Mobile Colistin Resistance (mcr) Gene-Containing Organisms in Poultry Sector in Low- and Middle-Income Countries: Epidemiology, Characteristics, and One Health Control Strategies
Abstract
1. Introduction
2. Causes of Colistin Selection Pressure and Development of mcr-Gene-Carrying Organisms in Poultry Sector in LMICs
3. Sources, Routes of Transmission, and One Health Impact of mcr Gene-Containing Organisms in the Poultry Sector in LMICs
3.1. Breeder Birds, Eggs, and Hatcheries
3.2. Poultry Feeds
3.3. Birds’ Drinking Water
3.4. Contaminated Farm Equipment and Environment
3.5. Vectors
3.5.1. Mammalian Vectors
3.5.2. Flies
3.5.3. Migratory/Free-Range Wild, Urban, and Aquatic Birds
3.6. Poultry Farm Litter/Manure/Slaughterhouse Sewage
3.7. Integrated Poultry–Fish Farms
3.8. Poultry Meat
3.9. Humans in Contact with Poultry Birds, Products, and Environment
Poultry Farm/Slaughterhouse Workers and Their Workwear
3.10. Poultry Bird Vendors
3.11. Persons in Proximity to Poultry Birds/Environments
3.12. Poultry Meat/Egg Handlers
3.13. Trade of Poultry Birds and Products
3.14. Travel
4. Regional and Country-Wise Prevalence and Characteristics of mcr Gene-Containing Organisms in the Poultry Sector in LMICs
4.1. Asia
4.1.1. Eastern Asia
China
4.1.2. Southern Asia
India
Pakistan
Bangladesh
Nepal
4.1.3. Western Asia (Middle East)
Lebanon
Iraq
Türkiye
4.1.4. Southeastern Asia
Lao People’s Democratic Republic
Thailand
Malaysia
Vietnam
Indonesia
4.2. Africa
4.2.1. Northern Africa
Tunisia
Algeria
Egypt
Morocco
4.2.2. Southern Africa
South Africa
Zimbabwe
4.2.3. Western Africa
Nigeria
4.2.4. East Africa
Tanzania
4.3. South America
4.3.1. Northern South America
- Ecuador
4.3.2. Western South America
Peru
4.3.3. Eastern South America
Brazil
4.3.4. Southern South America
- Argentina
4.3.5. Central South America
Paraguay
4.3.6. North America
Dominican Republic
4.3.7. Europe
Romania
Russia
Serbia
5. Control Strategies against the Spread of mcr Gene-Containing Organisms in the Poultry Sector in LMICs
6. Future Perspectives and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Gajdács, M.; Urbán, E.; Stájer, A.; Baráth, Z. Antimicrobial Resistance in the Context of the Sustainable Development Goals: A Brief Review. Eur. J. Investig. Health Psychol. Educ. 2021, 11, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. Rev. Antimicrob. Resist. Available online: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf (accessed on 10 December 2022).
- Erian, I.; Phillips, C.J.C. Public Understanding and Attitudes towards Meat Chicken Production and Relations to Consumption. Animals 2017, 7, 20. [Google Scholar] [CrossRef]
- Li, W.; Bai, X.; Sheng, H.; Chen, J.; Wang, Z.; Wang, T.; Sun, R.; Feng, Z.; Wang, Y.; Peng, K.; et al. Conjugative Transfer of Mcr-1-Bearing Plasmid from Salmonella to Escherichia coli in Vitro on Chicken Meat and in Mouse Gut. Food Res. Int. 2022, 157, 111263. [Google Scholar] [CrossRef] [PubMed]
- Mottet, A.; Tempio, G. Global Poultry Production: Current State and Future Outlook and Challenges. Worlds Poult. Sci. J. 2017, 73, 245–256. [Google Scholar] [CrossRef]
- De Mesquita Souza Saraiva, M.; Lim, K.; do Monte, D.F.M.; Givisiez, P.E.N.; Alves, L.B.R.; de Freitas Neto, O.C.; Kariuki, S.; Júnior, A.B.; de Oliveira, C.J.B.; Gebreyes, W.A. Antimicrobial Resistance in the Globalized Food Chain: A One Health Perspective Applied to the Poultry Industry. Braz. J. Microbiol. 2022, 53, 465–486. [Google Scholar] [CrossRef]
- Umair, M.; Hassan, B.; Farzana, R.; Ali, Q.; Sands, K.; Mathias, J.; Afegbua, S.; Haque, M.N.; Walsh, T.R.; Mohsin, M. International Manufacturing and Trade in Colistin, Its Implications in Colistin Resistance and One Health Global Policies: A Microbiological, Economic, and Anthropological Study. Lancet Microbe 2023, 4, e264–e276. [Google Scholar] [CrossRef]
- Cuong, N.V.; Padungtod, P.; Thwaites, G.; Carrique-Mas, J.J. Antimicrobial Usage in Animal Production: A Review of the Literature with a Focus on Low-and Middle-Income Countries. Antibiotics 2018, 7, 75. [Google Scholar] [CrossRef]
- The World Bank World Bank Country and Lending Groups. Available online: https://datahelpdesk.worldbank.org/knowledgebase/articles/906519-world-bank-country-and-lending-groups (accessed on 10 December 2022).
- Anyanwu, M.U.; Jaja, I.F.; Oguttu, J.W.; Jaja, C.J.; Chah, K.F.; Shodeinde Shoyinka, V. Is Africa Ready for Mobile Colistin Resistance Threat? Infect. Ecol. Epidemiol. 2021, 11, 1962781. [Google Scholar] [CrossRef]
- Njoga, E.O.; Ogugua, A.J.; Nwankwo, I.O.; Awoyomi, O.J.; Okoli, C.E.; Buba, D.M.; Oyeleye, F.A.; Ajibo, F.E.; Azor, N.; Ogunniran, T.M. Antimicrobial Drug Usage Pattern in Poultry Farms in Nigeria: Implications for Food Safety, Public Health and Poultry Disease Management. Vet. Ital. 2021, 57, 5–12. [Google Scholar] [CrossRef]
- Ikhimiukor, O.O.; Odih, E.E.; Donado-Godoy, P.; Okeke, I.N. A Bottom-up View of Antimicrobial Resistance Transmission in Developing Countries. Nat. Microbiol. 2022, 7, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Njoga, E.O.; Onunkwo, J.I.; Okoli, C.E.; Ugwuoke, W.I.; Nwanta, J.A.; Chah, K.F. Assessment of Antimicrobial Drug Administration and Antimicrobial Residues in Food Animals in Enugu State, Nigeria. Trop. Anim. Health Prod. 2018, 50, 897–902. [Google Scholar] [CrossRef] [PubMed]
- Coyne, L.; Arief, R.; Benigno, C.; Giang, V.N.; Huong, L.Q.; Jeamsripong, S.; Kalpravidh, W.; McGrane, J.; Padungtod, P.; Patrick, I.; et al. Characterizing Antimicrobial Use in the Livestock Sector in Three South East Asian Countries (Indonesia, Thailand, and Vietnam). Antibiotics 2019, 8, 33. [Google Scholar] [CrossRef]
- Tiseo, K.; Huber, L.; Gilbert, M.; Robinson, T.P.; Van Boeckel, T.P. Global Trends in Antimicrobial Use in Food Animals from 2017 to 2030. Antibiotics 2020, 9, 918. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Long, X.; Xu, X.; Ren, H.; Mao, D.; Alvarez, P.J.J.; Luo, Y. Global Increase of Antibiotic Resistance Genes in Conjugative Plasmids. Microbiol. Spectr. 2023, 11, e0447822. [Google Scholar] [CrossRef] [PubMed]
- Anyanwu, M.U.; Jaja, I.F.; Okpala, C.O.R.; Jaja, C.J.I.; Oguttu, J.W.; Chah, K.F.; Shoyinka, V.S. Potential Sources and Characteristic Occurrence of Mobile Colistin Resistance (Mcr) Gene-Harbouring Bacteria Recovered from the Poultry Sector: A Literature Synthesis Specific to High-Income Countries. PeerJ 2021, 9, e11606. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, R.; Schwarz, S.; Wu, C.; Shen, J.; Walsh, T.R.; Wang, Y. Farm Animals and Aquaculture: Significant Reservoirs of Mobile Colistin Resistance Genes. Environ. Microbiol. 2020, 22, 2469–2484. [Google Scholar] [CrossRef]
- Mulchandani, R.; Wang, Y.; Gilbert, M.; Van Boeckel, T.P. Global trends in antimicrobial use in food-producing animals: 2020 to 2030. PLOS Global Public Health 2023, 3, e0001305. [Google Scholar] [CrossRef]
- Hamame, A.; Davoust, B.; Cherak, Z.; Rolain, J.M.; Diene, S.M. Mobile Colistin Resistance (Mcr) Genes in Cats and Dogs and Their Zoonotic Transmission Risks. Pathogens 2022, 11, 698. [Google Scholar] [CrossRef]
- Abavisani, M.; Bostanghadiri, N.; Ghahramanpour, H.; Kodori, M.; Akrami, F.; Fathizadeh, H.; Hashemi, A.; Rastegari-Pouyani, M. Colistin Resistance Mechanisms in Gram-Negative Bacteria: A Focus on Escherichia coli. Lett. Appl. Microbiol. 2023, 76, ovad023. [Google Scholar] [CrossRef]
- Anyanwu, M.U.; Jaja, I.F.; Nwobi, O.C. Occurrence and Characteristics of Mobile Colistin Resistance (Mcr) Gene-Containing Isolates from the Environment: A Review. Int. J. Environ. Res. Public Health 2020, 17, 1028. [Google Scholar] [CrossRef]
- Anyanwu, M.U.; Anyogu, D.C.; Chah, K.F.; Shoyinka, V.S. Mobile Colistin Resistance (Mcr-1) Gene-Positive Escherichia coli from Chickens in Nigeria Is Potentially Pathogenic and Transfers Colistin Resistance to Other Organisms. Comp. Clin. Path 2022, 31, 323–332. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Anyanwu, M.U.; Jaja, I.F.; Nwobi, O.C.; Mgbeahuruike, A.C.; Ikpendu, C.N.; Okafor, N.A.; Oguttu, J.W. Epidemiology and Traits of Mobile Colistin Resistance (Mcr) Gene-Bearing Organisms from Horses. Microorganisms 2022, 10, 1499. [Google Scholar] [CrossRef] [PubMed]
- Martiny, H.-M.; Munk, P.; Brinch, C.; Szarvas, J.; Aarestrup, F.M.; Petersen, T.N. Global Distribution of Mcr Gene Variants in 214K Metagenomic Samples. mSystems 2022, 7, e00105-22. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Zeng, F.; Li, R.; Liu, Y.; Wang, Z. Subinhibitory Concentration of Colistin Promotes the Conjugation Frequencies of Mcr-1- and Bla NDM-5 -Positive Plasmids. Microbiol. Spectr. 2022, 10, e02160-21. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Li, W.; Zhao, B.; Xu, H.; Pan, Y.; He, D.; Hu, G.; Wu, H.; Yuan, L. Spreading Advantages of Coresident Plasmids Bla CTX-M-Bearing IncFII and Mcr-1-Bearing IncI2 in Escherichia coli. Antimicrob. Chemother. 2022, 10, e01706-21. [Google Scholar] [CrossRef]
- Anyanwu, M.U.; Okpala, C.O.R.; Chah, K.F.; Shoyinka, V.S. Prevalence and Traits of Mobile Colistin Resistance Gene Harbouring Isolates from Different Ecosystems in Africa. Biomed. Res. Int. 2021, 2021, 6630379. [Google Scholar] [CrossRef]
- Van, T.T.H.; Yidana, Z.; Smooker, P.M.; Coloe, P.J. Antibiotic Use in Food Animals Worldwide, with a Focus on Africa: Pluses and Minuses. J. Glob. Antimicrob. Resist. 2020, 20, 170–177. [Google Scholar] [CrossRef]
- Mead, A.; Richez, P.; Azzariti, S.; Pelligand, L. Pharmacokinetics of Colistin in the Gastrointestinal Tract of Poultry Following Dosing via Drinking Water and Its Bactericidal Impact on Enteric Escherichia coli. Front. Vet. Sci. 2021, 8, 698135. [Google Scholar] [CrossRef]
- Sharafi, T.; Ardebili, A. Plastic Binding Feature of Polymyxins: The Effect on MIC Susceptibility Measurements. Infect. Drug Resist. 2019, 12, 2649–2653. [Google Scholar] [CrossRef] [PubMed]
- Anyanwu, M.U.; Marrollo, R.; Paolucci, M.; Brovarone, F.; Nardini, P.; Chah, K.F.; Shoyinka, S.V.O.; Carretto, E. Isolation and Characterisation of Colistin-Resistant Enterobacterales from Chickens in Southeast Nigeria. J. Glob. Antimicrob. Resist. 2021, 26, 93–100. [Google Scholar] [CrossRef]
- Sia, C.M.; Greig, D.R.; Day, M.; Hartman, H.; Painset, A.; Doumith, M.; Meunier, D.; Jenkins, C.; Chattaway, M.A.; Hopkins, K.L.; et al. The characterization of mobile colistin resistance (mcr) genes among 33000 Salmonella enterica genomes from routine public health surveillance in England. Microb. Genom. 2020, 6, e000331. [Google Scholar] [CrossRef]
- Apostolakos, I.; Piccirillo, A. A Review on the Current Situation and Challenges of Colistin Resistance in Poultry Production. Avian Pathol. 2018, 47, 546–558. [Google Scholar] [CrossRef] [PubMed]
- Coppola, N.; Cordeiro, N.F.; Trenchi, G.; Esposito, F.; Fuga, B.; Fuentes-Castillo, D.; Lincopan, N.; Iriarte, A.; Bado, I.; Vignoli, R. Imported One-Day-Old Chicks as Trojan Horses for Multidrug-Resistant Priority Pathogens Harboring Mcr-9, RmtG, and Extended-Spectrum β-Lactamase Genes. Appl. Environ. Microbiol. 2022, 88, e01675-21. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.T.; Nguyen, H.M.; Nguyen, C.V.; Nguyen, T.v.; Nguyen, M.T.; Thai, H.Q.; Ho, M.H.; Thwaites, G.; Ngo, H.T.; Baker, S.; et al. Use of Colistin and Other Critical Antimicrobials on Pig and Chicken Farms in Southern Vietnam and Its Association with Resistance in Commensal Escherichia coli Bacteria. Appl. Environ. Microbiol. 2016, 82, 3727–3735. [Google Scholar] [CrossRef]
- Ahmed, S.; Das, T.; Islam, M.Z.; Herrero-Fresno, A.; Biswas, P.K.; Olsen, J.E. High Prevalence of Mcr-1-Encoded Colistin Resistance in Commensal Escherichia coli from Broiler Chicken in Bangladesh. Sci. Rep. 2020, 10, 18637. [Google Scholar] [CrossRef]
- Das, T.; Islam, M.Z.; Rana, E.A.; Dutta, A.; Ahmed, S.; Barua, H.; Biswas, P.K. Abundance of Mobilized Colistin Resistance Gene (Mcr-1) in Commensal Escherichia coli from Diverse Sources. Microb. Drug Resist. 2021, 27, 1585–1593. [Google Scholar] [CrossRef]
- Jalal, M.S.; Dutta, A.; Das, T.; Islam, M.Z. First Detection of Plasmid-Mediated Colistin-Resistance Gene (Mcr-1, Mcr-2 and Mcr-3) in Escherichia coli Isolated from Breeder Poultry of Bangladesh. Int. J. Infect. Dis. 2020, 101, 17. [Google Scholar] [CrossRef]
- Gantois, I.; Ducatelle, R.; Pasmans, F.; Haesebrouck, F.; Gast, R.; Humphrey, T.J.; Van Immerseel, F. Mechanisms of Egg Contamination by Salmonella Enteritidis: Review Article. FEMS Microbiol. Rev. 2009, 33, 718–738. [Google Scholar] [CrossRef]
- Hu, Y.; Fanning, S.; Nguyen, S.V.; Wang, W.; Liu, C.; Cui, X.; Dong, Y.; Gan, X.; Xu, J.; Li, F. Emergence of a Salmonella Enterica Serovar Typhimurium ST34 Isolate, CFSA629, Carrying a Novel Mcr-1.19 Variant Cultured from Egg in China. J. Antimicrob. Chemother. 2021, 76, 1776–1785. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Gu, X.; Zhang, L.; Liu, Y.; Li, Y.; Zou, M.; Liu, B. The Occurrence and Genomic Characteristics of Mcr-1-Harboring Salmonella from Retail Meats and Eggs in Qingdao, China. Foods 2022, 11, 3854. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, R.; Li, J.; Wu, Z.; Yin, W.; Schwarz, S.; Tyrrell, J.M.; Zheng, Y.; Wang, S.; Shen, Z.; et al. Comprehensive Resistome Analysis Reveals the Prevalence of NDM and MCR-1 in Chinese Poultry Production. Nat. Microbiol. 2017, 2, 16260. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liu, Z.; Zhang, Y.; Yuan, X.; Hu, M.; Liu, Y. Prevalence and Molecular Characteristics of Avian-Origin Mcr-1-Harboring Escherichia coli in Shandong Province, China. Front. Microbiol. 2020, 11, 255. [Google Scholar] [CrossRef] [PubMed]
- Al-Mir, H.; Osman, M.; Drapeau, A.; Hamze, M.; Madec, J.Y.; Haenni, M. WGS Analysis of Clonal and Plasmidic Epidemiology of Colistin-Resistance Mediated by Mcr Genes in the Poultry Sector in Lebanon. Front. Microbiol. 2021, 12, 624194. [Google Scholar] [CrossRef]
- Nesporova, K.; Jamborova, I.; Valcek, A.; Medvecky, M.; Literak, I.; Dolejska, M. Various Conjugative Plasmids Carrying the Mcr-5 Gene in Escherichia coli Isolates from Healthy Chickens in Paraguay. J. Antimicrob. Chemother. 2019, 74, 3394–3397. [Google Scholar] [CrossRef]
- Wang, R.; Liu, Y.; Zhang, Q.; Jin, L.; Wang, Q.; Zhang, Y.; Wang, X.; Hu, M.; Li, L.; Qi, J. The Prevalence of Colistin Resistance in Escherichia coli and Klebsiella pneumoniae Isolated from Food Animals in China: Coexistence of Mcr-1 and BlaNDM with Low Fitness Cost. Int. J. Antimicrob. Agents 2018, 51, 739–744. [Google Scholar] [CrossRef]
