOS Pathog. 2009, 5, e1000541. [Google Scholar] [CrossRef]
- Huseby, D.L.; Pietsch, F.; Brandis, G.; Garoff, L.; Tegehall, A.; Hughes, D. Mutation supply and relative fitness shape the genotypes of ciprofloxacin-resistant Escherichia coli. Mol. Biol. Evol. 2017, 34, 1029–1039. [Google Scholar] [CrossRef]
- Cavaco, L.M.; Aarestrup, F.M. Evaluation of Quinolones for Use in Detection of Determinants of Acquired Quinolone Resistance, Including the New Transmissible Resistance Mechanisms qnrA, qnrB, qnrS, and aac(6′)-Ib-cr, in Escherichia coli and Salmonella enterica and Determinations of Wild-Type Distributions. J. Clin. Microbiol. 2009, 47, 2751–2758. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Martínez, J.-M.; López-Cerero, L.; Díaz-De-Alba, P.; Chamizo-López, F.J.; Polo-Padillo, J.; Pascual, A. Assessment of a phenotypic algorithm to detect plasmid-mediated quinolone resistance in Enterobacteriaceae. J. Antimicrob. Chemother. 2016, 71, 845–847. [Google Scholar] [CrossRef]
- Strahilevitz, J.; Jacoby, G.A.; Hooper, D.C.; Robicsek, A. Plasmid-Mediated Quinolone Resistance: A Multifaceted Threat. Clin. Microbiol. Rev. 2009, 22, 664–689. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Action Plan on Antimicrobial Resistance; WHO: Geneva, Switzerland, 2015. [Google Scholar]
- Swiss Antibiotic Resistance Report 2018. Available online: https://www.bag.admin.ch/dam/bag/de/dokumente/mt/star/swiss-antibiotic-resistance-report-2018.pdf.download.pdf/swiss-antibiotic-resistance-report-2018.pdf (accessed on 19 May 2019).
- Chao, Y.S.; Farrah, K. Fluoroquinolones for the Treatment of Urinary Tract Infection: A Review of Clinical Effectiveness, Cost-Effectiveness, and Guidelines; Canadian Agency for Drugs and Technologies in Health: Ottawa, ON, Canda, 2019.
- Magri, V.; Stamatiou, K.; Trinchieri, A.; Perletti, G. Commentary: Pharmacological Interventions for Bacterial Prostatitis. Front. Pharmacol. 2020, 11, 573903. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Hooton, T.M.; Naber, K.G.; Wullt, B.; Colgan, R.; Miller, L.G.; Moran, G.J.; Nicolle, L.E.; Raz, R.; Schaeffer, A.J.; et al. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: A 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2011, 52, e103–e120. [Google Scholar] [CrossRef]
- Watkins, R.R.; Lemonovich, T.L. Diagnosis and management of community-acquired pneumonia in adults. Am. Fam. Physician 2011, 83, 1299–1306. [Google Scholar]
- Rafiq, M.; Farag, F.; Manley, K.; Ho, E. Ciprofloxacin: Single versus multiple doses in transrectal ultrasound guided prostate biopsy. Central Eur. J. Urol. 2020, 73, 91–93. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters Version 110. 2021. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_11.0_Breakpoint_Tables.pdf (accessed on 10 June 2021).
- Mancini, S.; Marchesi, M.; Imkamp, F.; Wagner, K.; Keller, P.M.; Quiblier, C.; Bodendoerfer, E.; Courvalin, P.; Böttger, E.C. Population-based inference of aminoglycoside resistance mechanisms in Escherichia coli. Ebiomedicine 2019, 46, 184–192. [Google Scholar] [CrossRef]
- Linde, H.-J.; Notka, F.; Metz, M.; Kochanowski, B.; Heisig, P.; Lehn, N. In Vivo Increase in Resistance to Ciprofloxacin in Escherichia coli Associated with Deletion of the C-Terminal Part of MarR. Antimicrob. Agents Chemother. 2000, 44, 1865–1868. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing. EUCAST Expert Rules v 32 on Enterobacterales. 2020. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Expert_Rules/2020/ExpertRules_V3.2_20190515_Enterobacterales.pdf (accessed on 10 May 2020).
