Characterization of the Antibacterial Activity of Quinone-Based Compounds Originating from the Alnumycin Biosynthetic Gene Cluster of a Streptomyces Isolate
Abstract
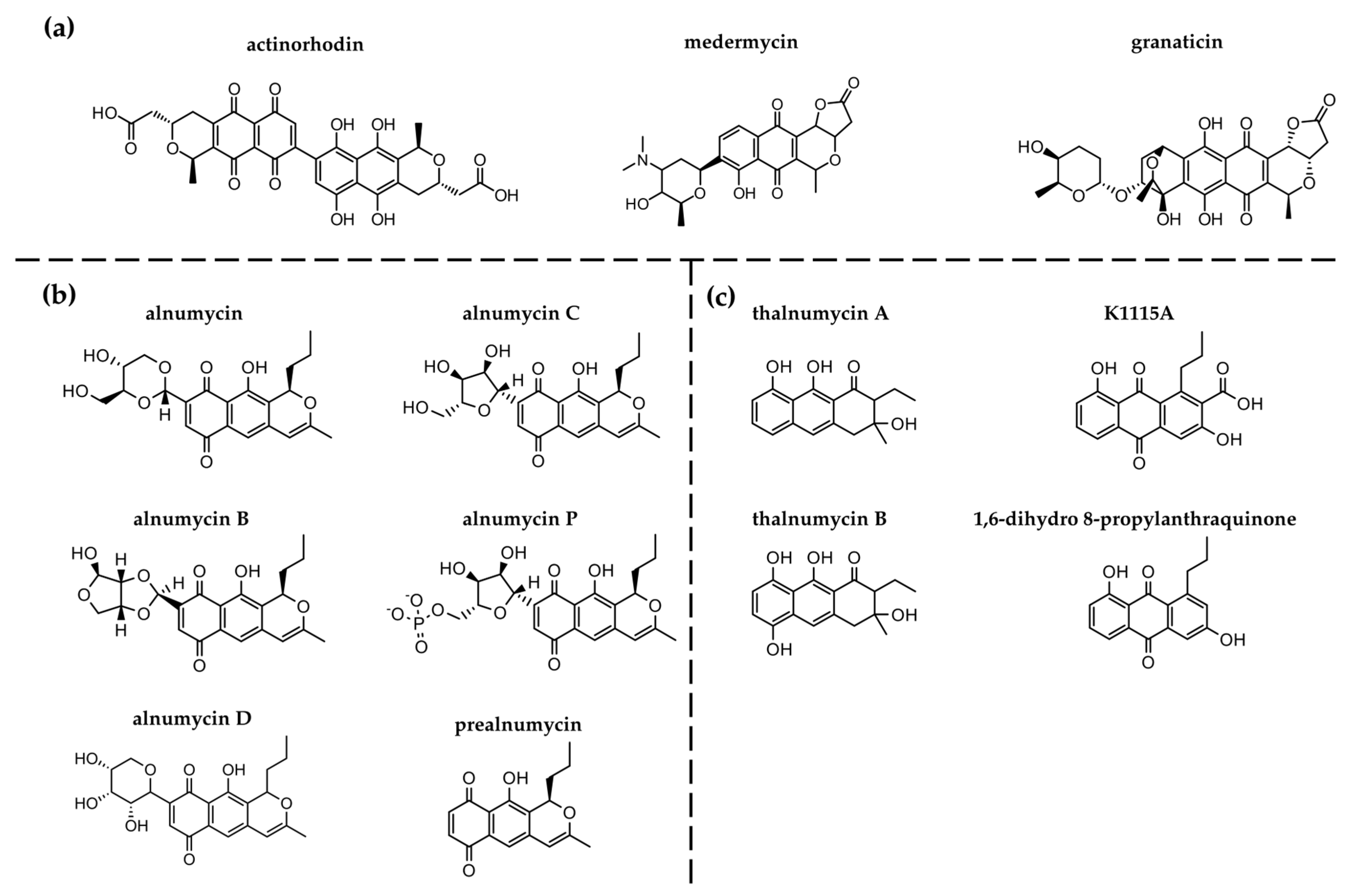
1. Introduction
2. Materials and Methods
2.1. Isolation of Strains
2.2. Genomic Analysis of Strains
2.3. Screening for Compound Production
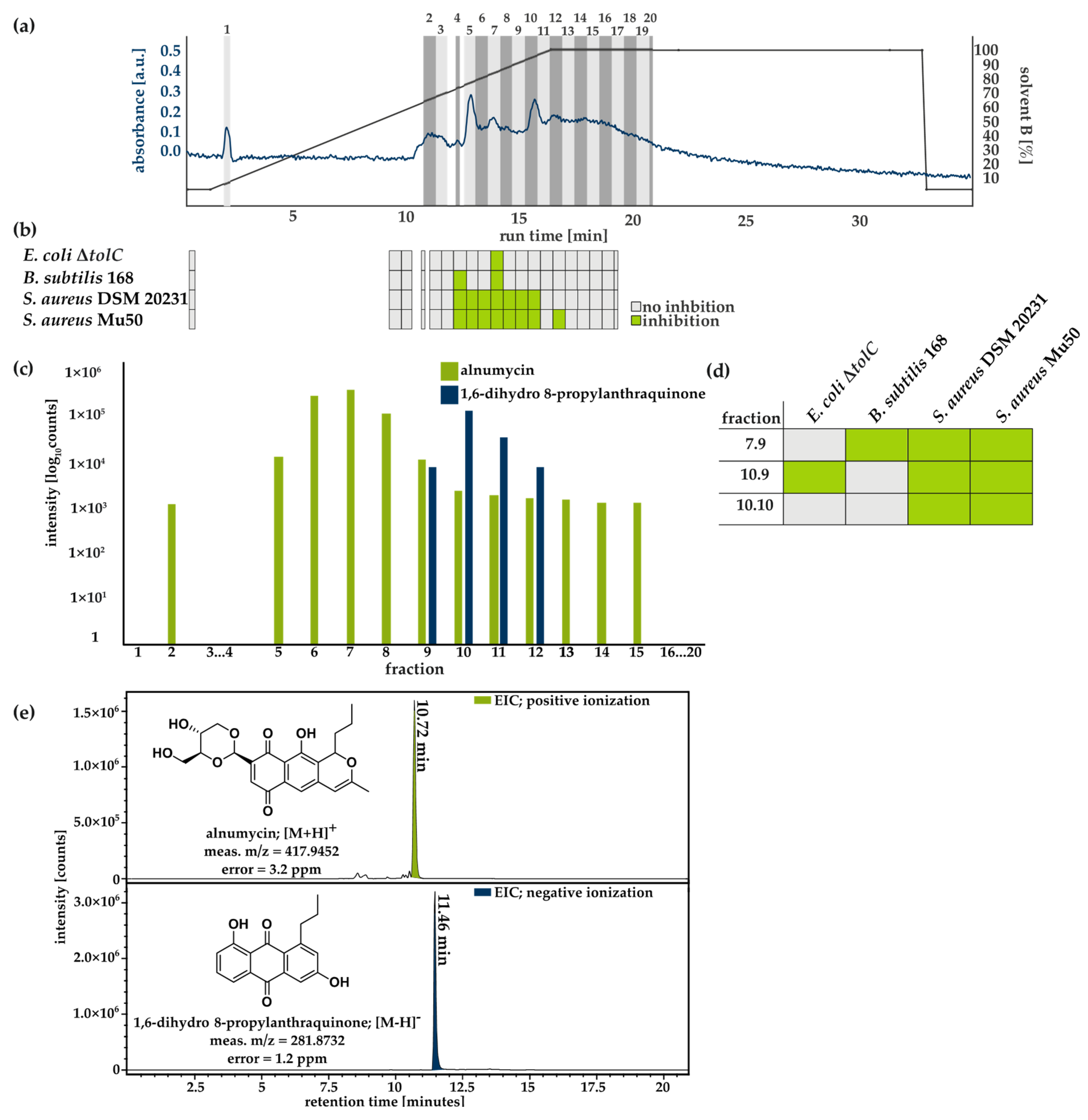
2.4. Activity and Mass Spectrometry-Guided Purification of Alnumycin and 1,6-Dihydro 8-Propylanthraquinone
2.5. Minimal Inhibitory Concentration
2.6. Structure Elucidation via Nuclear Magnetic Resonance Spectroscopy (NMR)
2.7. Gyrase Inhibition Assay
3. Results
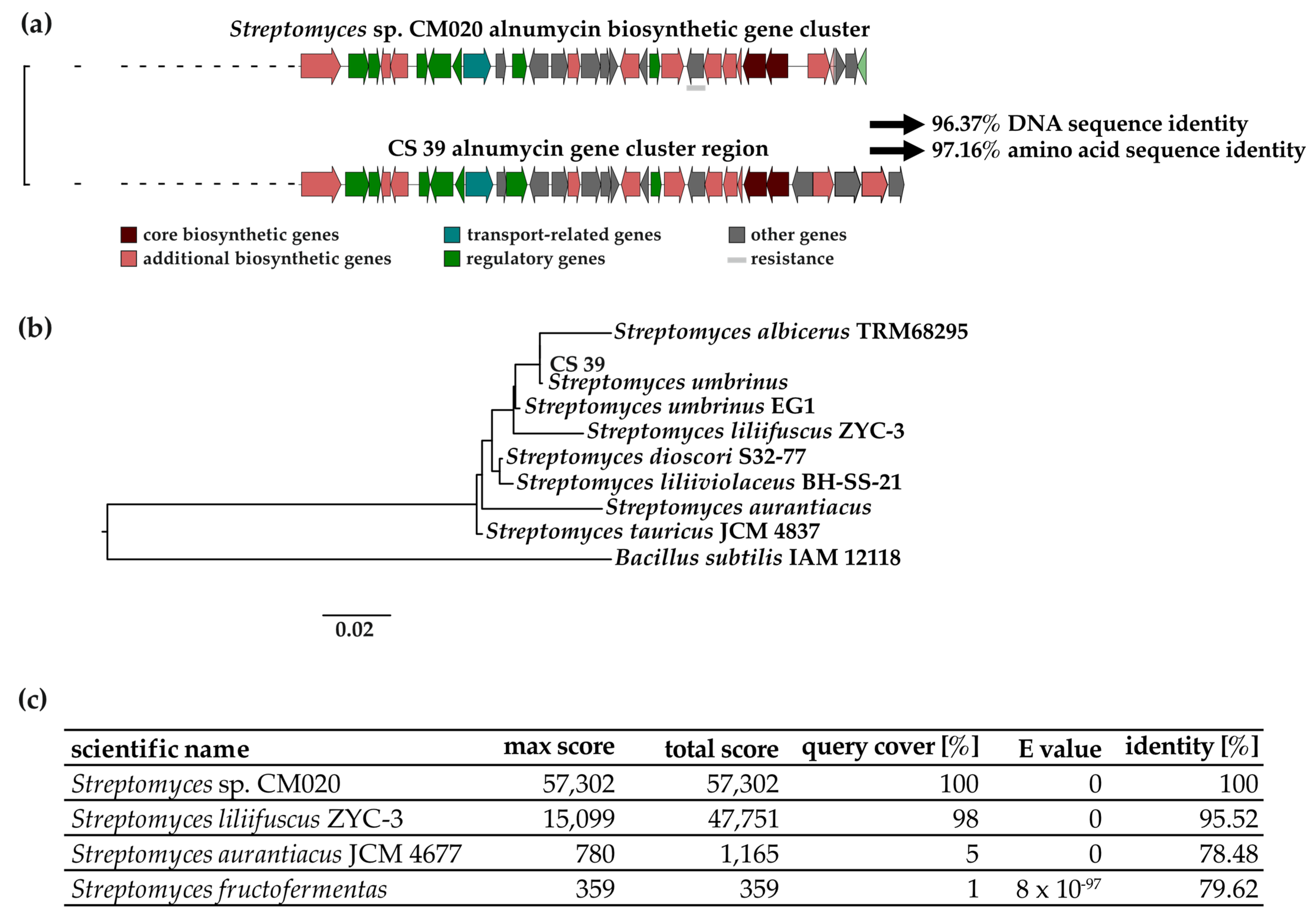
3.1. Genetic Analysis of the Alnumycin BGC
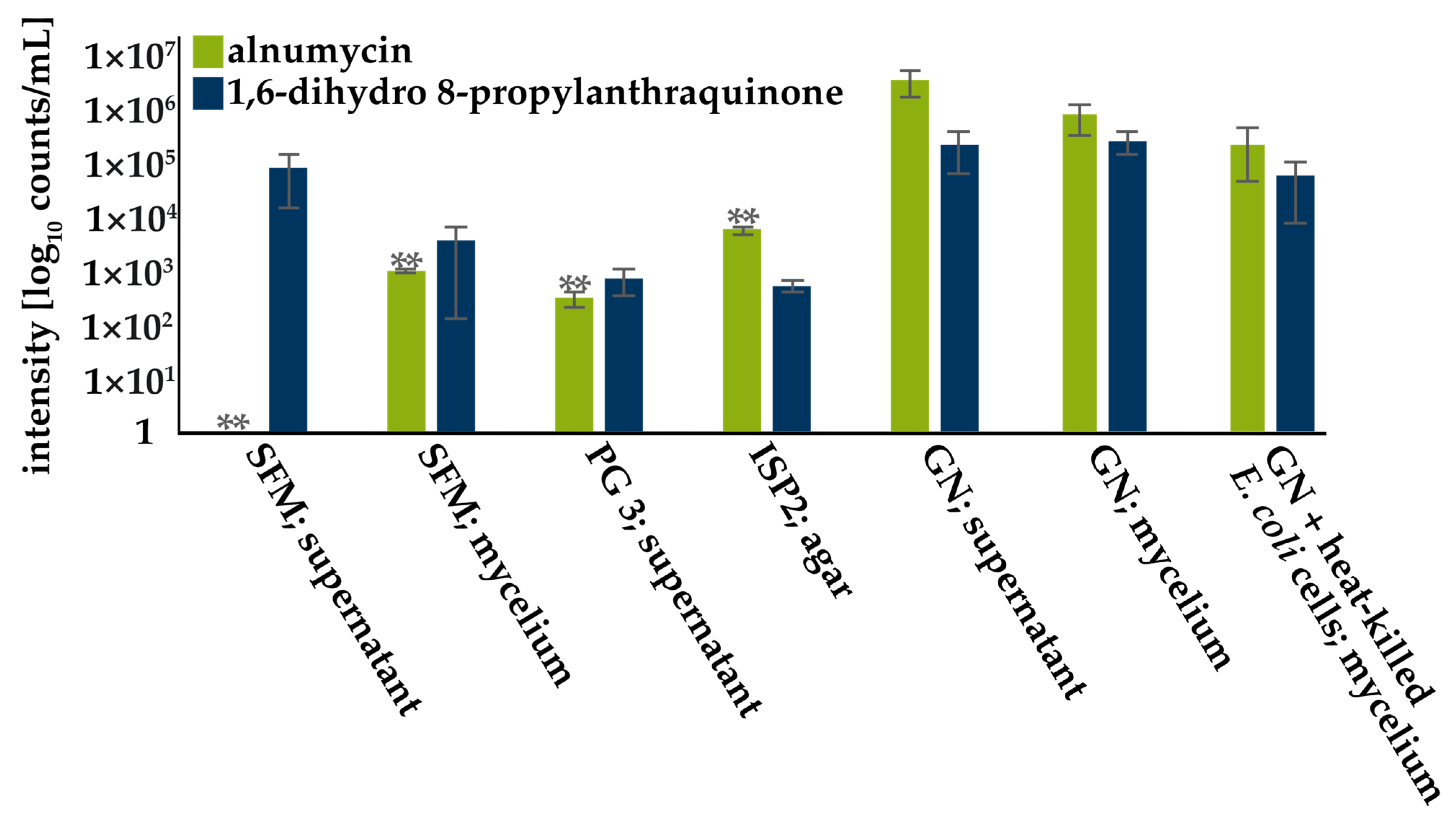
3.2. Screening for Compound Production