- Shen, Y.; Lv, Z.; Yang, L.; Liu, D.; Ou, Y.; Xu, C.; Liu, W.; Yuan, D.; Hao, Y.; He, J.; et al. Integrated Aquaculture Contributes to the Transfer of Mcr-1 between Animals and Humans via the Aquaculture Supply Chain. Environ. Int. 2019, 130, 104708. [Google Scholar] [CrossRef]
- Yang, X.; Peng, K.; Zhang, Y.; Liu, L.; Li, R. Characterization of a Novel Mcr-8.2-Bearing Plasmid in ST395 Klebsiella pneumoniae of Chicken Origin. Infect. Drug Resist. 2020, 13, 1781–1784. [Google Scholar] [CrossRef]
- Yang, Y.Q.; Li, Y.X.; Lei, C.W.; Zhang, A.Y.; Wang, H.N. Novel Plasmid-Mediated Colistin Resistance Gene Mcr-7.1 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2018, 73, 1791–1795. [Google Scholar] [CrossRef]
- Ahmed, S.; Das, T.; Islam, M.; Herrero-Fresno, A.; Biswas, P.; Olsen, J. Abundance of Mobilized Colistin Resistance Gene Mcr-1 in Genetically Diverse Commensal Escherichia coli in Broiler Chicken in Bangladesh. Int. J. Infect. Dis. 2020, 101, 77–78. [Google Scholar] [CrossRef]
- Yu, C.Y.; Ang, G.Y.; Chin, P.S.; Ngeow, Y.F.; Yin, W.F.; Chan, K.G. Emergence of Mcr-1-Mediated Colistin Resistance in Escherichia coli in Malaysia. Int. J. Antimicrob. Agents 2016, 47, 504–505. [Google Scholar] [CrossRef] [PubMed]
- Palupi, M.F.; Wibawan, I.W.T.; Sudarnika, E.; Maheshwari, H.; Darusman, H.S. Prevalence of Mcr-1 Colistin Resistance Gene in Escherichia coli along 19 Broiler Meat Supply Chain in Indonesia. Biotropia 2019, 26, 143–153. [Google Scholar] [CrossRef]
- Bich, V.T.N.; Thanh, L.V.; Thai, P.D.; Van Phuong, T.T.; Oomen, M.; Driessen, C.; Beuken, E.; Hoang, T.H.; Van Doorn, H.R.; Penders, J.; et al. An Exploration of the Gut and Environmental Resistome in a Community in Northern Vietnam in Relation to Antibiotic Use. Antimicrob. Resist. Infect. Control 2019, 8, 194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, L.; Wang, J.; Yassin, A.K.; Butaye, P.; Kelly, P.; Gong, J.; Guo, W.; Li, J.; Li, M.; et al. Molecular Detection of Colistin Resistance Genes (Mcr-1, Mcr-2 and Mcr-3) in Nasal/Oropharyngeal and Anal/Cloacal Swabs from Pigs and Poultry. Sci. Rep. 2018, 8, 3705. [Google Scholar] [CrossRef]
- Shi, X.; Li, Y.; Yang, Y.; Shen, Z.; Cai, C.; Wang, Y.; Walsh, T.R.; Shen, J.; Wu, Y.; Wang, S. High Prevalence and Persistence of Carbapenem and Colistin Resistance in Livestock Farm Environments in China. J. Hazard. Mater. 2021, 417, 125951, Corrigendum to J. Hazard. Mater. 2021, 406, 124298. [Google Scholar] [CrossRef]
- Ding, S.; Han, X.; Li, J.; Gao, W.; Chen, Z.; Feng, Y. Discovery of Multi-Drug Resistant, MCR-1 and ESBL-Coproducing ST117 Escherichia coli from Diseased Chickens in Northeast China. Sci. Bull. 2018, 63, 1059–1066. [Google Scholar] [CrossRef]
- Liu, X.; Li, R.; Dong, N.; Ye, L.; Chan, E.W.-C.; Chen, S. Complete Genetic Analysis of Plasmids Carried by Two Nonclonal Bla NDM-5—And Mcr-1 -Bearing Escherichia coli Strains: Insight into Plasmid Transmission among Foodborne Bacteria. Microbiol. Spectr. 2021, 9, e00217-21. [Google Scholar] [CrossRef]
- Song, Y.; Yu, L.; Zhang, Y.; Dai, Y.; Wang, P.; Feng, C.; Liu, M.; Sun, S.; Xie, Z.; Wang, F. Prevalence and Characteristics of Multidrug-Resistant Mcr-1-Positive Escherichia coli Isolates from Broiler Chickens in Tai’an, China. Poult. Sci. 2020, 99, 1117–1123. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Y.; Qi, X.; Wang, R.; Jin, L.; Zhao, M.; Zhang, Y.; Wang, Q.; Chen, H.; Wang, H. Molecular Epidemiology of Colistin-Resistant Enterobacteriaceae in Inpatient and Avian Isolates from China: High Prevalence of Mcr-Negative Klebsiella pneumoniae. Int. J. Antimicrob. Agents 2017, 50, 536–541. [Google Scholar] [CrossRef]
- Zhuge, X.; Ji, Y.; Tang, F.; Sun, Y.; Jiang, M.; Hu, W.; Wu, Y.; Xue, F.; Ren, J.; Zhu, W.; et al. Population Structure and Antimicrobial Resistance Traits of Avian-Origin Mcr-1-Positive Escherichia coli in Eastern China, 2015 to 2017. Transbound. Emerg. Dis. 2019, 66, 1920–1929. [Google Scholar] [CrossRef]
- Xiang, Y.; Liu, Z.; Yu, G.; Song, Y.; Li, Y.; Geng, X.; Ma, L.; Guo, J.; Tan, L.; Chen, P. Genetic characteristic of coexisting of mcr-1 and blaNDM-5 in Escherichia coli isolates from lesion bearing animal organs. Front. Microbiol. 2023, 14, 1116413. [Google Scholar] [CrossRef]
- Amin, M.B.; Sraboni, A.S.; Hossain, M.I.; Roy, S.; Mozmader, T.A.U.; Unicomb, L.; Rousham, E.K.; Islam, M.A. Occurrence and Genetic Characteristics of Mcr-1-Positive Colistin-Resistant E. coli from Poultry Environments in Bangladesh. J. Glob. Antimicrob. Resist. 2020, 22, 546–552. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.; Barua, H.; Jalal, M.S.; Dhar, P.K.; Biswas, S.K.; Biswas, P.K. An Investigation of Plasmid-Mediated Colistin Resistance Mechanism, MCR in Escherichia coli of Human, Veterinary and Environmental Origin in Bangladesh. Int. Journal. Infect. Dis. 2018, 73, 54. [Google Scholar] [CrossRef]
- Usman, M.; Rasool, M.H.; Khurshid, M.; Aslam, B.; Baloch, Z. Co-Occurrence of Mcr-1 and Carbapenem Resistance in Avian Pathogenic E. coli Serogroup O78 ST95 from Cololibacillosis-Infected Broiler Chickens. Antibiotics 2023, 12, 812. [Google Scholar] [CrossRef]
- Petrillo, M.; Angers-Loustau, A.; Kreysa, J. Possible Genetic Events Producing Colistin Resistance Gene mcr-1. Lancet Infect. Dis. 2016, 16, 280. [Google Scholar] [CrossRef]
- Joshi, P.R.; Thummeepak, R.; Paudel, S.; Acharya, M.; Pradhan, S.; Banjara, M.R.; Leungtongkam, U.; Sitthisak, S. Molecular Characterization of Colistin-Resistant Escherichia coli Isolated from Chickens: First Report from Nepal. Microb. Drug Resist. 2019, 25, 846–854. [Google Scholar] [CrossRef]
- Bista, S.; Shrestha, U.T.; Dhungel, B.; Koirala, P.; Gompo, T.R.; Shrestha, N.; Adhikari, N.; Joshi, D.R.; Banjara, M.R.; Adhikari, B.; et al. Detection of Plasmid-Mediated Colistin Resistant Mcr-1 Gene in Escherichia coli Isolated from Infected Chicken Livers in Nepal. Animals 2020, 10, 2060. [Google Scholar] [CrossRef] [PubMed]
- Muktan, B.; Thapa Shrestha, U.; Dhungel, B.; Mishra, B.C.; Shrestha, N.; Adhikari, N.; Banjara, M.R.; Adhikari, B.; Rijal, K.R.; Ghimire, P. Plasmid Mediated Colistin Resistant Mcr-1 and Co-Existence of OXA-48 among Escherichia coli from Clinical and Poultry Isolates: First Report from Nepal. Gut Pathog. 2020, 12, 44. [Google Scholar] [CrossRef] [PubMed]
- Dhaouadi, S.; Soufi, L.; Hamza, A.; Fedida, D.; Zied, C.; Awadhi, E.; Mtibaa, M.; Hassen, B.; Cherif, A.; Torres, C.; et al. Co-Occurrence of Mcr-1 Mediated Colistin Resistance and β-Lactamase-Encoding Genes in Multidrug-Resistant Escherichia coli from Broiler Chickens with Colibacillosis in Tunisia. J. Glob. Antimicrob. Resist. 2020, 22, 538–545. [Google Scholar] [CrossRef]
- Grami, R.; Mansour, W.; Mehri, W.; Bouallègue, O.; Boujaâfar, N.; Madec, J.; Haenni, M. Impact of Food Animal Trade on the Spread of Mcr-1-Mediated Colistin Resistance, Tunisia, July. Eurosurveillance 2016, 21, 30144. [Google Scholar] [CrossRef] [PubMed]
- Hassen, B.; Abbassi, M.S.; Ruiz-Ripa, L.; Mama, O.M.; Hassen, A.; Torres, C.; Hammami, S. High Prevalence of Mcr-1 Encoding Colistin Resistance and First Identification of BlaCTX-M-55 in ESBL/CMY-2-Producing Escherichia coli Isolated from Chicken Faeces and Retail Meat in Tunisia. Int. J. Food Microbiol. 2020, 318, 108478. [Google Scholar] [CrossRef] [PubMed]
- Saidani, M.; Messadi, L.; Chaouechi, A.; Tabib, I.; Saras, E.; Soudani, A.; Daaloul-Jedidi, M.; Mamlouk, A.; ben Chehida, F.; Chakroun, C.; et al. High Genetic Diversity of Enterobacteriaceae Clones and Plasmids Disseminating Resistance to Extended-Spectrum Cephalosporins and Colistin in Healthy Chicken in Tunisia. Microb. Drug Resist. 2019, 25, 195. [Google Scholar] [CrossRef]
- Lima Barbieri, N.; Nielsen, D.W.; Wannemuehler, Y.; Cavender, T.; Hussein, A.; Yan, S.; Nolan, L.K.; Logue, C.M. Mcr-1 Identified in Avian Pathogenic Escherichia coli (APEC). PLoS ONE 2017, 12, e0172997. [Google Scholar] [CrossRef]
- Elmonir, W.; Abd El-Aziz, N.K.; Tartor, Y.H.; Moustafa, S.M.; Abo Remela, E.M.; Eissa, R.; Saad, H.A.; Tawab, A.A. Emergence of Colistin and Carbapenem Resistance in Extended-Spectrum β-Lactamase Producing Klebsiella pneumoniae Isolated from Chickens and Humans in Egypt. Biology 2021, 10, 373. [Google Scholar] [CrossRef] [PubMed]
- Sadek, M.; Ortiz de la Rosa, J.M.; Abdelfattah Maky, M.; Korashe Dandrawy, M.; Nordmann, P.; Poirel, L. Genomic Features of MCR-1 and Extended-Spectrum β-Lactamase-Producing Enterobacterales from Retail Raw Chicken in Egypt. Microorganisms 2021, 9, 195. [Google Scholar] [CrossRef]
- Soliman, A.M.; Ramadan, H.; Zarad, H.; Sugawara, Y.; Yu, L.; Sugai, M.; Shimamoto, T.; Hiott, L.M.; Frye, J.G.; Jackson, C.R.; et al. Coproduction of Tet(X7) Conferring High-Level Tigecycline Resistance, Fosfomycin FosA4, and Colistin Mcr-1.1 in Escherichia coli Strains from Chickens in Egypt. Antimicrob. Agents Chemother. 2021, 65, e02084-20. [Google Scholar] [CrossRef] [PubMed]
- Badr, H.; Samir, A.; El-Tokhi, E.I.; Shahein, M.A.; Rady, F.M.; Hakim, A.S.; Fouad, E.A.; El-Sady, E.F.; Ali, S.F. Phenotypic and Genotypic Screening of Colistin Resistance Associated with Emerging Pathogenic Escherichia coli Isolated from Poultry. Vet. Sci. 2022, 9, 282. [Google Scholar] [CrossRef] [PubMed]
- Rahmatallah, N.; El Rhaffouli, H.; Laraqui, A.; Sekhsokh, Y.; Lahlou-Amine, I.; El Houadfi, M.; Fihri, O.F. Detection of Colistin Encoding Resistance Genes MCR-1 in Isolates Recovered from Broiler Chickens in Morocco. Saudi J. Pathol. Microbiol. 2018, 3, 520–521. [Google Scholar]
- Ramatla, T.; Mileng, K.; Ndou, R.; Mphuti, N.; Syakalima, M.; Lekota, K.E.; Thekisoe, O.M.M. Molecular Detection of Integrons, Colistin and β-Lactamase Resistant Genes in Salmonella Enterica Serovars Enteritidis and Typhimurium Isolated from Chickens and Rats Inhabiting Poultry Farms. Microorganisms 2022, 10, 313. [Google Scholar] [CrossRef]
- Sudatip, D.; Mostacci, N.; Tiengrim, S.; Thamlikitkul, V.; Chasiri, K.; Kritiyakan, A.; Phanprasit, W.; Thinphovong, C.; Abdallah, R.; Baron, S.A.; et al. The Risk of Pig and Chicken Farming for Carriage and Transmission of Escherichia coli Containing Extended-Spectrum Beta-Lactamase (ESBL) and Mobile Colistin Resistance (Mcr) Genes in Thailand. Microb. Genom. 2023, 9, 951. [Google Scholar] [CrossRef]
- Nguyen, P.T.L.; Hung, T.T.M.; Anh, T.H.; Thai, P.D.; Tan, L.M.; Thanh, N.H.; Lien, N.T.P.; Tho, N.T.T.; Ha, H.T.A.; Nguyen, C.; et al. Carriage of Plasmid-Mediated Colistin Resistance-1-Positive Escherichia coli in Humans, Animals, and Environment on Farms in Vietnam. Am. J. Trop. Med. Hyg. 2022, 107, 65–71. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Wang, Y.; Zhang, S.; Shen, Z.; Wang, S. Emergence of the Colistin Resistance Gene Mcr-1 and Its Variant in Several Uncommon Species of Enterobacteriaceae from Commercial Poultry Farm Surrounding Environments. Vet. Microbiol. 2018, 219, 161–164. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, Y.; Zhou, Y.; Wang, Z.; Wang, Y.; Zhang, S.; Shen, Z. Emergence of Colistin Resistance Gene Mcr-8 and Its Variant in Raoultella ornithinolytica. Front. Microbiol. 2019, 10, 228. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Farzana, R.; Sihalath, S.; Rattanavong, S.; Vongsouvath, M.; Mayxay, M.; Sands, K.; Newton, P.N.; Dance, D.A.B.; Hassan, B. A One-Health Sampling Strategy to Explore the Dissemination and Relationship between Colistin Resistance in Human, Animal, and Environmental Sectors in Laos. Engineering 2022, 15, 45–56. [Google Scholar] [CrossRef]
- Sobur, A.M.; Ievy, S.; Haque, Z.F.; Nahar, A.; Zaman, S.B.; Rahman, M.T. Emergence of Colistin-Resistant Escherichia coli in Poultry, House Flies, and Pond Water in Mymensingh, Bangladesh. J. Adv. Vet. Anim. Res. 2019, 6, 50–53. [Google Scholar]
- Hu, Z.; Chen, X.; Wang, Z.; Guo, G.; Xu, Z.; Zhou, Q.; Wei, X.; Liu, Y.; Zhou, L.; Tan, Z.; et al. Whole-Genome Analyses of APEC Carrying Mcr-1 in Some Coastal Areas of China from 2019 to 2020. J. Glob. Antimicrob. Resist. 2022, 30, 370–376. [Google Scholar] [CrossRef]
- Lu, X.; Xiao, X.; Liu, Y.; Li, Y.; Li, R.; Wang, Z. Chromosome-Mediated Mcr-1 in Escherichia coli Strain L73 from a Goose. Int. J. Antimicrob. Agents 2019, 54, 99–101. [Google Scholar] [CrossRef]
- Lu, X.; Du, Y.; Peng, K.; Zhang, W.; Li, J.; Wang, Z.; Li, R. Coexistence of Tet (X4), Mcr-1, and Bla NDM-5 in ST6775 Escherichia coli Isolates of Animal Origin in China. Microbiol. Spectr. 2022, 10, e00196-22. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, W.; Mohsin, M.; Wang, M.; Li, J.; Wang, Z.; Li, R. The Prevalence of Plasmid-Mediated Colistin Resistance Gene Mcr-1 and Different Transferability and Fitness of Mcr-1 -Bearing IncX4 Plasmids in Escherichia coli from Pigeons. Microbiol. Spectr. 2023, 11, e0363922. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.; Urmi, U.L.; Rana, M.; Sultana, F.; Jahan, N.; Hossain, B.; Iqbal, S.; Hossain, M.M.; Mosaddek, A.S.M.; Nahar, S. High Abundance of the Colistin Resistance Gene Mcr-1 in Chicken Gut-Bacteria in Bangladesh. Sci. Rep. 2020, 10, 17292. [Google Scholar] [CrossRef]
- Hu, J.; Yang, J.; Chen, W.; Liu, Z.; Zhao, Q.; Yang, H.; Sun, Z.; Chen, X.; Li, J. Prevalence and Characteristics of Mcr-1-Producing Escherichia coli in Three Kinds of Poultry in Changsha, China. Front. Microbiol. 2022, 13, 840520. [Google Scholar] [CrossRef] [PubMed]
- Yassin, A.K.; Zhang, J.; Wang, J.; Chen, L.; Kelly, P.; Butaye, P.; Lu, G.; Gong, J.; Li, M.; Wei, L.; et al. Identification and Characterization of Mcr Mediated Colistin Resistance in Extraintestinal Escherichia coli from Poultry and Livestock in China. FEMS Microbiol. Lett. 2017, 364, fnx242. [Google Scholar] [CrossRef]
- Hu, Y.; Nguyen, S.V.; Liu, C.; Wang, W.; Dong, Y.; Fanning, S.; Li, F. Complete Genome and Plasmid Sequences of Seven Isolates of Salmonella enterica Subsp. Enterica Harboring the Mcr-1 Gene Obtained from Food in China. Microbiol. Resour. Announc. 2019, 8, e00114-19. [Google Scholar] [CrossRef] [PubMed]
- Li, X.P.; Fang, L.X.; Song, J.Q.; Xia, J.; Huo, W.; Fang, J.T.; Liao, X.P.; Liu, Y.H.; Feng, Y.; Sun, J. Clonal Spread of Mcr-1 in PMQR-Carrying ST34 Salmonella Isolates from Animals in China. Sci. Rep. 2016, 6, 38511. [Google Scholar] [CrossRef] [PubMed]
- Lyu, N.; Feng, Y.; Pan, Y.; Huang, H.; Liu, Y.; Xue, C.; Zhu, B.; Hu, Y. Genomic Characterization of Salmonella Enterica Isolates from Retail Meat in Beijing, China. Front. Microbiol. 2021, 12, 636332. [Google Scholar] [CrossRef]
- Sheng, H.; Ma, J.; Yang, Q.; Li, W.; Zhang, Q.; Feng, C.; Chen, J.; Qin, M.; Su, X.; Wang, P.; et al. Prevalence and Characteristics of Mcr-9-Positive Salmonella Isolated from Retail Food in China. LWT 2022, 160, 113261. [Google Scholar] [CrossRef]