- European Committee on Antimicrobial Susceptibility Testing. ATU–The Area of Technical Uncertainty–Guidance to Laboratories on How to Deal with the ATU in Antimicrobial Susceptibility Testing. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/ATU/Area_of_Technical_Uncertainty_-_guidance_2019.pdf (accessed on 10 May 2020).
- Fisher, H.; Oluboyede, Y.; Chadwick, T.; Abdel-Fattah, M.; Brennand, C.; Fader, M.; Harrison, S.; Hilton, P.; Larcombe, J.; Little, P.; et al. Continuous low-dose antibiotic prophylaxis for adults with repeated urinary tract infections (AnTIC): A randomised, open-label trial. Lancet Infect. Dis. 2018, 18, 957–968. [Google Scholar] [CrossRef]
- Kelesidis, T.; Kelesidis, I.; Rafailidis, P.I.; Falagas, M.E. Counterfeit or substandard antimicrobial drugs: A review of the scientific evidence. J. Antimicrob. Chemother. 2007, 60, 214–236. [Google Scholar] [CrossRef] [PubMed]
- Martín-Gutiérrez, G.; Rodríguez-Beltrán, J.; Rodríguez-Martínez, J.M.; Costas, C.; Aznar, J.; Pascual, Á.; Blázquez, J. Urinary Tract Physiological Conditions Promote Ciprofloxacin Resistance in Low-Level-Quinolone-Resistant Escherichia coli. Antimicrob. Agents Chemother. 2016, 60, 4252–4258. [Google Scholar] [CrossRef] [PubMed]
- Machuca, J.; Ortiz, M.; Recacha, E.; Díaz-De-Alba, P.; Docobo-Perez, F.; Rodríguez-Martínez, J.-M.; Pascual, Á. Impact of AAC(6′)-Ib-cr in combination with chromosomal-mediated mechanisms on clinical quinolone resistance in Escherichia coli. J. Antimicrob. Chemother. 2016, 71, 3066–3071. [Google Scholar] [CrossRef]
- Kraychete, G.B.; Botelho, L.A.B.; Campana, E.H.; Picão, R.C.; Bonelli, R.R. Updated Multiplex PCR for Detection of All Six Plasmid-Mediated qnr Gene Families. Antimicrob. Agents Chemother. 2016, 60, 7524–7526. [Google Scholar] [CrossRef] [PubMed]
- Balloux, F.; Brynildsrud, O.B.; van Dorp, L.; Shaw, L.P.; Chen, H.; Harris, K.A.; Wang, H.; Eldholm, V. From Theory to Practice: Translating Whole-Genome Sequencing (WGS) into the Clinic. Trends Microbiol. 2018, 26, 1035–1048. [Google Scholar] [CrossRef]
- Lautenbach, E.; Strom, B.L.; Bilker, W.B.; Patel, J.B.; Edelstein, P.H.; Fishman, N.O. Epidemiological investigation of fluoroquinolone resistance in infections due to extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Clin. Infect. Dis. 2001, 33, 1288–1294. [Google Scholar] [CrossRef]
- Tayebi, Z.; Heidari, H.; Kazemian, H.; Ghafoori, S.M.; Boroumandi, S.; Houri, H. Comparison of quinolone and beta-lactam resistance among Escherichia coli strains isolated from urinary tract infections. Infez. Med. 2016, 24, 326–330. [Google Scholar]
- Zurfluh, K.; Abgottspon, H.; Hächler, H.; Nüesch-Inderbinen, M.; Stephan, R. Quinolone Resistance Mechanisms among Extended-Spectrum Beta-Lactamase (ESBL) Producing Escherichia coli Isolated from Rivers and Lakes in Switzerland. PLoS ONE 2014, 9, e95864. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing. EUCAST Disk Diffusion Method for Antimicrobial Susceptibility Testing, Version 80; EUCAST: Växjö, Sweden, 2020; Available online: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/_manuals/Manual_v_8.0_EUCAST_Disk_Test_.pdf (accessed on 21 October 2020).
- Hombach, M.; Zbinden, R.; Böttger, E.C. Standardisation of disk diffusion results for antibiotic susceptibility testing using the sirscan automated zone reader. BMC Microbiol. 2013, 13, 225. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing. Routine and Extended Internal Quality Control for MIC Determination and Disk Diffusion as Recommended by EUCAST. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/QC/v_10.0_EUCAST_QC_tables_routine_and_extended_QC.pdf (accessed on 11 May 2023).