3.3. Purification and Structural Validation of the Compound
3.4. Spectrum of Activity of Alnumycin and 1,6-Dihydro 8-Propylanthraquinone
4. Discussion
4.1. Alnumycin and 1,6-Dihydro 8-Propylanthraquinone Production
4.2. Antibacterial Activity of Alnumycin and 1,6-Dihydro 8-Propylanthraquinone
4.3. Relevance of 1,6-Dihydro 8-Propylanthraquinone in Nature and Drug Discovery
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural Products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Tenebro, C.P.; Trono, D.J.V.L.; Vicera, C.V.B.; Sabido, E.M.; Ysulat, J.A.; Macaspac, A.J.M.; Tampus, K.A.; Fabrigar, T.A.P.; Saludes, J.P.; Dalisay, D.S. Multiple strain analysis of Streptomyces species from Philippine marine sediments reveals intraspecies heterogeneity in antibiotic activities. Sci. Rep. 2021, 11, 17544–17558. [Google Scholar] [CrossRef]
- Guo, X.; Liu, N.; Li, X.; Ding, Y.; Shang, F.; Gao, Y.; Ruan, J.; Huang, Y. Red soils harbor diverse culturable actinomycetes that are promising sources of novel secondary metabolites. Appl. Environ. Microbiol. 2015, 81, 3086–3103. [Google Scholar] [CrossRef]
- Wen, Y.; Zhang, G.; Bahadur, A.; Xu, Y.; Liu, Y.; Tian, M.; Ding, W.; Chen, T.; Zhang, W.; Liu, G. Genomic investigation of desert Streptomyces huasconensis D23 reveals its environmental adaptability and antimicrobial activity. Microorganisms 2022, 10, 2408–2423. [Google Scholar] [CrossRef]
- Salehghamari, E.; Moradi, M.; Sardabi, M.; Etesami, S.; Hassani, G.; Hosseini, M.; Taheri, F.; Eshrati, Z.; Tahmaseb, M.; Irian, S.; et al. Diversity of actinomycete and their metabolites isolated from Howz Soltan Lake, Iran. Arch. Microbiol. 