- Tang, B.; Chang, J.; Zhang, L.; Liu, L.; Xia, X.; Hassan, B.H.; Jia, X.; Yang, H.; Feng, Y. Carriage of Distinct Mcr-1-Harboring Plasmids by Unusual Serotypes of Salmonella. Adv. Biosyst. 2020, 4, 1900219. [Google Scholar] [CrossRef]
- Anyanwu, M.U.; Nwobi, O.C.; Okpala, C.O.R.; Ezeonu, I.M. Mobile Tigecycline Resistance: An Emerging Health Catastrophe Requiring Urgent One Health Global Intervention. Front. Microbiol. 2022, 13, 808744. [Google Scholar] [CrossRef]
- Oliveira, C.C.; Lopes, E.S.; Barbosa, D.R.; Pimenta, R.L.; Sobrinho, N.M.B.A.; Coelho, S.M.O.; Souza, M.M.S.; Coelho, I.S. Occurrence of the Colistin Resistance Mcr-1 Gene in Soils from Intensive Vegetable Production and Native Vegetation. Eur. J. Soil. Sci. 2019, 70, 876–881. [Google Scholar] [CrossRef]
- He, Y.; Yuan, Q.; Mathieu, J.; Stadler, L.; Senehi, N.; Sun, R.; Alvarez, P.J.J. Antibiotic Resistance Genes from Livestock Waste: Occurrence, Dissemination, and Treatment. NPJ Clean Water 2020, 3, 4. [Google Scholar] [CrossRef]
- Lv, Z.; Shen, Y.; Liu, W.; Ye, H.; Liu, D.; Liu, J.; Fu, Y.; Peng, C.; Chen, K.; Deng, X.; et al. Prevalence and Risk Factors of Mcr-1-Positive Volunteers after Colistin Banning as Animal Growth Promoter in China: A Community-Based Case–Control Study. Clin. Microbiol. Infect. 2022, 28, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Yu, Y.; Kuang, X.; Li, X.; Wang, M.; Sun, R.; Sun, J.; Liu, Y.; Liao, X. Molecular Characteristics of Antimicrobial Resistance and Virulence in Klebsiella pneumoniae Strains Isolated from Goose Farms in Hainan, China. Appl. Environ. Microbiol. 2022, 88, e02457-21. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.P.; Lin, Q.Q.; He, W.Y.; Wang, J.; Yi, M.Y.; Lv, L.C.; Yang, J.; Liu, J.H.; Guo, J.Y. Co-Selection May Explain the Unexpectedly High Prevalence of Plasmid-Mediated Colistin Resistance Gene Mcr-1 in a Chinese Broiler Farm. Zool. Res. 2020, 41, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yu, L.; Chen, X.; Zhi, C.; Yao, X.; Liu, Y.; Wu, S.; Guo, Z.; Yi, L.; Zeng, Z.; et al. High Prevalence of Colistin Resistance and mcr-1 Gene in Escherichia coli Isolated from Food Animals in China. Front. Microbiol. 2017, 8, 562. [Google Scholar] [CrossRef] [PubMed]
- Shafiq, M.; Huang, J.; Shah, J.M.; Ali, I.; Rahman, S.U.; Wang, L. Characterization and Resistant Determinants Linked to Mobile Elements of ESBL-Producing and Mcr-1-Positive Escherichia coli Recovered from the Chicken Origin. Microb. Pathog. 2021, 150, 104722. [Google Scholar] [CrossRef]
- Wang, J.; Tang, B.; Lin, R.; Zheng, X.; Ma, J.; Xiong, X.; Jiang, H.; Yang, H.; Ding, B. Emergence of Mcr-1- and BlaNDM-5-Harbouring IncHI2 Plasmids in Escherichia coli Strains Isolated from Meat in Zhejiang, China. J. Glob. Antimicrob. Resist. 2022, 30, 103–106. [Google Scholar] [CrossRef]
- Ghafur, A.; Shankar, C.; GnanaSoundari, P.; Venkatesan, M.; Mani, D.; Thirunarayanan, M.A.; Veeraraghavan, B. Detection of Chromosomal and Plasmid-Mediated Mechanisms of Colistin Resistance in Escherichia coli and Klebsiella pneumoniae from Indian Food Samples. J. Glob. Antimicrob. Resist. 2019, 16, 48–52. [Google Scholar] [CrossRef]
- Kurekci, C.; Aydin, M.; Nalbantoglu, O.U.; Gundogdu, A. First Report of Escherichia coli Carrying the Mobile Colistin Resistance Gene Mcr-1 in Turkey. J. Glob. Antimicrob. Resist. 2018, 15, 169–170. [Google Scholar] [CrossRef]
- Adiguzel, M.C.; Baran, A.; Wu, Z.; Cengiz, S.; Dai, L.; Oz, C.; Ozmenli, E.; Goulart, D.B.; Sahin, O. Prevalence of Colistin Resistance in Escherichia coli in Eastern Turkey and Genomic Characterization of an Mcr-1 Positive Strain from Retail Chicken Meat. Microb. Drug Resist. 2021, 27, 424–432. [Google Scholar] [CrossRef]
- Le, P.Q.; Awasthi, S.P.; Hatanaka, N.; Hinenoya, A.; Hassan, J.; Ombarak, R.A.; Iguchi, A.; Tran, N.T.T.; Dao, K.V.T.; Vien, M.Q.; et al. Prevalence of Mobile Colistin Resistance (Mcr) Genes in Extended-Spectrum β-Lactamase-Producing Escherichia coli Isolated from Retail Raw Foods in Nha Trang, Vietnam. Int. J. Food Microbiol. 2021, 346, 109164, Corrigendum to Int. J. Food Microbiol. 2022, 370, 109540. [Google Scholar] [CrossRef] [PubMed]
- Malhotra-Kumar, S.; Xavier, B.B.; Das, A.J.; Lammens, C.; Hoang, H.T.T.; Pham, N.T.; Goossens, H. Colistin-Resistant Escherichia coli Harbouring Mcr-1 Isolated from Food Animals in Hanoi, Vietnam. Lancet Infect. Dis. 2016, 16, P286–P287. [Google Scholar] [CrossRef]
- Nakayama, T.; le Thi, H.; Thanh, P.N.; Minh, D.T.N.; Hoang, O.N.; Hoai, P.H.; Yamaguchi, T.; Jinnai, M.; Do, P.N.; Van, C.D.; et al. Abundance of Colistin-Resistant Escherichia coli Harbouring Mcr-1 and Extended-Spectrum β-Lactamase-Producing E. coli Co-Harbouring Bla CTX-M-55 or -65 with Bla TEM Isolates from Chicken Meat in Vietnam. Arch. Microbiol. 2022, 204, 137. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.N.; Khong, D.T.; Le, H.v.; Tran, H.T.; Phan, Q.N.; Le, H.T.T.; Kawahara, R.; Yamamoto, Y. Quantitative Analysis of Colistin-Resistant Escherichia coli in Retail Meat from Local Vietnamese Markets. Biomed. Res. Int. 2021, 2021, 6678901. [Google Scholar] [CrossRef]
- Trung, N.V.; Matamoros, S.; Carrique-Mas, J.J.; Nghia, N.H.; Nhung, N.T.; Chieu, T.T.B.; Mai, H.H.; van Rooijen, W.; Campbell, J.; Wagenaar, J.A.; et al. Zoonotic Transmission of Mcr-1 Colistin Resistance Gene from Small-Scale Poultry Farms, Vietnam. Emerg. Infect. Dis. 2017, 23, 529–532. [Google Scholar] [CrossRef] [PubMed]
- Vounba, P.; Rhouma, M.; Arsenault, J.; Bada Alambédji, R.; Fravalo, P.; Fairbrother, J.M. Prevalence of Colistin Resistance and Mcr-1/Mcr-2 Genes in Extended-Spectrum β-Lactamase/AmpC-Producing Escherichia coli Isolated from Chickens in Canada, Senegal and Vietnam. J. Glob. Antimicrob. Resist. 2019, 19, 222–227. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Kawahara, R.; Harada, K.; Teruya, S.; Nakayama, T.; Motooka, D.; Nakamura, S.; do Nguyen, P.; Kumeda, Y.; van Dang, C.; et al. The Presence of Colistin Resistance Gene Mcr-1 and -3 in ESBL Producing Escherichia coli Isolated from Food in Ho Chi Minh City, Vietnam. FEMS Microbiol. Lett. 2018, 365, fny100. [Google Scholar] [CrossRef]
- Aklilu, E.; Raman, K. MCR-1 Gene Encoded Colistin-Resistant Escherichia coli in Raw Chicken Meat and Bean Sprouts in Malaysia. Int. J. Microbiol. 2020, 2020, 8853582. [Google Scholar] [CrossRef]
- Ghaffoori Kanaan, M.H.; Al-Shadeedi, S.M.J.; Al-Massody, A.J.; Ghasemian, A. Drug Resistance and Virulence Traits of Acinetobacter Baumannii from Turkey and Chicken Raw Meat. Comp. Immunol. Microbiol. Infect. Dis. 2020, 70, 101451. [Google Scholar] [CrossRef]
- Moser, A.I.; Kuenzli, E.; Campos-Madueno, E.I.; Büdel, T.; Rattanavong, S.; Vongsouvath, M.; Hatz, C.; Endimiani, A. Antimicrobial-Resistant Escherichia coli Strains and Their Plasmids in People, Poultry, and Chicken Meat in Laos. Front. Microbiol. 2021, 12, 708182. [Google Scholar] [CrossRef]
- Chaalal, N.; Touati, A.; Yahiaoui-Martinez, A.; Aissa, M.A.; Sotto, A.; Lavigne, J.P.; Pantel, A. Colistin-Resistant Enterobacterales Isolated from Chicken Meat in Western Algeria. Microb. Drug Resist. 2021, 27, 991–1002. [Google Scholar] [CrossRef]
- Cyoia, P.S.; Koga, V.L.; Nishio, E.K.; Houle, S.; Dozois, C.M.; de Brito, K.C.T.; de Brito, B.G.; Nakazato, G.; Kobayashi, R.K.T. Distribution of ExPEC Virulence Factors, BlaCTX-M, FosA3, and Mcr-1 in Escherichia coli isolated from Commercialized Chicken Carcasses. Front. Microbiol. 2019, 10, 3254. [Google Scholar] [CrossRef]
- Monte, D.F.; Mem, A.; Fernandes, M.R.; Cerdeira, L.; Esposito, F.; Galvão, J.A.; Franco, B.D.G.M.; Lincopan, N.; Landgraf, M. Chicken Meat as a Reservoir of Colistin-Resistant Escherichia coli Strains Carrying Mcr-1 Genes in South America. Antimicrob. Agents Chemother. 2017, 61, e02718-16. [Google Scholar] [CrossRef]
- Perdomo, A.; Webb, H.E.; Bugarel, M.; Friedman, C.R.; Francois Watkins, L.K.; Loneragan, G.H.; Calle, A. First Known Report of Mcr-Harboring Enterobacteriaceae in the Dominican Republic. Int. J. Environ. Res. Public Health 2023, 20, 5123. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, K.V.; Pavlova, A.S.; Shedko, E.D.; Mikhaylova, Y.V.; Margos, G.; Hepner, S.; Chebotar, I.V.; Korneenko, E.V.; Podkolzin, A.T.; Akimkin, V.G. Mobile Colistin Resistance Genetic Determinants of Non-Typhoid Salmonella enterica Isolates from Russia. Microorganisms 2021, 9, 2515. [Google Scholar] [CrossRef] [PubMed]
- Johura, F.T.; Tasnim, J.; Barman, I.; Biswas, S.R.; Jubyda, F.T.; Sultana, M.; George, C.M.; Camilli, A.; Seed, K.D.; Ahmed, N.; et al. Colistin-resistant Escherichia coli carrying mcr-1 in food, water, hand rinse, and healthy human gut in Bangladesh. Gut Pathog 2020, 12, 5. [Google Scholar] [CrossRef]
- Yang, C.; Han, J.; Berglund, B.; Zou, H.; Gu, C.; Zhao, L.; Meng, C.; Zhang, H.; Ma, X.; Li, X. Dissemination of BlaNDM-5 and Mcr-8.1 in Carbapenem-Resistant Klebsiella pneumoniae and Klebsiella quasipneumoniae in an Animal Breeding Area in Eastern China. Front. Microbiol. 2022, 13, 1030490. [Google Scholar] [CrossRef]
- Sakdinun, P.; Sriwongsa, N.; Wongmuk, S. Detection of Colistin Resistance and Mcr-1 Gene in Salmonella Isolated from Feces of Poultry in Western Thailand during 2013–2016. KKU Vet. J. 2018, 28, 1–10. [Google Scholar]
- Dandachi, I.; Fayad, E.; Sleiman, A.; Daoud, Z.; Rolain, J.M. Dissemination of Multidrug-Resistant and Mcr-1 Gram-Negative Bacilli in Broilers, Farm Workers, and the Surrounding Environment in Lebanon. Microb. Drug Resist. 2020, 26, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Al-Mir, H.; Osman, M.; Azar, N.; Madec, J.Y.; Hamze, M.; Haenni, M. Emergence of Clinical Mcr-1-Positive Escherichia coli in Lebanon. J. Glob. Antimicrob. Resist. 2019, 19, 83–84. [Google Scholar] [CrossRef]
- Wang, X.; Sun, N.; Liu, X.; Li, F.; Sun, J.; Huang, J.; Li, R.; Wang, L. Small Clone Dissemination of TmexCD1-ToprJ1–Carrying Klebsiella pneumoniae Isolates in a Chicken Farm. J. Glob. Antimicrob. Resist. 2022, 29, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yang, Y.; Li, Y.; Liu, D.; Tuo, H.; Wang, H.; Call, D.R.; Davis, M.; Zhang, A. Isolation of an IncP-1 plasmid harbouring mcr-1 from a chicken isolate of citrobacter braakii in China. Int. J. Antimicrob. Agents 2018, 51, 936–940. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Cheng, P.; Li, X.; Liu, R.; Liu, H.; Zhang, X. Molecular Epidemiology and Colistin-Resistant Mechanism of Mcr-Positive and Mcr-Negative Escherichia coli Isolated from Animal in Sichuan Province, China. Front. Microbiol. 2022, 13, 818548. [Google Scholar] [CrossRef]
- Yang, L.; Shen, Y.; Jiang, J.; Wang, X.; Shao, D.; Lam, M.M.C.; Holt, K.E.; Shao, B.; Wu, C.; Shen, J.; et al. Distinct Increase in Antimicrobial Resistance Genes among Escherichia coli during 50 Years of Antimicrobial Use in Livestock Production in China. Nat. Food 2022, 3, 197–205. [Google Scholar] [CrossRef]
- Tang, B.; Chang, J.; Chen, Y.; Lin, J.; Xiao, X.; Xia, X.; Lin, J.; Yang, H.; Zhao, G. Escherichia fergusonii, an Underrated Repository for Antimicrobial Resistance in Food Animals. Microbiol. Spectr. 2022, 10, e01617-21. [Google Scholar] [CrossRef]
- Tang, B.; Chen, Y.; Zhang, L.; Chang, J.; Xia, X.; Yang, H. Complete Genome Sequence of Colistin-Resistant Escherichia Fergusonii Strain EFCF056. Microbiol. Resour. Announc. 2020, 9, e01571-19. [Google Scholar] [CrossRef]
- Lei, C.W.; Zhang, Y.; Wang, Y.T.; Wang, H.N. Detection of Mobile Colistin Resistance Gene Mcr-10.1 in a Conjugative Plasmid from Enterobacter Roggenkampii of Chicken Origin in China. Antimicrob. Agents Chemother. 2020, 64, e01191-20. [Google Scholar] [CrossRef]
- Shen, Z.; Wang, Y.; Shen, Y.; Shen, J.; Wu, C. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect. Dis. 2016, 16, 293. [Google Scholar] [CrossRef]
- Sun, J.; Li, X.P.; Yang, R.S.; Fang, L.X.; Huo, W.; Li, S.M.; Jiang, P.; Liao, X.P.; Liu, Y.H. Complete Nucleotide Sequence of an IncI2 Plasmid Coharboring blaCTX-M-55 and mcr-1. Antimicrob. Agents Chemother. 2016, 60, 5014–5017. [Google Scholar] [CrossRef]
- Li, X.; Li, L.; Yu, L.; Liu, S.; Liu, L.; Wei, X.; Song, Y.; Liu, C.; Jiang, M.; Wang, F. Prevalence of Avian-Origin Mcr-1–Positive Escherichia coli with a Potential Risk to Humans in Tai’an, China. Poult. Sci. 2020, 99, 5118–5126. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Wang, Y.; Shi, X.; Wang, S.; Ren, H.; Shen, Z.; Wang, Y.; Lin, J.; Wang, S. Rapid Rise of the ESBL and Mcr-1 Genes in Escherichia coli of Chicken Origin in China, 2008–2014. Emerg. Microbes Infect. 2018, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Q.; Li, Y.X.; Song, T.; Yang, Y.X.; Jiang, W.; Zhang, A.Y.; Guo, X.Y.; Liu, B.H.; Wang, Y.X.; Lei, C.W.; et al. Colistin Resistance Gene Mcr-1 and Its Variant in Escherichia coli Isolates from Chickens in China. Antimicrob. Agents Chemother. 2017, 61, e01204-16. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, J.; Wang, J.; Butaye, P.; Kelly, P.; Li, M.; Yang, F.; Gong, J.; Yassin, A.K.; Guo, W.; et al. Newly Identified Colistin Resistance Genes, Mcr-4 and Mcr-5, from Upper and Lower Alimentary Tract of Pigs and Poultry in China. PLoS ONE 2018, 13, e0193957. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wang, J.; Wang, X.; Bai, X.; Ma, J.; Dang, R.; Xiong, Y.; Fanning, S.; Bai, L.; Yang, Z. Characterization of Five Escherichia coli Isolates Co-Expressing ESBL and Mcr-1 Resistance Mechanisms from Different Origins in China. Front. Microbiol. 2019, 10, 1994. [Google Scholar] [CrossRef]
- Zou, M.; Ma, P.P.; Liu, W.S.; Liang, X.; Li, X.Y.; Li, Y.Z.; Liu, B.T. Prevalence and Antibiotic Resistance Characteristics of Extraintestinal Pathogenic Escherichia coli among Healthy Chickens from Farms and Live Poultry Markets in China. Animals 2021, 11, 1112. [Google Scholar] [CrossRef]
- Yang, R.S.; Feng, Y.; Lv, X.Y.; Duan, J.H.; Chen, J.; Fang, L.X.; Xia, J.; Liao, X.P.; Sun, J.; Liu, Y.H. Emergence of NDM-5- and MCR-1-Producing Escherichia coli Clones ST648 and ST156 from a Single Muscovy Duck (Cairina moschata). Antimicrob. Agents Chemother. 2016, 60, 6899–6902. [Google Scholar] [CrossRef]
- Yao, X.; Doi, Y.; Zeng, L.; Lv, L.; Liu, J.H. Carbapenem-Resistant and Colistin-Resistant Escherichia coli Co-Producing NDM-9 and MCR-1. Lancet Infect. Dis. 2016, 16, 288–289. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, Y.; Xi, W.; Liu, S.; Liu, J.; Mu, H.; Chen, B.; He, H.; Fan, Y.; Ma, W.; et al. Genetic Features of Plasmid- and Chromosome-Mediated Mcr-1 in Escherichia coli Isolates from Animal Organs with Lesions. Front. Microbiol. 2021, 12, 707332. [Google Scholar] [CrossRef]