- Hunt, M.; Mather, A.E.; Sánchez-Busó, L.; Page, A.J.; Parkhill, J.; Keane, J.A.; Harris, S.R. ARIBA: Rapid antimicrobial resistance genotyping directly from sequencing reads. Microb. Genom. 2017, 3, e000131. [Google Scholar] [CrossRef]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a New Bioinformatic Tool To Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef] [PubMed]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015; Available online: https://www.r-project.org/ (accessed on 2 May 2019).
- Praski Alzrigat, L.; Huseby, D.L.; Brandis, G.; Hughes, D. Fitness cost constrains the spectrum of marR mutations in ciprofloxacin-resistant Escherichia coli. J. Antimicrob. Chemother. 2017, 72, 3016–3024. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Dzink-Fox, J.L.; Chen, M.; Levy, S.B. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: Role of acrR mutations. Antimicrob. Agents Chemother. 2001, 45, 1515–1521. [Google Scholar] [CrossRef] [PubMed]
- Yamane, K.; Wachino, J.I.; Suzuki, S.; Kimura, K.; Shibata, N.; Kato, H.; Shibayama, K.; Konda, T.; Arakawa, Y. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 2007, 51, 3354–3360. [Google Scholar] [CrossRef]
- van der Putten, B.C.; Remondini, D.; Pasquini, G.; Janes, V.A.; Matamoros, S.; Schultsz, C. Quantifying the contribution of four resistance mechanisms to ciprofloxacin MIC in Escherichia coli: A systematic review. J. Antimicrob. Chemother 2019, 74, 298–310. [Google Scholar] [CrossRef]
- Herrera, G.; Aleixandra, V.; Urios, A.; Blanco, M. Quinolone action in Escherichia coli cells carrying gyrA and gyrB mutations. FEMS Microbiol. Lett. 1993, 106, 187–191. [Google Scholar] [CrossRef][Green Version]
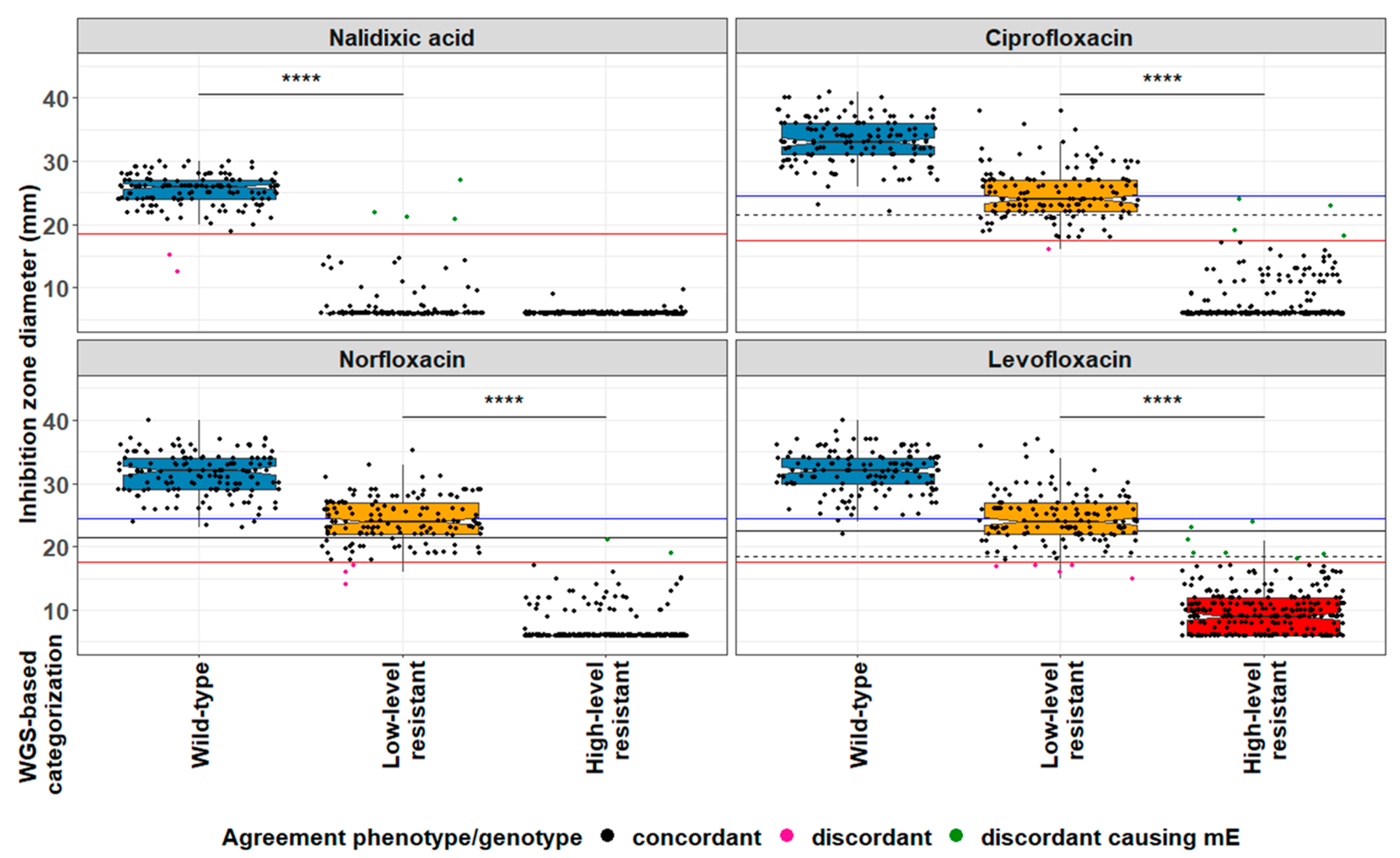