- Sun, S.; Gao, H.; Liu, Y.; Jin, L.; Wang, R.; Wang, X.; Wang, Q.; Yin, Y.; Zhang, Y.; Wang, H. Co-existence of a novel plasmid-mediated efflux pump with colistin resistance gene mcr in one plasmid confers transferable multidrug resistance in Klebsiella pneumoniae. Emerg. Microbes Infect. 2020, 9, 1102–1113. [Google Scholar] [CrossRef]
- Wang, Y.; Lyu, N.; Liu, F.; Liu, W.J.; Bi, Y.; Zhang, Z.; Ma, S.; Cao, J.; Song, X.; Wang, A.; et al. More diversified antibiotic resistance genes in chickens and workers of the live poultry markets. Environ. Int. 2021, 153, 106534. [Google Scholar] [CrossRef]
- Mei, C.-Y.; Jiang, Y.; Ma, Q.-C.; Lu, M.-J.; Wu, H.; Wang, Z.-Y.; Jiao, X.; Wang, J. Chromosomally and Plasmid-Located Mcr in Salmonella from Animals and Food Products in China. Microbiol. Spectr. 2022, 10, e02773-22. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, Y.; Jia, Y.; Sun, H.; Zhang, C.; Hu, G.; Yuan, L. Genomic Characteristics of Mcr-1 and BlaCTX-M-Type in a Single Multidrug-Resistant Escherichia coli ST93 from Chicken in China. Poult. Sci. 2021, 100, 101074. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Feng, Y.; Liu, F.; Jiang, H.; Qu, Z.; Lei, M.; Wang, J.; Zhang, B.; Hu, Y.; Ding, J.; et al. A Phage-like IncY Plasmid Carrying the Mcr-1 Gene in Escherichia coli from a Pig Farm in China. Antimicrob. Agents Chemother. 2017, 61, e02035-16. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; van Dorp, L.; Shaw, L.P.; Bradley, P.; Wang, Q.; Wang, X.; Jin, L.; Zhang, Q.; Liu, Y.; Rieux, A.; et al. The Global Distribution and Spread of the Mobilized Colistin Resistance Gene Mcr-1. Nat. Commun. 2018, 9, 38. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, R.; Yang, Y.; Li, H.; Wang, J.; Lan, J.; Li, P.; Zhu, Y.; Xie, Z.; Jiang, S. Occurrence and Molecular Characteristics of Mcr-1- Positive Escherichia coli from Healthy Meat Ducks in Shandong Province of China. Animals 2020, 10, 1299. [Google Scholar] [CrossRef]
- Liu, L.; Feng, Y.; Zhang, X.; McNally, A.; Zong, Z. New Variant of Mcr-3 in an Extensively Drug-Resistant Escherichia coli Clinical Isolate Carrying Mcr-1 and BlaNDM-5. Antimicrob. Agents Chemother. 2017, 61, e01757-17. [Google Scholar] [CrossRef]
- Liu, B.T.; Song, F.J.; Zou, M.; di Zhang, Q.; Shan, H. High Incidence of Escherichia coli Strains Coharboring Mcr-1 and BlaNDM from Chickens. Antimicrob. Agents Chemother. 2017, 61, e02347-16. [Google Scholar] [CrossRef]
- Liu, X.; Geng, S.; Chan, E.W.-C.; Chen, S. Increased Prevalence of Escherichia coli Strains from Food Carrying BlaNDM and Mcr-1-Bearing Plasmids That Structurally Resemble Those of Clinical Strains, China, 2015 to 2017. Eurosurveillance 2019, 24, 1800113. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, C.; Zhang, R.; Chen, Y.; Shen, Y.; Hu, F.; Liu, D.; Lu, J.; Guo, Y.; Xia, X.; et al. Changes in Colistin Resistance and Mcr-1 Abundance in Escherichia coli of Animal and Human Origins Following the Ban of Colistin-Positive Additives in China: An Epidemiological Comparative Study. Lancet Infect. Dis. 2020, 20, 1161–1171. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, T.; Wang, C.; Liang, G.; Lu, Q.; Wen, G.; Guo, Y.; Cheng, Y.; Wang, Z.; Shao, H.; et al. Prevalence of Colistin Resistance Gene Mcr-1 in Escherichia coli Isolated from Chickens in Central China, 2014 to 2019. J. Glob. Antimicrob. Resist. 2022, 29, 241–246. [Google Scholar] [CrossRef]
- Fan, R.; Li, C.; Duan, R.; Qin, S.; Liang, J.; Xiao, M.; Lv, D.; Jing, H.; Wang, X. Retrospective Screening and Analysis of Mcr-1 and BlaNDM in Gram-Negative Bacteria in China, 2010. Front. Microbiol. 2020, 11, 121. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Tang, B.; Zheng, X.; Chang, J.; Ma, J.; He, Y.; Yang, H.; Wu, Y. Emergence of Incl2 Plasmid-Mediated Colistin Resistance in Avian Escherichia Fergusonii. FEMS Microbiol. Lett. 2022, 369, fnac016. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Wang, J.; Zheng, X.; Chang, J.; Ma, J.; Wang, J.; Ji, X.; Yang, H.; Ding, B. Antimicrobial Resistance Surveillance of Escherichia coli from Chickens in the Qinghai Plateau of China. Front. Microbiol. 2022, 13, 885132. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Q.; Zhang, A.Y.; Ma, S.Z.; Kong, L.H.; Li, Y.X.; Liu, J.X.; Davis, M.A.; Guo, X.Y.; Liu, B.H.; Lei, C.W.; et al. Co-Occurrence of Mcr-1 and ESBL on a Single Plasmid in Salmonella enterica. J. Antimicrob. Chemother. 2016, 71, 2336–2338. [Google Scholar] [CrossRef]
- Sun, C.; Cui, M.; Zhang, S.; Wang, H.; Song, L.; Zhang, C.; Zhao, Q.; Liu, D.; Wang, Y.; Shen, J.; et al. Plasmid-Mediated Tigecycline-Resistant Gene Tet(X4) in Escherichia coli from Food-Producing Animals, China, 2008–2018. Emerg. Microbes Infect. 2019, 8, 1524–1527. [Google Scholar] [CrossRef]
- Xu, L.; Wan, F.; Fu, H.; Tang, B.; Ruan, Z.; Xiao, Y.; Luo, Q. Emergence of Colistin Resistance Gene Mcr-10 in Enterobacterales Isolates Recovered from Fecal Samples of Chickens, Slaughterhouse Workers, and a Nearby Resident. Microbiol. Spectr. 2022, 10, e00418-22. [Google Scholar] [CrossRef]
- Wang, X.; Zhai, W.; Wang, S.; Shen, Z.; Wang, Y.; Zhang, Q. A Novel Transposon, Tn6518, Mediated Transfer of Mcr-3 Variant in ESBL-Producing Aeromonas Veronii. Infect. Drug Resist. 2020, 13, 893–899. [Google Scholar] [CrossRef]
- Wang, X.; Zhai, W.; Li, J.; Liu, D.; Zhang, Q.; Shen, Z.; Wang, S.; Wang, Y. Presence of an Mcr-3 Variant in Aeromonas Caviae, Proteus Mirabilis, and Escherichia coli from One Domestic Duck. Antimicrob. Agents Chemother. 2018, 62, e02106-17. [Google Scholar] [CrossRef]
- Ling, Z.; Yin, W.; Li, H.; Zhang, Q.; Wang, X.; Wang, Z.; Ke, Y.; Wang, Y.; Shena, J. Chromosome-Mediated Mcr-3 Variants in Aeromonas Veronii from Chicken Meat. Antimicrob. Agents Chemother. 2017, 61, e01272-17. [Google Scholar] [CrossRef]
- Lv, D.; Duan, R.; Fan, R.; Mu, H.; Liang, J.; Xiao, M.; He, Z.; Qin, S.; Yang, J.; Jing, H.; et al. BlaNDM and Mcr-1 to Mcr-5 Gene Distribution Characteristics in Gut Specimens from Different Regions of China. Antibiotics 2021, 10, 233. [Google Scholar] [CrossRef]
- Haroon, A.; Abbas, M.A.; Rahim, A.; Siddique, N. The Mcr-1 Gene Transformation through Conjugation Assay in Avian Pathogenic E. coli from Pakistan. Int. J. Biosci. 2019, 15, 100–109. [Google Scholar]
- Azam, M.; Ehsan, I.; Sajjad-ur-Rahman; Saleemi, M.K.; Javed, M.R.; Mohsin, M. Detection of the Colistin Resistance Gene Mcr-1 in Avian Pathogenic Escherichia coli in Pakistan. J. Glob. Antimicrob. Resist. 2017, 11, 152–153. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Mohsin, M.; Lei, S.; Srinivas, S.; Wiqar, R.T.; Lin, J.; Feng, Y. Discovery of a Mcr-1-Bearing Plasmid in Commensal Colistin-Resistant Escherichia coli from Healthy Broilers in Faisalabad, Pakistan. Virulence 2018, 9, 994–999. [Google Scholar] [CrossRef] [PubMed]
- Javed, H.; Saleem, S.; Zafar, A.; Ghafoor, A.; Bin Shahzad, A.; Ejaz, H.; Junaid, K.; Jahan, S. Emergence of Plasmid-Mediated Mcr Genes from Gram-Negative Bacteria at the Human-Animal Interface. Gut Pathog. 2020, 12, 54. [Google Scholar] [CrossRef] [PubMed]
- Rafique, M.; Potter, R.F.; Ferreiro, A.; Wallace, M.A.; Rahim, A.; Ali Malik, A.; Siddique, N.; Abbas, M.A.; D’Souza, A.W.; Burnham, C.A.D.; et al. Genomic Characterization of Antibiotic Resistant Escherichia coli Isolated from Domestic Chickens in Pakistan. Front. Microbiol. 2020, 10, 3052. [Google Scholar] [CrossRef]
- Zulqarnain, M.; Sarwar, N.; Anjum, A.A.; Firyal, S.; Yaqub, T.; Rabbani, M. Molecular Detection of Colistin Resistance Gene (MCR-1) in E. coli Isolated from Cloacal Swabs of Broilers. Pak. Vet. J. 2021, 41, 284–288. [Google Scholar]
- Li, R.; Lu, X.; Munir, A.; Abdullah, S.; Liu, Y.; Xiao, X.; Wang, Z.; Mohsin, M. Widespread Prevalence and Molecular Epidemiology of Tet(X4) and Mcr-1 Harboring Escherichia coli Isolated from Chickens in Pakistan. Sci. Total Environ. 2022, 806, 150689. [Google Scholar] [CrossRef]
- Shafiq, M.; Rahman, S.U.; Bilal, H.; Ullah, A.; Noman, S.M.; Zeng, M.; Yuan, Y.; Xie, Q.; Li, X.; Jiao, X. Incidence and Molecular Characterization of ESBL-Producing and Colistin-Resistant Escherichia coli Isolates Recovered from Healthy Food-Producing Animals in Pakistan. J. Appl. Microbiol. 2022, 133, 1169–1182. [Google Scholar] [CrossRef]
- Ali, M.W.; Utsho, K.S.; Karmakar, S.; Hoque, M.N.; Rahman, M.T.; Hassan, J. First Report on the Molecular Characteristics of Mcr-1 Colistin Resistant E. coli Isolated from Retail Broiler Meat in Bangladesh. Int. J. Food Microbiol. 2023, 388, 110065. [Google Scholar] [CrossRef]
- Jamil, A.; Zahoor, M.A.; Nawaz, Z.; Siddique, A.B.; Khurshid, M. Genetic Diversity of Escherichia coli Coharboring mcr-1 and Extended Spectrum Beta-Lactamases from Poultry. Biomed Res. Int. 2022, 2022, 8224883. [Google Scholar] [CrossRef]
- Uddin, M.B.; Alam, M.N.; Hasan, M.; Hossain, S.M.B.; Debnath, M.; Begum, R.; Samad, M.A.; Hoque, S.F.; Chowdhury, M.S.R.; Rahman, M.M.; et al. Molecular Detection of Colistin Resistance Mcr-1 Gene in Multidrug-Resistant Escherichia coli Isolated from Chicken. Antibiotics 2022, 11, 97. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.B.; Hossain, S.B.; Hasan, M.; Alam, M.N.; Debnath, M.; Begum, R.; Roy, S.; Harun-Al-rashid, A.; Chowdhury, M.S.R.; Rahman, M.M.; et al. Multidrug Antimicrobial Resistance and Molecular Detection of MCR-1 Gene in Salmonella Species Isolated from Chicken. Animals 2021, 11, 206. [Google Scholar] [CrossRef] [PubMed]
- Hmede, Z.; Kassem, I.I. The Colistin Resistance Gene Mcr-1 Is Prevalent in Commensal Escherichia coli Isolated from Preharvest Poultry in Lebanon. Antimicrob. Agents Chemother. 2018, 62, e01304-18. [Google Scholar] [CrossRef]
- Kassem, I.I.; Mann, D.; Li, S.; Deng, X. Draft Genome Sequences and Resistome Analysis of Multidrug-Resistant Mcr-1-Harbouring Escherichia coli Isolated from Pre-Harvest Poultry in Lebanon. J. Glob. Antimicrob. Resist. 2021, 25, 114–116. [Google Scholar] [CrossRef]
- Mikhayel, M.; Leclercq, S.O.; Sarkis, D.K.; Doublet, B. Occurrence of the Colistin Resistance Gene Mcr-1 and Additional Antibiotic Resistance Genes in ESBL/AmpC-Producing Escherichia coli from Poultry in Lebanon: A Nationwide Survey. Microbiol. Spectr. 2021, 9, e02773-22. [Google Scholar] [CrossRef] [PubMed]
- Abou Fayad, A.; el Azzi, M.; Sleiman, A.; Kassem, I.I.; Bawazeer, R.A.; Okdah, L.; Doumith, M.; Alghoribi, M.F.; Matar, G.M. Acquired Resistome and Plasmid Sequencing of Mcr-1 Carrying MDR Enterobacteriaceae from Poultry and Their Relationship to STs Associated with Humans. JAC Antimicrob. Resist. 2022, 4, dlab198. [Google Scholar] [CrossRef] [PubMed]
- Aklilu, E.; Harun, A.; Singh, K.K.B. Molecular characterization of blaNDM, blaOXA-48, mcr-1 and blaTEM-52 positive and concurrently carbapenem and colistin resistant and extended spectrum beta-lactamase producing Escherichia coli in chicken in Malaysia. BMC Vet Res. 2022, 18, 190. [Google Scholar] [CrossRef]
- Hu, Y.; Liu, F.; Lin, I.Y.C.; Gao, G.F.; Zhu, B. Dissemination of the Mcr-1 Colistin Resistance Gene. Lancet Infect. Dis. 2016, 16, 146–147. [Google Scholar] [CrossRef] [PubMed]
- Lemlem, M.; Aklilu, E.; Mohammed, M.; Kamaruzzaman, F.; Zakaria, Z.; Harun, A.; Devan, S.S. Molecular detection and antimicrobial resistance profiles of Extended-Spectrum Beta-Lactamase (ESBL) producing Escherichia coli in broiler chicken farms in Malaysia. PLoS ONE 2023, 18, e0285743. [Google Scholar] [CrossRef]
- Karim, M.R.; Zakaria, Z.; Hassan, L.; Mohd Faiz, N.; Ahmad, N.I. Antimicrobial Resistance Profiles and Co-Existence of Multiple Antimicrobial Resistance Genes in mcr-Harbouring Colistin-Resistant Enterobacteriaceae Isolates Recovered from Poultry and Poultry Meats in Malaysia. Antibiotics 2023, 12, 1060. [Google Scholar] [CrossRef]
- Maamar, E.; Alonso, C.A.; Hamzaoui, Z.; Dakhli, N.; Abbassi, M.S.; Ferjani, S.; Saidani, M.; Boutiba-Ben Boubaker, I.; Torres, C. Emergence of Plasmid-Mediated Colistin-Resistance in CMY-2-Producing Escherichia coli of Lineage ST2197 in a Tunisian Poultry Farm. Int. J. Food Microbiol. 2018, 269, 60–63. [Google Scholar] [CrossRef]
- Oueslati, W.; Rjeibi, M.R.; Benyedem, H.; Mamlouk, A.; Souissi, F.; Selmi, R.; Ettriqui, A. Prevalence, Risk Factors, Antimicrobial Resistance and Molecular Characterization of Salmonella in Northeast Tunisia Broiler Flocks. Vet. Sci. 2022, 9, 12. [Google Scholar] [CrossRef]
- Hadjadj, L.; Riziki, T.; Zhu, Y.; Li, J.; Diene, S.M.; Rolain, J.M. Study of Mcr-1 Gene-Mediated Colistin Resistance in Enterobacteriaceae Isolated from Humans and Animals in Different Countries. Genes 2017, 8, 394. [Google Scholar] [CrossRef] [PubMed]
- Moawad, A.A.; Hotzel, H.; Neubauer, H.; Ehricht, R.; Monecke, S.; Tomaso, H.; Hafez, H.M.; Roesler, U.; El-Adawy, H. Antimicrobial Resistance in Enterobacteriaceae from Healthy Broilers in Egypt: Emergence of Colistin-Resistant and Extended-Spectrum β-Lactamase-Producing. Escherichia coli. Gut Pathog. 2018, 10, 39. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, H.; Soliman, A.M.; Hiott, L.M.; Elbediwi, M.; Woodley, T.A.; Chattaway, M.A.; Jenkins, C.; Frye, J.G.; Jackson, C.R. Emergence of Multidrug-Resistant Escherichia coli Producing CTX-M, MCR-1, and FosA in Retail Food from Egypt. Front. Cell. Infect. Microbiol. 2021, 11, 681588. [Google Scholar] [CrossRef]
- Moawad, A.A.; Hotzel, H.; Hafez, H.M.; Ramadan, H.; Tomaso, H.; Braun, S.D.; Ehricht, R.; Diezel, C.; Gary, D.; Engelmann, I.; et al. Occurrence, Phenotypic and Molecular Characteristics of Extended-Spectrum Beta-Lactamase-Producing Escherichia coli in Healthy Turkeys in Northern Egypt. Antibiotics 2022, 11, 1075. [Google Scholar] [CrossRef]
- Perreten, V.; Strauss, C.; Collaud, A.; Gerber, D. Colistin Resistance Gene Mcr-1 in Avian-Pathogenic Escherichia coli in South Africa. Antimicrob. Agents Chemother. 2016, 60, 4414–4415. [Google Scholar] [CrossRef]
- Hassan, I.Z.; Wandrag, B.; Gouws, J.J.; Qekwana, D.N.; Naidoo, V. Antimicrobial Resistance and Mcr-1 Gene in Escherichia coli Isolated from Poultry Samples Submitted to a Bacteriology Laboratory in South Africa. Vet. World 2021, 14, 2662–2669. [Google Scholar] [CrossRef] [PubMed]
- Takawira, F.T.; Pitout, J.D.D.; Thilliez, G.; Mashe, T.; Gutierrez, A.V.; Kingsley, R.A.; Peirano, G.; Matheu, J.; Midzi, S.M.; Mwamakamba, L.W.; et al. Faecal Carriage of ESBL Producing and Colistin Resistant Escherichia coli in Avian Species over a 2-Year Period (2017–2019) in Zimbabwe. Front. Cell. Infect. Microbiol. 2022, 12, 1035145. [Google Scholar] [CrossRef]
- Ngbede, E.O.; Poudel, A.; Kalalah, A.; Yang, Y.; Adekanmbi, F.; Adikwu, A.A.; Adamu, A.M.; Mamfe, L.M.; Daniel, S.T.; Useh, N.M.; et al. Identification of Mobile Colistin Resistance Genes (Mcr-1.1, Mcr-5 and Mcr-8.1) in Enterobacteriaceae and Alcaligenes Faecalis of Human and Animal Origin, Nigeria. Int. J. Antimicrob. Agents 2020, 56, 106108. [Google Scholar] [CrossRef]
- Vinueza-Burgos, C.; Ortega-Paredes, D.; Narvaéz, C.; de Zutter, L.; Zurita, J. Characterization of Cefotaxime Resistant Escherichia coli Isolated from Broiler Farms in Ecuador. PLoS ONE 2019, 14, e0207567. [Google Scholar] [CrossRef] [PubMed]
- Aworh, M.K.; Kwaga, J.K.P.; Hendriksen, R.S.; Okolocha, E.C.; Thakur, S. Genetic Relatedness of Multidrug Resistant Escherichia coli Isolated from Humans, Chickens and Poultry Environments. Antimicrob. Resist. Infect. Control 2021, 10, 58. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Calvopina, M.; Izurieta, R.; Villacres, I.; Kawahara, R.; Sasaki, M.; Yamamoto, M. Colistin-Resistant Escherichia coli with Mcr Genes in the Livestock of Rural Small-Scale Farms in Ecuador. BMC Res. Notes 2019, 12, 519. [Google Scholar] [CrossRef]
- Loayza-Villa, F.; Salinas, L.; Tijet, N.; Villavicencio, F.; Tamayo, R.; Salas, S.; Rivera, R.; Villacis, J.; Satan, C.; Ushiña, L.; et al. Diverse Escherichia coli Lineages from Domestic Animals Carrying Colistin Resistance Gene Mcr-1 in an Ecuadorian Household. J. Glob. Antimicrob. Resist. 2020, 22, 63–67. [Google Scholar] [CrossRef]
- Bastidas-Caldes, C.; Guerrero-Freire, S.; Ortuño-Gutiérrez, N.; Sunyoto, T.; Gomes-Dias, C.A.; Ramírez, M.S.; Calero-Cáceres, W.; Harries, A.D.; Rey, J.; de Waard, J.H.; et al. Colistin Resistance in Escherichia coli and Klebsiella pneumoniae in Humans and Backyard Animals in Ecuador. Rev. Panam. Salud Pública 2023, 47, e48. [Google Scholar] [CrossRef]
- Bastidas-Caldes, C.; Cisneros-Vásquez, E.; Zambrano, A.; Mosquera-Maza, A.; Calero-Cáceres, W.; Rey, J.; Yamamoto, Y.; Yamamoto, M.; Calvopiña, M.; de Waard, J.H. Co-Harboring of Beta-Lactamases and Mcr-1 Genes in Escherichia coli and Klebsiella pneumoniae from Healthy Carriers and Backyard Animals in Rural Communities in Ecuador. Antibiotics 2023, 12, 856. [Google Scholar] [CrossRef]
- Murray, M.; Salvatierra, G.; Dávila-Barclay, A.; Ayzanoa, B.; Castillo-Vilcahuaman, C.; Huang, M.; Pajuelo, M.J.; Lescano, A.G.; Cabrera, L.; Calderón, M.; et al. Market Chickens as a Source of Antibiotic-Resistant Escherichia coli in a Peri-Urban Community in Lima, Peru. Front. Microbiol. 2021, 12, 635871. [Google Scholar] [CrossRef]
- Carhuaricra, D.; Duran Gonzales, C.G.; Rodríguez Cueva, C.L.; Ignacion León, Y.; Silvestre Espejo, T.; Marcelo Monge, G.; Rosadio Alcántara, R.H.; Lincopan, N.; Espinoza, L.L.; Maturrano Hernández, L. Occurrence and Genomic Characterization of Mcr-1-Harboring Escherichia coli Isolates from Chicken and Pig Farms in Lima, Peru. Antibiotics 2022, 11, 1781. [Google Scholar] [CrossRef]
- Fernandes, M.; Moura, Q.; Sartori, L.; Silva, K.; Cunha, M.; Esposito, F.; Lopes, R.; Otutumi, L.; Gonçalves, D.; Dropa, M.; et al. Silent Dissemination of Colistin-Resistant Escherichia coli in South America Could Contribute to the Global Spread of the Mcr-1 Gene. Eurosurveillance 2016, 21, 30214. [Google Scholar] [CrossRef] [PubMed]
- Lentz, S.A.; de Lima-Morales, D.; Cuppertino, V.M.; de Nunes, L.S.; da Motta, A.S.; Zavascki, A.P.; Barth, A.L.; Martins, A.F. Letter to the Editor: Escherichia coli Harbouring Mcr-1 Gene Isolated from Poultry Not Exposed to Polymyxins in Brazil. Eurosurveillance 2016, 21, 6. [Google Scholar] [CrossRef] [PubMed]
- Moreno, L.Z.; Gomes, V.T.M.; Moreira, J.; de Oliveira, C.H.; Peres, B.P.; Silva, A.P.S.; Thakur, S.; La Ragione, R.M.; Moreno, A.M. First Report of Mcr-1-Harboring Salmonella Enterica Serovar Schwarzengrund Isolated from Poultry Meat in Brazil. Diagn. Microbiol. Infect. Dis. 2019, 93, 376–379. [Google Scholar] [CrossRef]
- Barbieri, N.L.; Pimenta, R.L.; de Melo, D.A.; Nolan, L.K.; de Souza, M.M.S.; Logue, C.M. Mcr-1 Identified in Fecal Escherichia coli and Avian Pathogenic E. coli (APEC) From Brazil. Front. Microbiol. 2021, 12, 659613. [Google Scholar] [CrossRef]
- Pontes, L.D.S.; Pimenta, R.; Silveira, M.C.; Tavares-Teixeira, C.B.; Pereira, N.F.; da Conceiçao Neto, O.C.; de Oliveira Santos, I.C.; da Costa, B.S.; Carvalho-Assef, A.P.D.A.; de Souza, M.M.S.; et al. Letter to the Editor: Escherichia Fergusonii Harboring IncHI2 Plasmid Containing Mcr-1 Gene-A Novel Reservoir for Colistin Resistance in Brazil. Microb. Drug Resist. 2021, 27, 721–725. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, J.E.; Figueroa Espinosa, R.A.; Redondo, L.M.; Cejas, D.; Gutkind, G.O.; Chacana, P.A.; Di Conza, J.A.; Fernández-Miyakawa, M.E. Plasmid-Mediated Colistin Resistance in Escherichia coli Recovered from Healthy Poultry. Rev. Argent. Microbiol. 2017, 49, 297–298. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, J.E.; Faccone, D.; Tijet, N.; Gomez, S.; Corso, A.; Fernández-Miyakawa, M.E.; Melano, R.G. Characterization of Escherichia coli carrying Mcr-1-Plasmids Recovered from Food Animals from Argentina. Front. Cell. Infect. Microbiol. 2019, 9, 41. [Google Scholar] [CrossRef] [PubMed]
- Nesporova, K.; Valcek, A.; Papagiannitsis, C.; Kutilova, I.; Jamborova, I.; Davidova-Gerzova, L.; Bitar, I.; Hrabak, J.; Literak, I.; Dolejska, M. Multi-Drug Resistant Plasmids with Esbl/Ampc and Mcr-5.1 in Paraguayan Poultry Farms: The Linkage of Antibiotic Resistance and Hatcheries. Microorganisms 2021, 9, 866. [Google Scholar] [CrossRef]
- Maciuca, I.E.; Cummins, M.L.; Cozma, A.P.; Rimbu, C.M.; Guguianu, E.; Panzaru, C.; Licker, M.; Szekely, E.; Flonta, M.; Djordjevic, S.P.; et al. Genetic Features of Mcr-1 Mediated Colistin Resistance in CMY-2-Producing Escherichia coli From Romanian Poultry. Front. Microbiol. 2019, 10, 2267. [Google Scholar] [CrossRef]
- Bortolaia, V.; Ronco, T.; Romascu, L.; Nicorescu, I.; Milita, N.M.; Vaduva, A.M.; Leekitcharoenphon, P.; Kjeldgaard, J.S.; Hansen, I.M.; Svendsen, C.A.; et al. Co-Localization of Carbapenem (BlaOXA-162) and Colistin (Mcr-1) Resistance Genes on a Transferable IncHI2 Plasmid in Escherichia coli of Chicken Origin. J. Antimicrob. Chemother. 2021, 76, 3063–3065. [Google Scholar] [CrossRef]
- Mišić, D.; Kiskaroly, F.; Szostak, M.P.; Cabal, A.; Ruppitsch, W.; Bernreiter-Hofer, T.; Milovanovic, V.; Feßler, A.T.; Allerberger, F.; Spergser, J.; et al. The First Report of Mcr-1-Carrying Escherichia coli Originating from Animals in Serbia. Antibiotics 2021, 10, 1063. [Google Scholar] [CrossRef]
- Ortega-Paredes, D.; Barba, P.; Zurita, J. Colistin-Resistant Escherichia coli Clinical Isolate Harbouring the Mcr-1 Gene in Ecuador. Epidemiol. Infect. 2016, 144, 2967–2970. [Google Scholar] [CrossRef]
- Gelbíčová, T.; Baráková, A.; Florianová, M.; Jamborová, I.; Zelendová, M.; Pospíšilová, L.; Koláčková, I.; Karpíšková, R. Dissemination and Comparison of Genetic Determinants of Mcr-Mediated Colistin Resistance in Enterobacteriaceae via Retailed Raw Meat Products. Front. Microbiol. 2019, 10, 2824. [Google Scholar] [CrossRef] [PubMed]
- Nishino, Y.; Shimojima, Y.; Suzuki, Y.; Ida, M.; Fukui, R.; Kuroda, S.; Hirai, A.; Sadamasu, K. Detection of the Mcr-1 Gene in Colistin-Resistant Escherichia coli from Retail Meat in Japan. Microbiol. Immunol. 2017, 61, 554–557. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, F.; Zhu, B.; Gao, G.F. Metagenomic data screening reveals the distribution of mobilized resistance genes tet (X), mcr and carbapenemase in animals and humans. J. Infect. 2020, 80, 121–142. [Google Scholar] [CrossRef] [PubMed]
- Skov, R.L.; Monnet, D.L. Plasmid-Mediated Colistin Resistance (Mcr-1 Gene): Three Months Later, the Story Unfolds. Eurosurveillance 2016, 21, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Von Wintersdorff, C.J.H.; Wolffs, P.F.G.; Van Niekerk, J.M.; Beuken, E.; Van Alphen, L.B.; Stobberingh, E.E.; Oude Lashof, A.M.L.; Hoebe, C.J.P.A.; Savelkoul, P.H.M.; Penders, J. Detection of the Plasmid-Mediated Colistin-Resistance Gene Mcr-1 in Faecal Metagenomes of Dutch Travellers. J. Antimicrob. Chemother. 2016, 71, 3416–3419. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global Trends in Antimicrobial Use in Food Animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef]
- Yam, E.L.Y.; Hsu, L.Y.; Yap, E.P.H.; Yeo, T.W.; Lee, V.; Schlundt, J.; Lwin, M.O.; Limmathurotsakul, D.; Jit, M.; Dedon, P.; et al. Antimicrobial Resistance in the Asia Pacific Region: A Meeting Report. Antimicrob. Resist. Infect. Control 2019, 8, 202. [Google Scholar] [CrossRef]
- Gao, Y.; Lu, C.; Shen, D.; Liu, J.; Ma, Z.; Yang, B.; Ling, W.; Waigi, M.G. Elimination of the Risks of Colistin Resistance Gene (Mcr-1) in Livestock Manure during Composting. Environ. Int. 2019, 126, 61–68. [Google Scholar] [CrossRef]
- Li, J.; Shi, X.; Yin, W.; Wang, Y.; Shen, Z.; Ding, S.; Wang, S. A Multiplex SYBR Green Real-Time PCR Assay for the Detection of Three Colistin Resistance Genes from Cultured Bacteria, Feces, and Environment Samples. Front. Microbiol. 2017, 8, 2078. [Google Scholar] [CrossRef]
- Valiakos, G.; Kapna, I. Colistin Resistant Mcr Genes Prevalence in Livestock Animals (Swine, Bovine, Poultry) from a Multinational Perspective. A Systematic Review. Vet. Sci. 2021, 8, 265. [Google Scholar] [CrossRef]
- Binsker, U.; Käsbohrer, A.; Hammerl, J.A. Global Colistin Use: A Review of the Emergence of Resistant Enterobacterales and the Impact on Their Genetic Basis. FEMS Microbiol. Rev. 2022, 46, fuab049. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.R.; Wu, Y. China Bans Colistin as a Feed Additive for Animals. Lancet Infect. Dis. 2016, 16, 1102–1103. [Google Scholar] [CrossRef] [PubMed]
- Webb, H.E.; Angulo, F.J.; Granier, S.A.; Scott, H.M.; Loneragan, G.H. Illustrative Examples of Probable Transfer of Resistance Determinants from Food Animals to Humans: Streptothricins, Glycopeptides, and Colistin. F1000Research 2017, 6, 1805. [Google Scholar] [CrossRef]
- Shen, Y.; Xu, C.; Sun, Q.; Schwarz, S.; Ou, Y.; Yang, L.; Huang, Z.; Eichhorn, I.; Walsh, T.R.; Wang, Y.; et al. Prevalence and Genetic Analysis of Mcr-3-Positive Aeromonas Species from Humans, Retail Meat, and Environmental Water Samples. Antimicrob. Agents Chemother. 2018, 62, e00404-18. [Google Scholar] [CrossRef] [PubMed]
- Borowiak, M.; Baumann, B.; Fischer, J.; Thomas, K.; Deneke, C.; Hammerl, J.A.; Szabo, I.; Malorny, B. Development of a Novel Mcr-6 to Mcr-9 Multiplex PCR and Assessment of Mcr-1 to Mcr-9 Occurrence in Colistin-Resistant Salmonella enterica Isolates from Environment, Feed, Animals and Food (2011–2018) in Germany. Front. Microbiol. 2020, 11, 80. [Google Scholar] [CrossRef] [PubMed]
- Dandachi, I.; Chaddad, A.; Hanna, J.; Matta, J.; Daoud, Z. Understanding the Epidemiology of Multi-Drug Resistant Gram-Negative Bacilli in the Middle East Using a One Health Approach. Front. Microbiol. 2019, 10, 1941. [Google Scholar] [CrossRef]
- Feng, S.; Liang, W.; Li, J.; Chen, Y.; Zhou, D.; Liang, L.; Lin, D.; Li, Y.; Zhao, H.; Du, H.; et al. MCR-1-Dependent Lipid Remodelling Compromises the Viability of Gram-Negative Bacteria. Emerg. Microbes Infect. 2022, 11, 1236–1249. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Zhong, L.L.; Yang, Y.; Doi, Y.; Paterson, D.L.; Stoesser, N.; Ma, F.; El-Sayed Ahmed, M.A.E.G.; Feng, S.; Huang, S.; et al. Dynamics of Mcr-1 Prevalence and Mcr-1-Positive Escherichia coli after the Cessation of Colistin Use as a Feed Additive for Animals in China: A Prospective Cross-Sectional and Whole Genome Sequencing-Based Molecular Epidemiological Study. Lancet Microbe 2020, 1, e34–e43. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, Y.; Lu, J.; Wang, Q.; Cui, Y.; Wang, Y.; Quan, J.; Zhao, D.; Du, X.; Liu, H.; et al. Clinical Relevance and Plasmid Dynamics of Mcr-1-Positive Escherichia coli in China: A Multicentre Case-Control and Molecular Epidemiological Study. Lancet Microbe 2020, 1, e24–e33. [Google Scholar] [CrossRef]
- Snesrud, E.; McGann, P.; Chandler, M. The Birth and Demise of the ISApl1-Mcr-1-ISApl1 Composite Transposon: The Vehicle for Transferable Colistin Resistance. mBio 2018, 9, e02381-17. [Google Scholar] [CrossRef]
- Shen, C.; Zhong, L.L.; Ma, F.; El-Sayed Ahmed, M.A.E.G.; Doi, Y.; Zhang, G.; Liu, Y.; Huang, S.; Li, H.Y.; Zhang, L.; et al. Genomic Patterns and Characterizations of Chromosomally-Encoded Mcr-1 in Escherichia coli Populations. Gut Pathog. 2020, 12, 55. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, Y.; Ji, R.Y.; Wang, Z.Y.; Lu, M.J.; Wu, H.; Mei, C.Y.; Li, Q.C.; Jiao, X. Colistin- and Tigecycline-Resistant CTX-M-14-Producing Salmonella enterica Serovar Kentucky ST198 from Retail Chicken Meat, China. Int. J. Antimicrob. Agents 2022, 59, 106504. [Google Scholar] [CrossRef]
- Chang, M.X.; Zhang, J.; Zhang, J.F.; Ding, X.M.; Lu, Y.; Zhang, J.; Li, R.; Jiang, H.X. Formation, Transmission, and Dynamic Evolution of a Multidrug-Resistant Chromosomally Integrated Plasmid in Salmonella spp. Front. Microbiol. 2022, 13, 846954. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, A.; Ugarte-Ruiz, M.; Hernández, M.; Miguela-Villoldo, P.; Rodríguez-Lázaro, D.; Domínguez, L.; Quesada, A. Involvement of Hpap2 and Dgka Genes in Colistin Resistance Mediated by Mcr Determinants. Antibiotics 2020, 9, 531. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, L.; Wang, Z.; Bai, L.; Li, R. Emergence of Mcr-8.2-Bearing Klebsiella quasipneumoniae of Animal Origin. J. Antimicrob. Chemother. 2019, 74, 2814–2817. [Google Scholar] [CrossRef] [PubMed]
- Rhouma, M.; Letellier, A. Extended-Spectrum β-Lactamases, Carbapenemases and the Mcr-1 Gene: Is There a Historical Link? Int. J. Antimicrob. Agents 2017, 49, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Zhang, J.; Lu, T.; Zhang, K. Metagenomics Revealed the Mobility and Hosts of Antibiotic Resistance Genes in Typical Pesticide Wastewater Treatment Plants. Sci. Total Environ. 2022, 817, 153033. [Google Scholar] [CrossRef]
- Lu, X.; Xiao, X.; Liu, Y.; Li, R.; Wang, Z. Emerging Opportunity and Destiny of Mcr-1—And Tet (X4)-Coharboring Plasmids in Escherichia coli. Microbiol. Spectr. 2021, 9, e01520-21. [Google Scholar] [CrossRef]
- Kieffer, N.; Royer, G.; Decousser, J.-W.; Bourrel, A.-S.; Palmieri, M.; Ortiz De La Rosa, J.-M.; Jacquier, H.; Denamur, E.; Nordmann, P.; Poirel, L. Mcr-9, an Inducible Gene Encoding an Acquired Phosphoethanolamine Transferase in Escherichia coli, and Its Origin. Antimicrob. Agents Chemother. 2019, 63, e00965-19. [Google Scholar] [CrossRef]