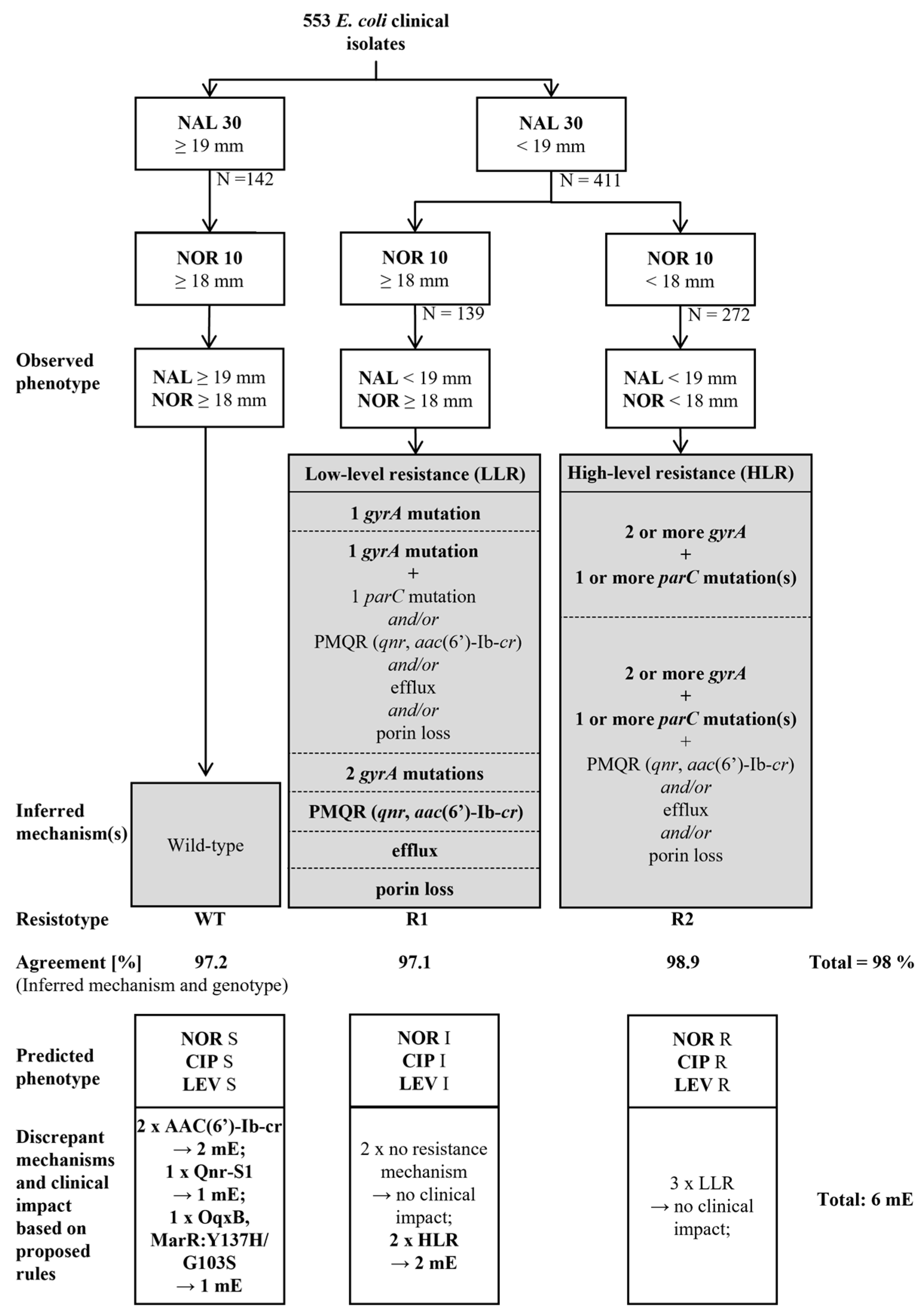
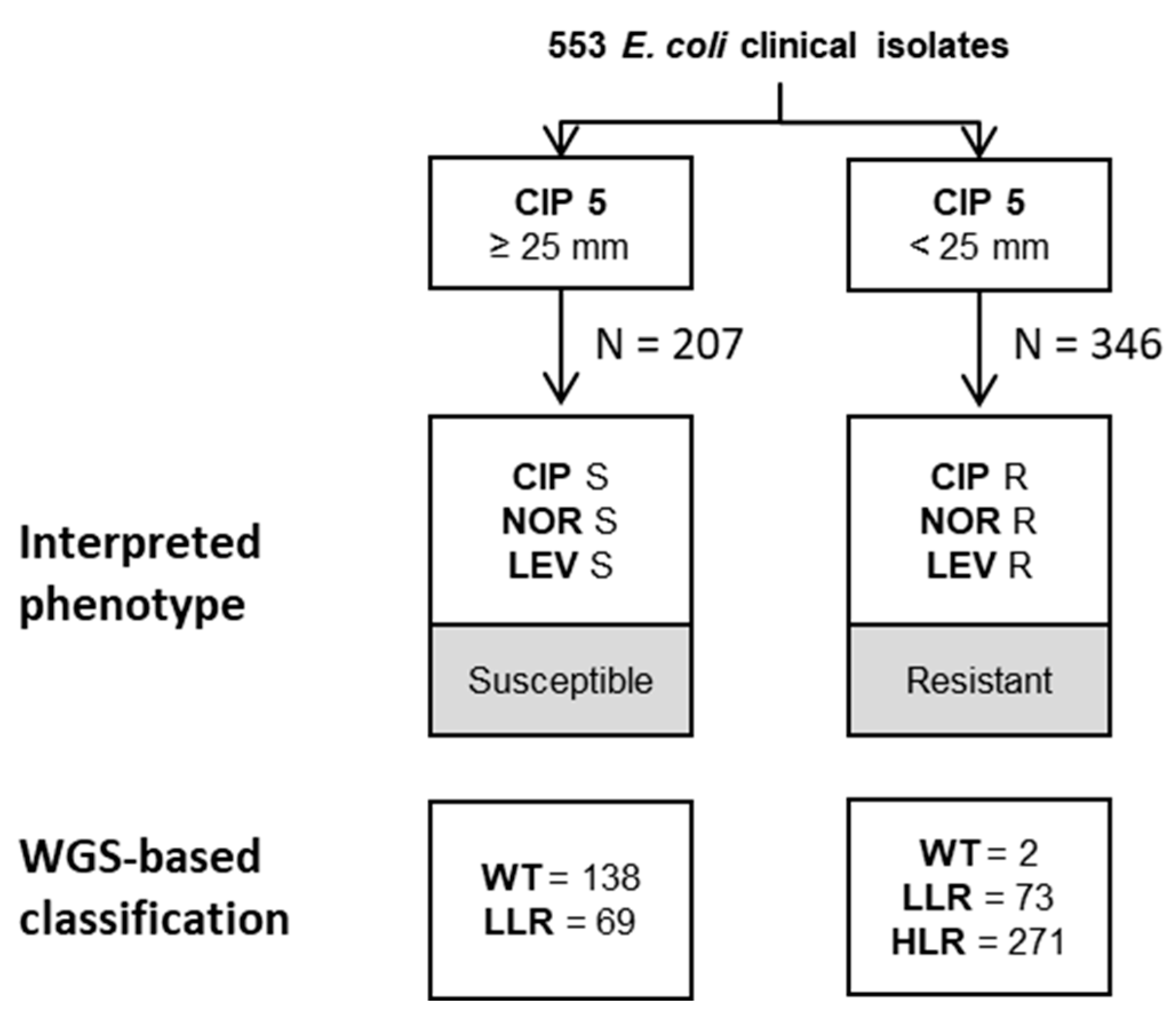
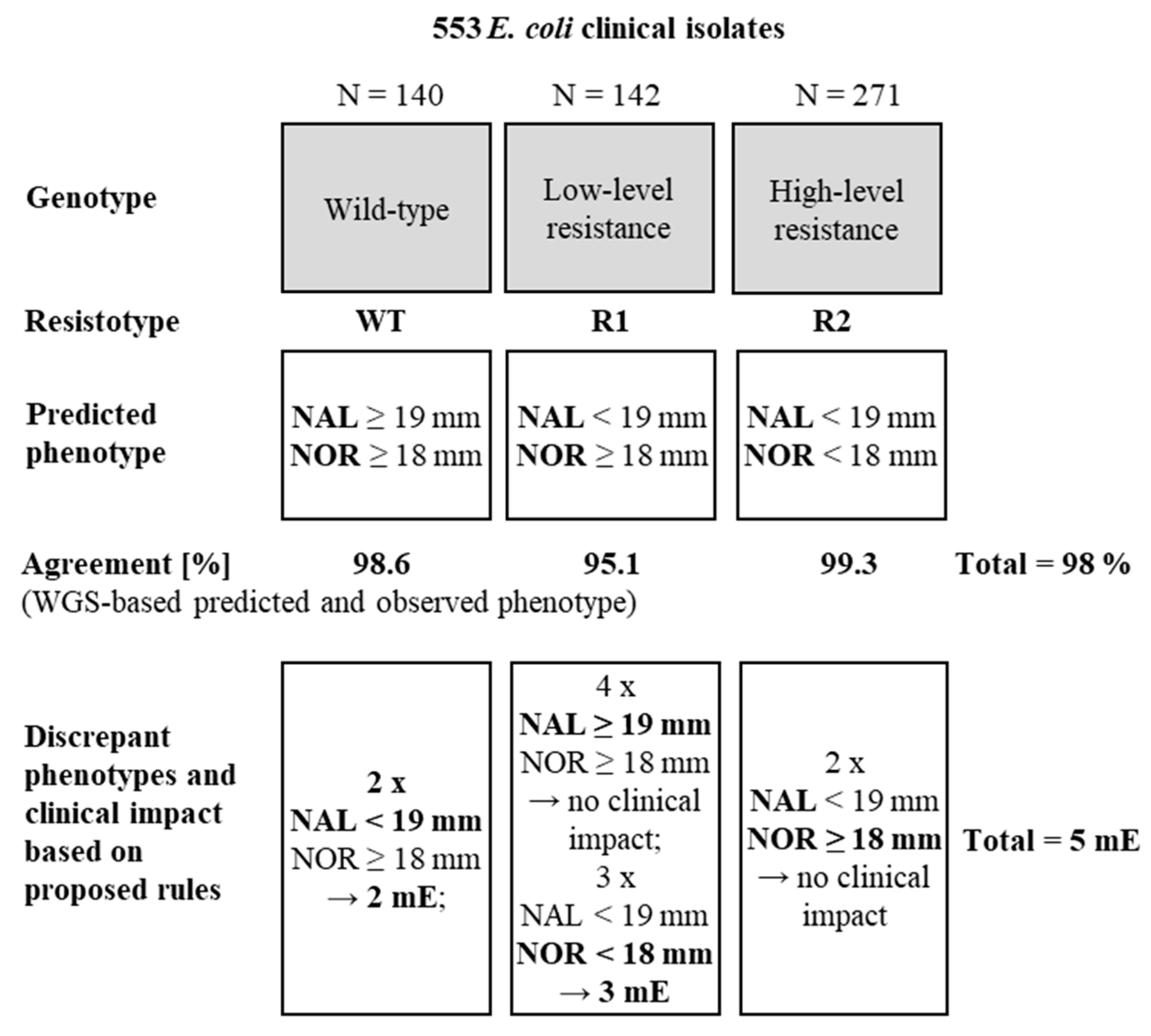
| Algorithm | Cut-Offs | Number of Isolates Analysed | Wildtype | Low Level Resistance | High Level Resistance | Total | Clinical Impact after Applying Internal Rules 1 |
|---|---|---|---|---|---|---|---|
| Agreement Inferred Mechanisms/Genotype | |||||||
| Phenotype-based algorithm | ECOFF (NAL = 19 mm) ECOFF (NOR = 18 mm) | 553 | 138/142 (97.2%) | 135/139 (97.1%) | 269/272 (98.9%) | 542/553 (98%) | 6 mE 2 |
| ECOFF (NAL = 19 mm) ECOFF (CIP = 18 mm) | 138/142 (97.2%) | 137/143 (95.8%) | 267/268 (99.6%) | 542/553 (98%) | 8 mE | ||
| ECOFF (NAL = 19 mm) ECOFF (LEV = 18 mm) | 138/142 (97.2%) | 133/142 (91.7%) | 264/269 (98.1%) | 532/553 (96.2%) | 11 mE | ||
| WGS-based algorithm | ECOFF (NAL = 19 mm) ECOFF (NOR = 18 mm) | Agreement genotype/phenotype | |||||