- Ohji, G.; Doi, A.; Yamamoto, S.; Iwata, K. Is De-Escalation of Antimicrobials Effective? A Systematic Review and Meta-Analysis. Int. J. Infect. Dis. 2016, 49, 71–79. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, H.; Lao, G.; Zhou, Z.; Liu, Z.; Cai, J.; Sun, Q. Identification of Mobile Colistin Resistance Gene Mcr-10 in Disinfectant and Antibiotic Resistant Escherichia coli from Disinfected Tableware. Antibiotics 2022, 11, 883. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, G.R.; Zhao, K.; He, X.; Chen, S.; Liu, S.; Mustafa, A.; He, L.; Yang, Y.; Yu, X.; Penttinen, P. Heavy Metal Resistance in Salmonella Typhimurium and Its Association with Disinfectant and Antibiotic Resistance. Front. Microbiol. 2021, 12, 2120. [Google Scholar] [CrossRef]
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Rasool, M.H.; Shahid, A.; et al. Antibiotic Resistance: One Health One World Outlook. Front. Cell. Infect. Microbiol. 2021, 11, 771510. [Google Scholar] [CrossRef] [PubMed]
- Virolle, C.; Goldlust, K.; Djermoun, S.; Bigot, S.; Lesterlin, C. Plasmid Transfer by Conjugation in Gram-Negative Bacteria: From the Cellular to the Community Level. Genes 2020, 11, 1239. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Yi, L.X.; Yu, L.F.; Wang, J.; Liu, Y.; Chen, X.; Lv, L.; Yang, J.; Liu, J.H. Fitness Advantage of Mcr-1-Bearing IncI2 and IncX4 Plasmids in Vitro. Front. Microbiol. 2018, 9, 331. [Google Scholar] [CrossRef]
- Yang, Q.E.; Tansawai, U.; Andrey, D.O.; Wang, S.; Wang, Y.; Sands, K.; Kiddee, A.; Assawatheptawee, K.; Bunchu, N.; Hassan, B.; et al. Environmental Dissemination of Mcr-1 Positive Enterobacteriaceae by Chrysomya Spp. (Common Blowfly): An Increasing Public Health Risk. Environ. Int. 2019, 122, 281–290. [Google Scholar] [CrossRef]
- Yi, L.; Durand, R.; Grenier, F.; Yang, J.; Yu, K.; Burrus, V.; Liu, J.H. PixR, a Novel Activator of Conjugative Transfer of IncX4 Resistance Plasmids, Mitigates the Fitness Cost of Mcr-1 Carriage in Escherichia coli. mBio 2022, 13, e03209-21. [Google Scholar] [CrossRef]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J.D.D. Global Extraintestinal Pathogenic Escherichia coli (Expec) Lineages. Clin. Microbiol. Rev. 2019, 32, e00135-18. [Google Scholar] [CrossRef]
- Massella, E.; Giacometti, F.; Bonilauri, P.; Reid, C.J.; Djordjevic, S.P.; Merialdi, G.; Bacci, C.; Fiorentini, L.; Massi, P.; Bardasi, L.; et al. Antimicrobial Resistance Profile and Expec Virulence Potential in Commensal Escherichia coli of Multiple Sources. Antibiotics 2021, 10, 351. [Google Scholar] [CrossRef]
- Li, W.; Liu, Z.; Yin, W.; Yang, L.; Qiao, L.; Song, S.; Ling, Z.; Zheng, R.; Wu, C.; Wang, Y.; et al. Mcr Expression Conferring Varied Fitness Costs on Host Bacteria and Affecting Bacteria Virulence. Antibiotics 2021, 10, 872. [Google Scholar] [CrossRef]
- Lu, J.; Quan, J.; Zhao, D.; Wang, Y.; Yu, Y.; Zhu, J. Prevalence and Molecular Characteristics of Mcr-1 Gene in Salmonella Typhimurium in a Tertiary Hospital of Zhejiang Province. Infect. Drug Resist. 2019, 12, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Feng, Y. MCR-1-Producing Salmonella typhimurium ST34 Links Animal Foods to Human Community Infections. eBioMedicine 2019, 42, P10–P11. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, X.; Forcelledo, L.; Balboa-Palomino, S.; Fernández, J.; Rodicio, M.R. Nosocomial Pneumonia Caused in an Immunocompetent Patient by the Emergent Monophasic ST34 Variant of Salmonella enterica Serovar Typhimurium: Treatment-Associated Selection of Fluoroquinolone and Piperacillin/Tazobactam Resistance. Antibiotics 2022, 11, 303. [Google Scholar] [CrossRef]
- Ray, M.; Manjunath, A.; Halami, P.M. Prevalence of Polymyxin Resistance through the Food Chain, the Global Crisis. J. Antibiot. 2022, 75, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Pathak, A.; Kumar, A.; Rahman, M.; Singh, A.; Gonzalez-Zorn, B.; Prasad, K.N. Emergence of Chromosome-Borne Colistin Resistance Gene Mcr-1 in Clinical Isolates of Klebsiella pneumoniae from India. Antimicrob. Agents Chemother. 2018, 62, e01885-17. [Google Scholar] [CrossRef]
- Hameed, F.; Khan, M.A.; Muhammad, H.; Sarwar, T.; Bilal, H.; Rehman, T.U. Plasmid-Mediated Mcr-1 Gene in Acinetobacter Baumannii and Pseudomonas Aeruginosa: First Report from Pakistan. Rev. Soc. Bras. Med. Trop. 2019, 52, e20190237. [Google Scholar] [CrossRef]
- Kassem, I.I.; Osman, M. A Brewing Storm: The Impact of Economic Collapse on the Access to Antimicrobials in Lebanon. J. Glob. Antimicrob. Resist. 2022, 29, 313–315. [Google Scholar] [CrossRef]
- Al-Kadmy, I.M.S.; Ibrahim, S.A.; Al-Saryi, N.; Aziz, S.N.; Besinis, A.; Hetta, H.F. Prevalence of Genes Involved in Colistin Resistance in Acinetobacter Baumannii: First Report from Iraq. Microb. Drug Resist. 2020, 26, 616–622. [Google Scholar] [CrossRef]
- Güzel, M.; Avşaroğlu, M.D.; Soyer, Y. Determination of Colistin Resistance in Escherichia coli Isolates from Foods in Turkey, 2011–2015. Food and Health 2020, 6, 160–169. [Google Scholar] [CrossRef]
- Kansak, N.; Aksaray, S.; Aslan, M.; Adaleti, R.; Gönüllü, N. Detection of Colistin Resistance among Multidrug-Resistant Klebsiella pneumoniae and Escherichia coli Clinical Isolates in Turkey. Acta Microbiol. Immunol. Hung. 2021, 68, 99–106. [Google Scholar] [CrossRef]
- Pishnian, Z.; Haeili, M.; Feizi, A. Prevalence and Molecular Determinants of Colistin Resistance among Commensal Enterobacteriaceae Isolated from Poultry in Northwest of Iran. Gut Pathog. 2019, 11, 2. [Google Scholar] [CrossRef] [PubMed]
- Ilbeigi, K.; Askari Badouei, M.; Vaezi, H.; Zaheri, H.; Aghasharif, S.; Kafshdouzan, K. Molecular survey of mcr1 and mcr2 plasmid mediated colistin resistance genes in Escherichia coli isolates of animal origin in Iran. BMC Res. Notes 2021, 14, 107. [Google Scholar] [CrossRef] [PubMed]
- Nhung, N.T.; Cuong, N.v.; Thwaites, G.; Carrique-Mas, J. Antimicrobial Usage and Antimicrobial Resistance in Animal Production in Southeast Asia: A Review. Antibiotics 2016, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Thongmalayvong, B.; Akkhavong, K.; Somphavong, S.; Paboriboune, P.; Khounsy, S.; Morand, S.; Rolain, J.-M. Clonal Transmission of a Colistin-Resistant Escherichia coli from a Domesticated Pig to a Human in Laos. J. Antimicrob. Chemother. 2015, 70, 3402–3404. [Google Scholar]
- Sudatip, D.; Chasiri, K.; Kritiyakan, A.; Phanprasit, W.; Thinphovong, C.; Tiengrim, S.; Thamlikitkul, V.; Abdallah, R.; Baron, S.A.; Rolain, J.M.; et al. A One Health Approach to Assessing Occupational Exposure to Antimicrobial Resistance in Thailand: The FarmResist Project. PLoS ONE 2021, 16, e0245250. [Google Scholar] [CrossRef]
- Devan, S.S.; Aklilu, E.; Hamdan, R.H.; Lemlem, M.; Zakaria, Z. Detection of colistin-resistant Escherichia coli isolated from broiler chickens in Kelantan, Malaysia. Trop Biomed. 2022, 39, 197–202. [Google Scholar] [CrossRef]
- Umali, D.v. Antimicrobial Susceptibility Profiles and Detection of Colistin Resistance Mcr-1 Gene in Salmonella Enterica Isolates from Chicken Eggs in Metro Manila, Philippines. Philipp. J. Vet. Med. 2019, 56, 89–91. [Google Scholar]
- Founou, L.L.; Founou, R.C.; Essack, S.Y. Antibiotic Resistance in the Food Chain: A Developing Country-Perspective. Front. Microbiol. 2016, 7, 1881. [Google Scholar] [CrossRef]
- Chabou, S.; Leangapichart, T.; Okdah, L.; Le Page, S.; Hadjadj, L.; Rolain, J.M. Real-Time Quantitative PCR Assay with Taqman® Probe for Rapid Detection of MCR-1 Plasmid-Mediated Colistin Resistance. New Microbes New. Infect. 2016, 13, 71–74. [Google Scholar] [CrossRef]
- Di Francesco, A.; Salvatore, D.; Sakhria, S.; Bertelloni, F.; Catelli, E.; Ben Yahia, S.; Tlatli, A. Colistin Resistance Genes in Broiler Chickens in Tunisia. Animals 2023, 13, 1409. [Google Scholar] [CrossRef]
- Dhaouadi, S.; Romdhani, A.; Bouglita, W.; Chedli, S.; Chaari, S.; Soufi, L.; Cherif, A.; Mnif, W.; Abbassi, M.S.; Elandoulsi, R.B. High Biofilm-Forming Ability and Clonal Dissemination among Colistin-Resistant Escherichia coli Isolates Recovered from Cows with Mastitis, Diarrheic Calves, and Chickens with Colibacillosis in Tunisia. Life 2023, 13, 299. [Google Scholar] [CrossRef] [PubMed]
- Cummins, M.L.; Reid, C.J.; Chowdhury, P.R.; Bushell, R.N.; Esbert, N.; Tivendale, K.A.; Noormohammadi, A.H.; Islam, S.; Marenda, M.S.; Browning, G.F.; et al. Whole Genome Sequence Analysis of Australian Avian Pathogenic Escherichia coli That Carry the Class 1 Integrase Gene. Microb. Genom. 2019, 5, e000250. [Google Scholar] [CrossRef] [PubMed]
- Zakaria, A.S.; Edward, E.A.; Mohamed, N.M. Genomic Insights into a Colistin-Resistant Uropathogenic Escherichia coli Strain of O23:H4-St641 Lineage Harboring Mcr-1.1 on a Conjugative Inchi2 Plasmid from Egypt. Microorganisms 2021, 9, 799. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.A.A.; Hamed, E.A.; Abdelaty, M.F.; Sorour, H.K.; Badr, H.; Hassan, W.M.; Shalaby, A.G.; Mohamed, A.A.; Soliman, M.A.; Roshdy, H. Distribution pattern of antibiotic resistance genes in Escherichia coli isolated from colibacillosis cases in broiler farms of Egypt. Vet World 2023, 16, 1–11. [Google Scholar] [CrossRef]
- Theobald, S.; Etter, E.M.C.; Gerber, D.; Abolnik, C. Antimicrobial Resistance Trends in Escherichia coli in South African Poultry: 2009–2015. Foodborne Pathog. Dis. 2019, 16, 652–660. [Google Scholar]
- Labuschagne, Q.; Schellack, N.; Gous, A.; Bronkhorst, E.; Schellack, G.; Van Tonder, L.; Truter, A.; Smith, C.; Lancaster, R.; Kolman, S. COLISTIN: Adult and Paediatric Guideline for South Africa. S. Afr. J. Infect. Dis. 2016, 31, 3–7. [Google Scholar] [CrossRef]
- Ngbede, E.O.; Adekanmbi, F.; Poudel, A.; Kalalah, A.; Kelly, P.; Yang, Y.; Adamu, A.M.; Daniel, S.T.; Adikwu, A.A.; Akwuobu, C.A.; et al. Concurrent Resistance to Carbapenem and Colistin Among Enterobacteriaceae Recovered from Human and Animal Sources in Nigeria Is Associated with Multiple Genetic Mechanisms. Front. Microbiol. 2021, 12, 740348. [Google Scholar] [CrossRef]
- Mendelson, M.; Brink, A.; Gouws, J.; Mbelle, N.; Naidoo, V.; Pople, T.; Schellack, N.; van Vuuren, M.; Rees, H.; Banoo, S.; et al. The One Health Stewardship of Colistin as an Antibiotic of Last Resort for Human Health in South Africa. Lancet Infect. Dis. 2018, 18, e288–e294. [Google Scholar] [CrossRef]
- Büdel, T.; Kuenzli, E.; Clément, M.; Bernasconi, O.J.; Fehr, J.; Mohammed, A.H.; Hassan, N.K.; Zinsstag, J.; Hatz, C.; Endimiani, A. Polyclonal Gut Colonization with Extended-Spectrum Cephalosporin-and/or Colistin-Resistant Enterobacteriaceae: A Normal Status for Hotel Employees on the Island of Zanzibar, Tanzania. J. Antimicrob. Chemother. 2019, 74, 2880–2890. [Google Scholar] [CrossRef] [PubMed]
- Dávalos-Almeyda, M.; Guerrero, A.; Medina, G.; Dávila-Barclay, A.; Salvatierra, G.; Calderón, M.; Gilman, R.H.; Tsukayama, P. Antibiotic Use and Resistance Knowledge Assessment of Personnel on Chicken Farms with High Levels of Antimicrobial Resistance: A Cross-Sectional Survey in Ica, Peru. Antibiotics 2022, 11, 190. [Google Scholar] [CrossRef]
- Rabello, R.F.; Bonelli, R.R.; Penna, B.A.; Albuquerque, J.P.; Souza, R.M.; Cerqueira, A.M.F. Antimicrobial Resistance in Farm Animals in Brazil: An Update Overview. Animals 2020, 10, 552. [Google Scholar] [CrossRef] [PubMed]
- Rau, R.B.; De Lima-Morales, D.; Wink, P.L.; Ribeiro, A.R.; Martins, A.F.; Barth, A.L. Emergence of Mcr-1 Producing Salmonella Enterica Serovar Typhimurium from Retail Meat: First Detection in Brazil. Foodborne Pathog. Dis. 2018, 15, 58–59. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Zeng, X.; Hinenoya, A.; Lin, J. MCR-1 Confers Cross-Resistance to Bacitracin, a Widely Used In-Feed Antibiotic. mSphere 2018, 3, e00411-18. [Google Scholar] [CrossRef]
- EUCAST. EUCAST. Available online: https://eucast.org/ (accessed on 7 September 2020).
- Quiroga, C.; Nastro, M.; Di Conza, J. Current Scenario of Plasmid-Mediated Colistin Resistance in Latin America. Rev. Argent. Microbiol. 2019, 51, 93–100. [Google Scholar] [CrossRef]
- Webb, H.E.; Kim, J.Y.; Tagg, K.A.; Kapsak, C.J.; Tobolowsky, F.; Birhane, M.G.; Francois Watkins, L.; Folster, J.P. Fourteen Mcr-1-Positive Salmonella enterica Isolates Recovered from Travelers Returning to the United States from the Dominican Republic. Microbiol. Resour. Announc. 2022, 11, e00118-22. [Google Scholar] [CrossRef]
- Andrade, F.F.; Silva, D.; Rodrigues, A.; Pina-Vaz, C. Colistin Update on Its Mechanism of Action and Resistance, Present and Future Challenges. Microorganisms 2020, 8, 1716. [Google Scholar] [CrossRef]
- Miranda, C.; Igrejas, G.; Capita, R.; Alonso-Calleja, C.; Poeta, P. Worldwide Colistin Use and Spread of Resistant- Enterobacteriaceae in Animal Production. In The Global Antimicrobial Resistance Epidemic–Innovative Approaches and Cutting-Edge Solutions [200Working Title]; IntechOpen: London, UK, 2022. [Google Scholar]
- Ageevets, V.; Lazareva, I.; Mrugova, T.; Gostev, V.; Lobzin, Y.; Sidorenko, S. IncX4 Plasmids Harbouring Mcr-1 Genes: Further Dissemination. J. Glob. Antimicrob. Resist. 2019, 18, 166–167. [Google Scholar] [CrossRef] [PubMed]
- Tarabai, H.; Valcek, A.; Jamborova, I.; Vazhov, S.V.; Karyakin, I.V.; Raab, R.; Literak, I.; Dolejska, M. Plasmid-Mediated mcr-1 Colistin Resistance in Escherichia coli from a Black Kite in Russia. Antimicrob Agents Chemother 2019, 63, e01266-19. [Google Scholar] [CrossRef]
- Jovčić, B.; Novović, K.; Filipić, B.; Velhner, M.; Todorović, D.; Matović, K.; Rašić, Z.; Nikolić, S.; Kiškarolj, F.; Kojić, M. Genomic Characteristics of Colistin-Resistant Salmonella Enterica Subsp. Enterica Serovar Infantis from Poultry Farms in the Republic of Serbia. Antibiotics 2020, 9, 886. [Google Scholar] [CrossRef] [PubMed]
- Okpala, C.O.R.; Anyanwu, M.U.; Łukanko, S.; Nwobi, O.C.; Korzeniowska, M.; Ezeonu, I.M. Animal-Food-Human Antimicrobial Resistance Fundamentals, Prevention Mechanisms and Global Surveillance Trends: A Terse Review. Appl. Food Biotechnol. 2021, 8, 89–102. [Google Scholar] [CrossRef]
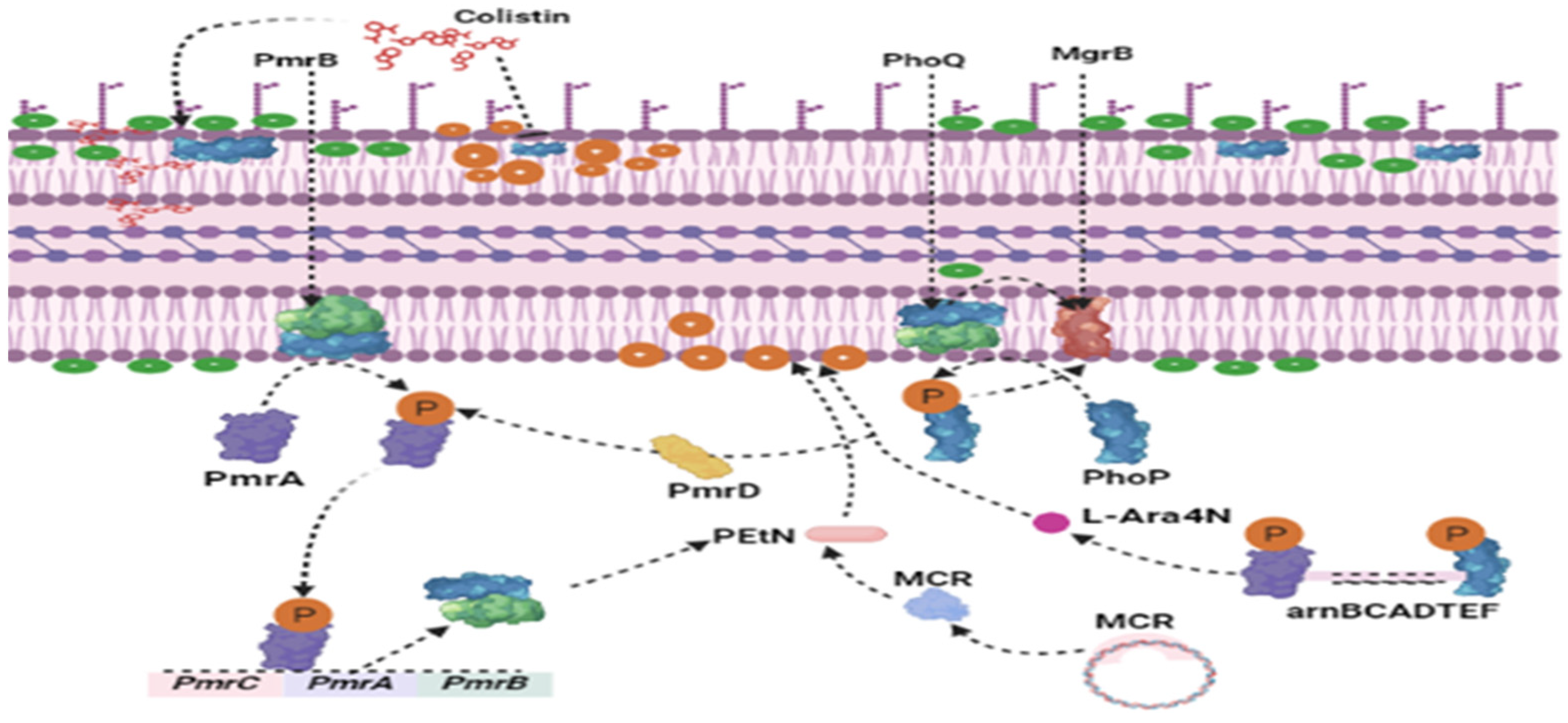
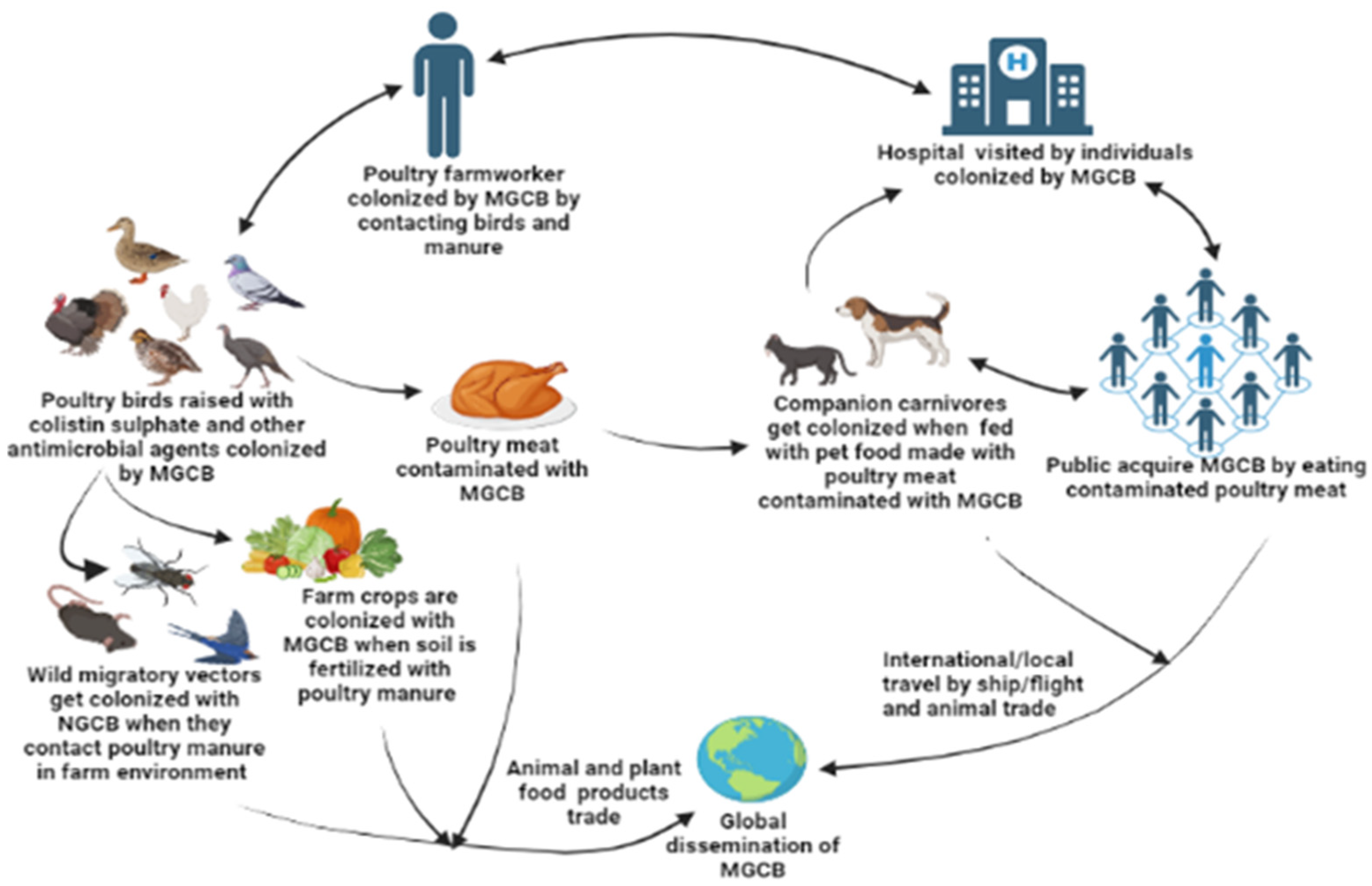
 : Poultry bird;
: Poultry bird;  : poultry meat;
: poultry meat;  : egg;
: egg;  : poultry feed;
: poultry feed;  : persons in contact with poultry bird/poultry product;
: persons in contact with poultry bird/poultry product;  : poultry environment (litter/manure, sewage, vectors, soil, or water). This map was created with https://www.mapchart.net/ (accessed on 30 May 2023).
: poultry environment (litter/manure, sewage, vectors, soil, or water). This map was created with https://www.mapchart.net/ (accessed on 30 May 2023).
 : Poultry bird;
: Poultry bird;  : poultry meat;
: poultry meat;  : egg;
: egg;  : poultry feed;
: poultry feed;  : persons in contact with poultry bird/poultry product;
: persons in contact with poultry bird/poultry product;  : poultry environment (litter/manure, sewage, vectors, soil, or water). This map was created with https://www.mapchart.net/ (accessed on 30 May 2023).
: poultry environment (litter/manure, sewage, vectors, soil, or water). This map was created with https://www.mapchart.net/ (accessed on 30 May 2023).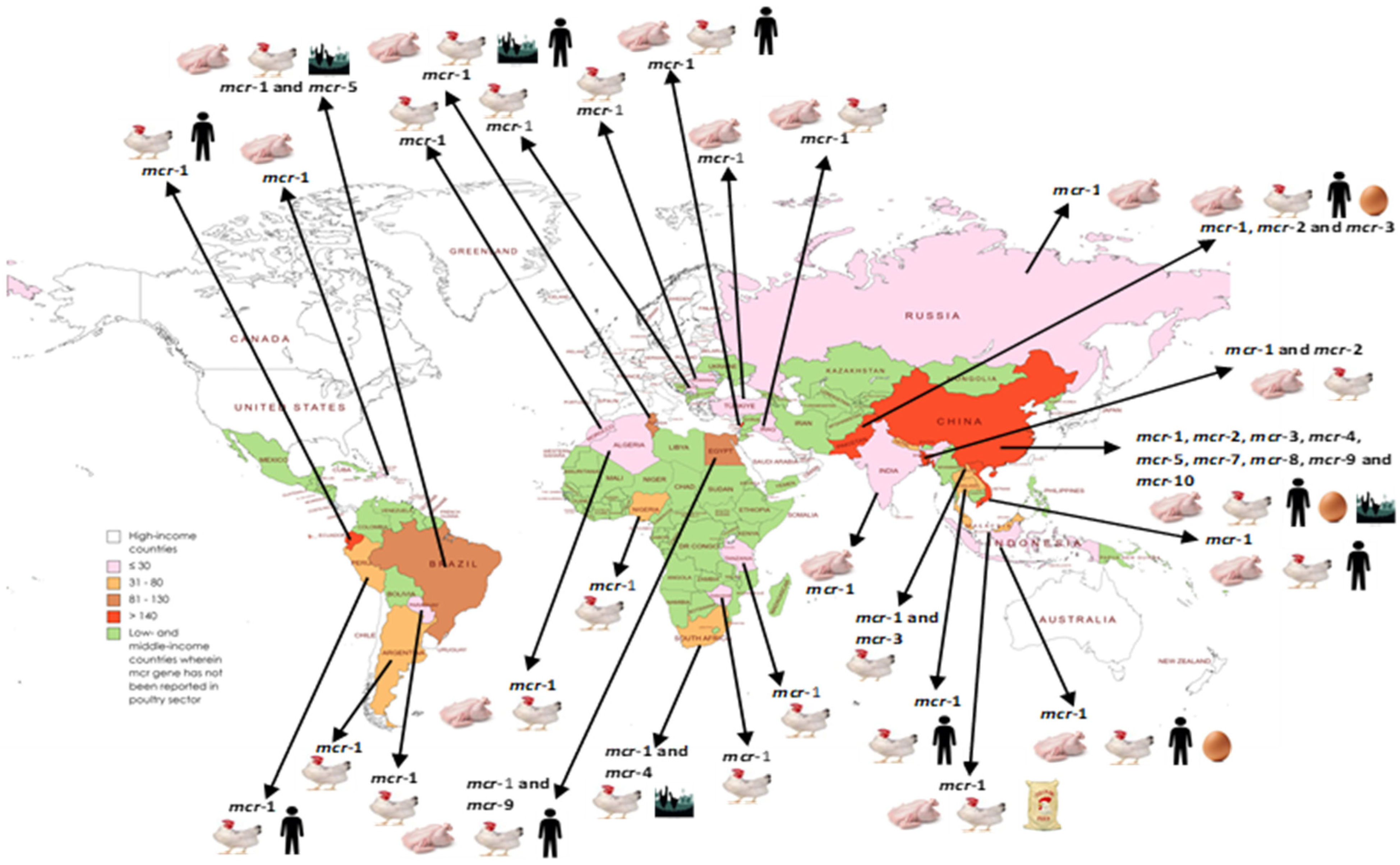
| Country | Source of Isolates | Date of Isolation (mcr Gene Assayed) | Number of Isolates Tested for mcr | Identified Gene (Number of Organisms) | Sequence Type | Virulence Genes (Phylogroup) | Plasmid (Associated Insertion Sequence and Integrons) | Additional Resistance Traits | References | |
|---|---|---|---|---|---|---|---|---|---|---|
| China | Poultry birds, meats, feed, organs, litter, sewage, bird market, and vectors in poultry environment | 1970–2022 (mcr-1 to mcr-10) | 16,288 | mcr-1 (2400 Escherichia coli, 33 Escherichia fergusonii, 21 Salmonella, 9 Klebsiella pneumoniae, 3 Citrobacter, 2 Cronobacter, 1 Enterobacter, 1 Rauoltella, and 1 Providencia) | E. coli: ST6775, ST189, ST156, ST5454, ST602, ST362, ST117, ST2944, ST10338, ST1403, ST1421, ST162, ST3941, ST6321, ST69, ST7153, ST93, ST12735, ST297, ST2973, ST2847, ST101, ST617, ST10, ST1011, ST767, ST155, ST457, ST48, ST4204, ST533, ST1638, ST6395, ST4408, ST1968, ST83, ST21, ST696, ST226, ST3519, ST359, ST2179, ST124, ST6706, ST175, ST6740, ST65, ST1296, ST4935, ST612, ST171, ST7115, ST1158, ST2732, ST354, ST82, ST7189, ST616, ST88, ST77, ST542, ST5879, ST5865, ST1431, ST1290, ST873, ST971, ST952, ST5851, ST761, ST6050, ST351, ST361, ST744, ST3044, ST2491, ST2345, ST1642, ST5909, ST601, ST3944, ST870, ST3133, ST215, ST178, ST58, ST3481, ST5542, ST2914, ST68, ST501, ST38, ST2085, ST1060, ST827, ST131, ST2323, ST167, ST4214, ST302, ST540, ST227, ST3014, ST6257, ST410, ST452, ST3499, ST6484, ST95, ST2171, ST29, ST294, ST723, ST109, ST678, ST17, ST62, ST3, ST5694, ST127, ST2018, ST3489, ST6388, ST2736, ST206, ST2599, ST648, ST5498, ST1564, ST1589, ST3041, ST1286, ST5229, ST29, ST46, ST165, ST7157, ST220, ST16, ST7454, ST2929, ST2223, ST1582, ST5259, ST1251, ST219, ST4710, ST4477, ST423, ST224, ST168, ST43, ST4969, ST7584, ST8900, ST271, ST4753, ST1266, ST4129, ST7108, ST349, ST448, ST23, ST770, and ST22 | cvaC, etsC, hlyF, gad, ireA, iss, iucC, iutA, terC, traT, Irea, iroN, aafII, eae, stx1, KpsM II, papA, papC, fimD, pefC, mchF, ompT, sitA, entB,C,E,F,S, acrB, fepA,D,G, altB, ompA, gndA, galF, rpos, ugD, rfBK, gad, hra, lpfA, astA, gad, lpfA, cma, neuC, ompT and sitA (A, B1, B2, and D) | IncI2, IncHI2, IncFIC, IncI, IncI1, IncFIA, IncFIB, IncFII, IncY, IncX2, IncX3, IncX4, IncHI2A, IncB/O/K/Z, IncP, IncN, Incp0111, IncA/C and IncL/M; and chromosomal (ISAplI, ISEcp1, ISCR1, ISKpn3, ISAs17, IS26s, IS186B, ISAs13, ISAeca6, ∆IS903B, ISAs2, ISAs20, ISEcl1, ISKpn26, Δ1ISAhy2 and Δ2ISAhy2, and Intl1 | tet(X4), tmexCD1-toprJ1, blaCTX-M-1, blaCTX-M-9, blaCTX-M-27, blaDHA-1, blaFOX-2, blaSHV-11, blaOXA-12, blaSHV, blaCEPH-A3-like, blaPSE, blaPER-3, blaCTX-M-14, blaCTX-M-15, blaCTX-M-28, blaSHV-28, blaCTX-M-44, blaTEM-99, blaCTX-M-3, blaCTX-M-55, blaCTX-M-64, blaCTX-M-137, blaCTX-M-65, blaCTX-M-66, blaCTX-M-82b, blaTEM-1A, blaTEM-1B, blaTEM-1D, blaCTX-M-27, blaSHV−73, blaTEM-141, blaTEM-122, blaMIR, blaCMY, blaCMY-2, ampC2, ampC1, blaNDM-1, blaNDM-4, blaTEM-198, blaOXA-1, blaOXA-10, blaNDM-5, erm(B), erm(C), erm(42), mef(B), mphI, lnu(F), rmtB, mdf(A), mph(A), armA, aac(6)-Ia, aac(3′)-Iv, aacCA5, aadA, aac(6′)-Iaa, aac(6′)-Ib, aadA5, aadA7, aac(6ʹ)-Ib-cr, aadA16, aph(6)-Id, aadA1, aph(3′)-Ia, aph(3′)-Ib, aph(39)-IId, aph(300)-Ib, aph(39)-Ia, aac(3)-IId, aadA2, ant(6)-Ia, aac(3)-IIa, aac(3′)-IId, aph(4)-Ia, aphA2, ant(3′)-Ia, ant(3″), strA, strB, armA, sat2a, arr, tet(A), tet(B), tet(C), tet(M), tet(D), I(E), tet(34), tet(O), qnrA, qepA, qnrS, qnrD, qnrS1, qnrS4, qnrB4, qnrB52, oqxA, oqxB, fosA, fosA3, fosA7, floR, catA1, catB3, cml, cmlA, catB, cmlA1, ul1, sul2, sul3, dfrA1, dfrA5, dfrA7, dfrA10, dfrA12, dfrA14, dfrA17, dfrA27, qacL, qacE, and mutations in gyrA and parC | [25,43,44,45,46,49,50,51,57,58,59,60,62,63,86,89,91,92,94,96,97,98,99,100,105,106,107,108,109,129,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171] | |
| mcr-2 (66 E. coli) | ||||||||||
| mcr-3 (5 Aeromonas, 1 E. coli, and 1 Proteus) | ||||||||||
| mcr-7 (3 K. pneumoniae) | ||||||||||
| mcr-8 (31 K. pneumoniae and 2 Rauoltella) | ||||||||||
| mcr-9 (131 Salmonella) | ||||||||||
| mcr-10 (2 K. pneumoniae, 2 Enterobacter, and 1 E. coli) | ||||||||||
| Enterobacter: ST1605 and ST1056 | ||||||||||
| K. pneumoniae: ST11, ST15, ST37, ST3332, and ST395 | ||||||||||
| Salmonella: ST292, ST399, ST34, and ST2529 | ||||||||||
| Aeromonas: ST51, ST514, and ST216 | ||||||||||
| India | Poultry meat | 2019 (mcr-1) | 12 | mcr-1 (2 E. coli) | ST50 | iss and gad | IncX1 and IncHI2 (ISApl1) | aadA1, aadA2, aph(6)-Id, aph(3′)-Ib,blaTEM-1B, qnrS1, tetA, dfrA14, dfrA15, floR, and sul3 | [110] | |
| Pakistan | Chickens, chicken meat, internal organs, and secretions | 2015–2020 (mcr-1 to mcr-10) | 703 | mcr-1 (228 E. coli, 12 K. pneumoniae, 1 Pseudomonas aeruginosa, and 1 Proteus mirabilis) | ST1035, ST131, ST1215, ST2279, ST88, ST1650, ST410, ST3059, ST10, ST95, ST2847, ST155, ST361, ST6395, ST244, ST23, ST7187, ST156, ST746 ST354, ST135, ST117, ST4085, ST761, ST744, ST58, ST1121, ST1267, ST221, ST11, ST392 ST6751, ST351, ST10009, ST38, ST617, ST694, ST3902, ST218, ST457, ST6635, ST2750, ST206, ST2690, ST1140, ST10010, ST8804, ST359, ST162, ST6635, ST93, ST4980, ST191, ST10011, ST1011, ST355, ST2224, ST2067, ST69, ST48, ST2253, ST2099, ST189, ST2207, ST2334, and ST6706 | ompT, hylF, iutA, iroN, iss, iucD, astA, tsh, papC, and cvi | IncX4, IncF, IncY, p0111, IncH, IncI, IncQ, ColPVC, IncP, IncY, and so on (ISCR1, ISApl1, and ISECP1) | tet(X4), blaCTX-M, blaCTX-M-15, blaTEM1B, blaTEM-1, blaSHV, blaCMY-2, blaNDM-1, blaKPC, blaOXA-48, blaIMP, tet(A), tet(B), sul1, floR, cmlA1, aadA1, strB, aac(6′)-Ib-cr, aph(3″)-Ib, aac(3)-IId, aph(3′)-Id, aph(3′)-Ia, aac(3)-IIa, aph(6′)-Ic, aph(6′)-Id, dfrA12, dfrA14, sul2, sul3, and so on | [8,67,172,173,174,175,176,177,178,179,180] | |
| Bangladesh | Poultry birds, meats, farm environment, water, and cages | 2017–2019 (mcr-1 to mcr-5) | 3307 | mcr-1 (471 E. coli, 17 Proteus, 10 Klebsiella, 12 Salmonella, 1 Shigella, and 2 Enterobacter) | ST1324, ST155, ST1818, ST354, ST178, ST43, ST4965, ST2705, ST1196, ST206, ST359, ST867, ST602, ST867, ST3107, ST48, and ST189 | iss, lpfA, eaeA, astA, etpD, air, eilA, cma, iroN, mchF, eilA, iha, ireA, and tssh | IncHI1, IncHI, IncHI2, IncFIB, IncQ1, IncFIB, IncX1, and IncI2, IncN, IncFIA, ColRNAI, ColE10, ColpVC, and p0111 (ISApl1) | blaCTX-M-55, blaDHA-1, blaTEM-106, blaTEM-126, blaTEM-135, blaTEM-176, blaTEM-1A, blaTEM-1B, blaTEM-220, blaTEM-57, blaCTX-M-group-1, blaTEM, blaOXA-1, blaOXA-47, blaCTX-M-group-9, rmtB aac(3)-IId, aadA1, aadA2, aadA2b, aadA8b,aaph(3″)-Ib,’aph(3′)-Ia, aph(6)-Id, mdf(A), mph(A), catA1, cml, cmlA1, floR, qepA4, sul1, sul2, sul3, tet(A), tet(B), tet(M), dfrA1, dfrA12, dfrA14, dfrA17, qnrB2, qnrB4, qnrB7, qnrS, qnrB. qnrD3, qnrS1, mutations in gyrA, parC parE, pmrA, and pmrB | [39,40,41,53,66,88,93,181,182,183] | |
| Salmonella: invA | ||||||||||
| mcr-2 (2 Proteus mirabilis, 1 E. coli, 1 K. pneumoniae, 1 Salmonella, and 1 Enterobacter) | ||||||||||
| mcr-3 (11 E. coli) | ||||||||||
| Nepal | Chickens and chicken vendors | 2017–2019 (mcr-1 to mcr-10) | 312 | mcr-1 (68 E. coli) | ST23 and ST10 | - | IncK/B and IncI2, IncI1, IncFIC(FII), and IncFIB | blaOXA-48, blaCTX-M, tet, sul, qnr, and dfr | [69,70,71] | |
| Lebanon | Chickens, chicken feed, litter, soil, and farmworkers | 2017–2018 (mcr-1 to mcr-10) | 617 | mcr-1 (315 E. coli and 31 K. pneumoniae) | ST1011, ST6856, ST93, ST744, ST388, ST359, ST752, ST1421, ST6844, ST6115, ST354, ST1638, ST2705, ST48, ST206, ST398, ST1626, ST101, ST1140, ST226, ST2705, ST162, ST2936, ST3288, ST6448, ST746, ST1196, ST359, ST2220, ST5687, ST248, ST7458, ST115, ST1589, and ST3941 | - | IncX4, IncI2, and IncHI2 | blaCMY-3, blaTEM-1B, blaTEM-1C, blaTEM-141, blaTEM, blaTEM-1, blaTEM-122, blaTEM-141, blaTEM-122, blaCTXM, blaCTXM-3, blaCTX-M-65, blaCTX-M-3, blaCTX-M-55, blaCMY-2, blaSHV, tet(A), tet(B), tet(M), tet(), qnrS1, gyrA and parC, sul1, sul2, sul3, dfrA15, dfrA17, dfrA1, dfrA14, dfrA17, dfrA5, floR, catA1, cmlA1, lnu(F), mdf(A), mph(A), mpH(A), fosA3, qnrS1, aad, aadA2, aadA5, aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib, ant(3″)-Ia, aph(3′)-Iia, aac(3)-IId, aph(3′)-Ib, ant(3′)-Ia, and IntI1 | [35,47,184,185,186,187] | |
| Iraq | Chicken and turkey meats | 2017–2019 (mcr-1) | 200 | mcr-1 (26 Acinetobacter baumannii) | - | fimH, afa/draBC, sfa/foc DE, cnfI, and cnf2 | - | blaCITM, blaIMP, blaSHV, blaSIM, blaVIM, blaOXA-58-like, blaOXA-24-like, blaOXA-23-like, blaOXA-51-like, aac(3)-I, aadA1, cmlA, cat1, sul1, dfrA1, tet(B), and tet(A) | [121] | |
| Indonesia | Chickens, litter and drinking water in farms, and chicken meat | 2017 (mcr-1) | 58 | mcr-1 (13 E. coli) | - | - | - | - | [55] | |
| Turkey | Chicken meat | 2016–2019 (mcr-1 to mcr-10) | 127 | mcr-1 (5 E. coli) | ST3941, ST1049, and ST6094 | ast and gad | IncX4 and IncI2 | blaTEM-1b, blaTEM-1c, blaCTX-M-8, qnrB19, mdf(A), tet(A), tet(B), sul1, sul2, dfrA1, floR, catA1, mphA, aph(3′)-Ib, aph(6)-Id, and aadA5 | [111,112] | |
| Lao Peoples Democratic Republic | Chickens and chicken meat | 2018 (mcr-1 to mcr-8) | 175 | mcr-1 (34 E. coli) | ST165, ST10, ST1630, ST11090, ST2179, ST69, ST48, ST7352, ST212, ST48, ST206, ST165, ST46, ST117, ST5229, ST373, ST641, ST2690, ST648, and ST1585 | entC,E,B, fepA,B,D, fes, fimF, fimG, chuU,V,W, shuA and shuX (A, B1, B2, C, D, E, F, G, and clade I) | IncX4, IncI2, IncP1, IncFII, IncFIA, IncFIB, IncR and IncHI1 (IS15DI,) ISKpn40, and IS26 | blaTEM-1, blaCTX-M-55, blaCTX-M-123, aac(3)-IId, aadA1, aadA2, aadA2b, aph(300)-Ib, aph(6)-Id, aac(3)-IV, aac(6′)-Ib-cr, aph(4)-Ia, aph(6)-Id, aph(3″)-Ib,’aph(3′)-Ia, mdf(A), mph(A), cmlA1, catB3, floR, sul2, sul3, dfrA12, dfrA1, tet(A), tet(M), fosA3, arr-3, qnrS1, oqxA, oqxB, and qnrS13 | [87,122] | |