| 138/140 (98.6%) | 135/142 (95.1%) | 269/271 (99.3%) | 542/553 (98%) | 5 mE 3 | |||
| Resistance Mechanism(s) | Number of Susceptible Isolates (CIP ≥ 25 mm) | Number of Resistant Isolates (CIP < 25 mm) |
|---|---|---|
| 1 gyrA mutation | 52 | 49 |
| 2 gyrA mutations | 1 | 3 |
| 1 gyrA mutation + 1 parC mutation | 2 | 7 |
| 1 gyrA mutation + Qnr-S1 | 1 | 4 |
| 1 gyrA mutation + Qnr-B4 | 2 | - |
| 1 gyrA mutation + 2 marR mutations | 5 | - |
| AAC(6′)-Ib-cr | 2 | 1 |
| Qnr-A1 | - | 1 |
| Qnr-B4 | - | 1 |
| Qnr-B19 | 1 | - |
| Qnr-S1 | 1 | 5 |
| Qnr-S2 | - | 2 |
| Qnr-S1+ 2 marR mutations | 1 | - |
| OqxB + 2 marR mutations | 1 | - |
| Total | 69 | 73 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imkamp, F.; Bodendoerfer, E.; Mancini, S. QUIRMIA—A Phenotype-Based Algorithm for the Inference of Quinolone Resistance Mechanisms in Escherichia coli. Antibiotics 2023, 12, 1119. https://doi.org/10.3390/antibiotics12071119
Imkamp F, Bodendoerfer E, Mancini S. QUIRMIA—A Phenotype-Based Algorithm for the Inference of Quinolone Resistance Mechanisms in Escherichia coli. Antibiotics. 2023; 12(7):1119. https://doi.org/10.3390/antibiotics12071119
Chicago/Turabian StyleImkamp, Frank, Elias Bodendoerfer, and Stefano Mancini. 2023. "QUIRMIA—A Phenotype-Based Algorithm for the Inference of Quinolone Resistance Mechanisms in Escherichia coli" Antibiotics 12, no. 7: 1119. https://doi.org/10.3390/antibiotics12071119
APA StyleImkamp, F., Bodendoerfer, E., & Mancini, S. (2023). QUIRMIA—A Phenotype-Based Algorithm for the Inference of Quinolone Resistance Mechanisms in Escherichia coli. Antibiotics, 12(7), 1119. https://doi.org/10.3390/antibiotics12071119