| mcr-3 (3 E. coli) | ||||||||||
| Thailand | Chickens and ducks | 2013–2019 (mcr-1) | 34 | mcr-1 (75 E. coli and 1 Salmonella) | ST2973 | - | - | blaCTXM-14 | [83,130] | |
| Malaysia | Chickens, chicken meat, litter and feed | 2013–2021 (mcr-1 to mcr-5) | 262 | mcr-1 (63 E. coli and 1 Salmonella enterica) | ST3489, ST93, ST540, ST69, ST154, ST345, ST196, ST1001, ST1638, ST155, ST2179, ST872, ST410, ST373, ST770, and ST117 | A, B1, B2, and D | IncI2, IncHI1A, IncHI1B IncQ1, IncFIA, IncFIB, IncI1, IncI2, IncFIC, and ColpVc (ISApl1) | aadA1, aadA2, blaTEM-1B, blaCTX-M-55, blaTEM-52,blaNDM, blaOXA-48, blaIMP, qnrS1, fosA, mph(A), mef(B), floR, sul1, sul3, tet(A), tet(M), tet(34), dfrA12, aph(3′)-Ia, aac-3-IV, aadA1, aac(6)-lb, strA, and strB | [54,68,120,188,189,190,191] | |
| mcr-5 (1 Salmonella) | ||||||||||
| Vietnam | Chickens and chicken meat | 2011–2019 (mcr-1 to mcr-10) | 872 | mcr-1 (278 E. coli) | ST156, ST50, ST162, ST155, ST93, ST 354, ST1158, ST2690, ST48, ST206, ST69, ST6726, ST648, ST656, and ST1602 | astA, csgG, entA,B,C,D,E,F,S, fepA,B,C,D,G, fes, ompA, aslA, csgB,D,F, fdeC, fimA,B,C,D,E,F,G,H,I, gspC,E,F,G,H,I,J,K,L,M, yagV/ecpE, yagW/ecpD, yagX/ecpC, yagY/ecpB, yagZ/ecpA, ykgk/ecpR, astA, chuS,T,U,V,W, chuY, fyuA, iroN, iutA, kpsD, kpsM, and papC | IncHI2, IncI2, IncHI2A IncN, IncX4, IncY, and IncP-1 (ISAplI) | blaTEM, blaCTX-M, blaCTX-M-2, blaCTX-M-65, blaCTX-M-9, blaCTX-M-1, blaOXA-1, blaCMY-2, blaCARB-3, blaCARB-2, blaCTX-M, ampC, blaOXA, blaTEM−1, tet(A), strA, strB, aac(3)-IIa, aac(6′)-IIa, ant(3″)-IIa, aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id, aadA, fosA, sul2, sul3, dfrA, floR, catA4, catB3, emr(E), mphA, mphB, qnrD1, qnrS, qnrA, and mutation in gyrA, | [38,84,113,114,115,116,117,118,119] |
| Country | Source of Isolate | Date of Isolation (mcr Gene Assayed) | Number of Isolates Tested for mcr | Identified Gene (Number of Organisms) | Sequence Type | Virulence Genes (Phylogroup) | Plasmid (Associated Insertion Sequence) | Additional Resistance Traits | References |
|---|---|---|---|---|---|---|---|---|---|
| Tunisia | Chickens and chicken meat | 2015–2018 (mcr-1to mcr-10) | 195 | mcr-1 (116 E. coli) | ST398, ST4187, ST2197, ST10, ST69, ST349, ST57, ST1011, ST3882, ST5693, ST8932, ST162, ST2220, ST5686, ST57, ST117, and ST6798 | fimA, stx1, stx2, papC and aer (A, B1, B2, D, and D2) | IncI1, IncI2, IncHI2, IncFIB, and IncP (ISApl1) | blaTEM, blaSHV, blaCMY-2, blaCTX-M-1, blaCTX-M-g-1, blaCTX-M-55, blaCTX-M-14, blaCMY, blaTEM-1b,blaCTX-M-1, tet(A), tet(B), aadA, aadA1 strA, strB, sul1, sul3, dfrA, floR, cmlA, and Int1 | [72,73,74,75,191,192,193] |
| Algeria | Chickens and chicken meat | 2015–2017 (mcr-1 to mcr-9) | 27 | mcr-1 (16 E. coli) | ST48, ST758, ST224, ST168, ST1241, ST260, ST 5161, ST155, ST10, ST744, ST 4654, and ST648 | fimH, iroN, iutA, iucD, traT, cva, iss, ompT, sfa, foc, afa, dr, kpsMTII, papA, papC, hlyA, ireA and malX (A, B1, and F) | IncHI2, IncFV, and IncFIIK (ISApl1) | blaTEM-1, blaSHV-1, sul1, sul2, and sul | [123,190,194] |
| Egypt | Chickens, chicken farm workers, environment, and meats | 2010–2020 (mcr-1 to mcr-10) | 448 | mcr-1 (111 E. coli and 1 Citrobacter freundii) | ST986, ST373, ST156, ST1011, ST5687, ST1125, ST371, ST398, ST196, ST101, ST115, and ST10 | cma, hemL, iroN, iss, pic, vat, hylE, ireA, mchF sitABCD operon, and iucABCD/iutA operon | IncHI2, IncI2, and IncX4 (ISApl1 and Tn6330) | tet(X7), blaTEM, blaTEM-1, blaCTXM, blaCTX-M-9, blaSHV-12, blaCTX-M-15, blaCTX-M-14, blaTEM-1B, blaCTX-M-1, blaOXA-1, blaCMY, blaNDM, blaVIM, blaSHV, sul1, sul2, sul3, dfrA1, dfrA14, dfrA7, dfrA12, dfrA15, fosA4, tet(A), aadA1, aphA, mphA, mrx, aadA2, aph(6)-Id, aac(3)- IIa, aph(39′)-Ib, aph(39)-Ia, strA, strB, lnu(F), ere(A), erm(B), erm(42), mph(A), mph(B), mdfA, catA1, floR, cmlA1, and arr-2 | [76,77,78,79,80,195,196,197] |
| mcr-9 (1 E. coli) | |||||||||
| Morocco | Chickens | 2012–2017 (mcr-1) | 12 | mcr-1 (3 E. coli) | - | - | - | - | [81] |
| South Africa | Chickens and rats in poultry farms | 2015-2019 (mcr-1 to mcr-5) | 115 | mcr-1 (39 E. coli) | - | - | IncI2 (ISApl1) | blaCTX-M-1, blaTEM, blaCTX-M-2, blaCTX-M-9, blaCTX-M-15, aadA1, strA, strB, tetA, sul3, dfrA12, IntI1, and IntI2 | [82,198,199] |
| mcr-4 (31 Salmonella) | |||||||||
| Zimbabwe | Chickens and ducks | 2017–2019 (mcr-1 to mcr-10) | 21 | mcr-1 (1 E. coli) | ST10 | fimH, csg, agn43, kpsD, kpsMll, ibeBC, fes, fepA, iucA, gspL, and hlyE | IncF | qnr, tet, blaTEM, blaOXA, and blaCTX-M | [200] |
| Nigeria | Chickens, chicken farmers, chicken farm water, and live bird market environment | 2018–2020 (mcr-1 to mcr-10) | 450 | mcr-1 (32 E. coli, 2 K. pneumoniae, and 1 Citrobacter werkmanii) | ST34, ST48, ST155, ST1286, ST226, ST10, ST656, ST4542, ST168, ST398, ST6836, ST746, and ST2485 | - | IncX4 | blaTEM-93, blaTEM-57, blaCTX-M-55, blaCMY-47, blaTEM-1, tet(G), tet(D), tet(C), qnrB17, qnrB19, qnrS1, mdfA, aph(3)-Ib, aph(3)-Ia, aph(6)-Id, aadA1, aac(3)-IId, sul2, and catA1 | [34,201,202] |
| mcr-5 (1 E. coli) | |||||||||
| mcr-8 (1 E. coli and 1 K. pneumoniae) |
| Country | Source of Isolate | Date of Isolation (mcr Gene Assayed) | Number of Isolates Tested for mcr | Identified Gene/Variant (Number of Organisms) | Sequence Type | Virulence Genes (Phylogroup) | Plasmid (Associated Insertion Sequence) | Additional Resistance Traits | References |
|---|---|---|---|---|---|---|---|---|---|
| Ecuador | Chickens | 2013–2020 (mcr-1) | 326 | mcr-1 (145 E. coli) | ST394, ST5855, ST6995, ST6940, ST10, ST98, and ST3856 | - | IncI | blaCTX-M-2, blaCTX-M-14, blaCTX-M-65, blaCMY-2, blaOXA-48, blaCTXM-1, blaTEM, blaCTXM-9, blaNDM, fosA3, aadA5, dfrA17, sul1, tet(B), and mutations in gyrA and parC | [203,204,205,206,207] |
| Peru | Chickens | 2018–2020 (mcr-1 to mcr-10) | 274 | mcr-1 (44 E. coli) | ST23 ST10, ST48, ST602, ST746, ST46, and ST345 | gad, iss, astA, cba, cma, iha, iroN, lpfA, capU, kpsE, kpsMII_K5, ompT, sepA, traT, cea, cib, hlyF, stb, toxB, cif, espABFJ, nleABC, terC, tir, eae, perA, tsh, fyuA, ireA, irp2, iucC, iutA, sitA, and hlyF | IncFIB, IncI1, IncFIC, IncX1, ColRNAI, and IncFII | blaCTX-M-65, blaTEM-30, aadA1, aadA2, aph(4)-Ia, aac(3)-Iva, aac(3)-IIa, aac(3)-IId, aadA5, aadA15, aadA17, aph(3′)-Ia, aph(3′)-IIa, aph(6)-Id, mph(A), catA, floR, cmlA1, sul2, sul3, dfrA17, tet(B), tet(A), dfrA1, and mutations in gyrA and parC | [208,209] |
| Brazil | Chickens and poultry meats | 2000–2016 (mcr-1to mcr-4) | 686 | mcr-1 (97 E. coli, 3 E. fergusonnii and 2 Salmonella enterica serovar Schwarzengrund) | ST132, ST48, ST4419, ST96, ST522, and ST10 | iss, iroN, lpfA, mchB, mchC, mchF, hlyF, ompT, iss, iron and iutA, ireA, and gad (A, B1, B2, and D) | IncX4, IncI2, IncHII, IncFIB, and IncB/O (ISEc12) | blaTEM, blaCTX-M-1, blaCTX-M-8, blaCTX-M-15, blaCTX-M-group 2, blaSHV, blaCMY-2, blaCTX-M-8, aadA, aadA1, aadB, aac(3)-VI, aadA2, aadA5, aph(3′)-Ic, aph(3)-IA, sul1, sul2, dfr17, tet(A), tet(B), pcoD, qac, and IntI1 | [124,125,210,211,212,213,214] |
| mcr-5 (5 E. coli) | |||||||||
| Argentina | Chickens | 2013–2014 (mcr-1) | 168 | mcr-1 (38 E. coli) | ST617, ST1141, ST410, ST155, ST1286, ST1011, ST10 and ST1408 | - | IncI2 (ISApl1) | blaCTX-M-14, blaCMY-2, blaCTX-M-2, blaCMY-2, qnrA, qnrB, qnrD, qnrS, oqxB, and qepA | [215,216] |
| Paraguay | Chickens | 2012 (mcr-1 to mcr-5) | 66 | mcr-5 (29 E. coli) | ST457, ST38, ST57, ST8061, ST224, ST580, ST6853, ST189, ST93, and ST2705 | F | IncI1, IncFII, IncHI1, IncI1-N, and IncFII-FIB | blaCTX-M-8, blaCMY-2, blaSHV-12, blaTEM-1A, aph(6)-Id, aph(3″)-Ib, aph(3″)-Ib, sul2, mdf(A), and dfrA8 | [48,217] |
| Country | Source of Isolate | Date of Isolation (mcr Gene Assayed) | Number of Isolates Tested for mcr | Identified Gene (Number of Organisms) | Sequence Type | Virulence Genes (Phylogroup) | Plasmid (Associated Insertion Sequence) | Additional Resistance Traits | References |
|---|---|---|---|---|---|---|---|---|---|
| Romania | Chickens | 2011–2017 (mcr-1 to mcr-10) | 96 | mcr-1 (18 E. coli) | ST744, ST57, ST156, ST10, and ST4980 | astA, gad, iss, ompT, and terC (A, B1 and D) | IncHI2, IncX, IncF, and IncI (ISApl1 and Tn6330) | blaTEM-1B,blaTEM-1, blaCMY, blaOXA-62, acc(3)-IIa,aph(3)-Ia, aph(3″)-Ib, aph(6)-Id,strA, strB, sul2, sul3,tet(A), floR, and mutations in gyrA and parC | [218,219] |
| Russia | Chicken meat | 2019 (mcr-1) | 3 | mcr-1 (1 Salmonella Enteritidis) | ST11 | - | IncX4 | - | [127] |
| Serbia | Turkeys | 2020 (mcr-1 to mcr-5) | 5 | mcr-1 (5 E. coli) | ST410, ST641, and ST58 | cfaA-H, ecpA-E, elfA,C,D,H, hpcA-C, fimA,C-I, eaeH, aec15-32, fliC, sitA-D, iroB-E,N, fyuA, irp1/2, ybtA,E,P,Q,S-U,X, cah, ehaA/B/G, upaG, agn43, espL1,L4,R1,X1,X4,X5, ibeB/C, hlyE/clyA, uge, wzc, flaA, ompT, and rmlD | IncX4 | blaTEM-1, aadA1, aadA2, strA, strB, sul2, sul3, dfrA12, qnrS1, tet(A), tet(M), cmlA1, floR, and mutations in gyrA and parC | [220] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anyanwu, M.U.; Jaja, I.F.; Okpala, C.O.R.; Njoga, E.O.; Okafor, N.A.; Oguttu, J.W. Mobile Colistin Resistance (mcr) Gene-Containing Organisms in Poultry Sector in Low- and Middle-Income Countries: Epidemiology, Characteristics, and One Health Control Strategies. Antibiotics 2023, 12, 1117. https://doi.org/10.3390/antibiotics12071117
Anyanwu MU, Jaja IF, Okpala COR, Njoga EO, Okafor NA, Oguttu JW. Mobile Colistin Resistance (mcr) Gene-Containing Organisms in Poultry Sector in Low- and Middle-Income Countries: Epidemiology, Characteristics, and One Health Control Strategies. Antibiotics. 2023; 12(7):1117. https://doi.org/10.3390/antibiotics12071117
Chicago/Turabian StyleAnyanwu, Madubuike Umunna, Ishmael Festus Jaja, Charles Odilichukwu R. Okpala, Emmanuel Okechukwu Njoga, Nnenna Audrey Okafor, and James Wabwire Oguttu. 2023. "Mobile Colistin Resistance (mcr) Gene-Containing Organisms in Poultry Sector in Low- and Middle-Income Countries: Epidemiology, Characteristics, and One Health Control Strategies" Antibiotics 12, no. 7: 1117. https://doi.org/10.3390/antibiotics12071117
APA StyleAnyanwu, M. U., Jaja, I. F., Okpala, C. O. R., Njoga, E. O., Okafor, N. A., & Oguttu, J. W. (2023). Mobile Colistin Resistance (mcr) Gene-Containing Organisms in Poultry Sector in Low- and Middle-Income Countries: Epidemiology, Characteristics, and One Health Control Strategies. Antibiotics, 12(7), 1117. https://doi.org/10.3390/antibiotics12071117