2023, 205, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Pipite, A.; Lockhart, P.J.; McLenachan, P.A.; Christi, K.; Kumar, D.; Prasad, S.; Subramani, R. Isolation, antibacterial screening, and identification of bioactive cave dwelling bacteria in Fiji. Front. Microbiol. 2022, 13, 1012867. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; de Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef]
- Bibb, M.J.; Biro, S.; Motamedi, H.; Collins, J.F.; Hutchinson, C.R. Analysis of the nucleotide sequence of the Streptomyces glaucescens tcmI genes provides key information about the enzymology of polyketide antibiotic biosynthesis. EMBO J. 1989, 8, 2727–2736. [Google Scholar] [CrossRef]
- Sayari, M.; Steenkamp, E.T.; van der Nest, M.A.; Wingfield, B.D. Diversity and evolution of polyketide biosynthesis gene clusters in the Ceratocystidaceae. Fungal. Biol. 2018, 122, 856–866. [Google Scholar] [CrossRef]
- Bieber, B.; Nüske, J.; Ritzau, M.; Gräfe, U. Alnumycin a new naphtoquinone antibiotic produced by an endophytic, Streptomyces sp. J. Antibiot. 1998, 51, 381–382. [Google Scholar] [CrossRef]
- Naruse, N.; Goto, M.; Watanabe, Y.; Terasawa, T.; Dobashi, K. K1115 A, a new anthraquinone that inhibits the binding of activator protein-1 (AP-1) to its recognition sites. J. Antibiot. 1998, 51, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Caballero, L.; Martinez, E.; Malpartida, F.; Hopwood, D.A. Organisation and functions of the actVA region of the actinorhodin biosynthetic gene cluster of Streptomyces coelicolor. Mol. Genet. Genom. 1991, 230, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Ichinose, K.; Ozawa, M.; Itou, K.; Kunieda, K.; Ebizuka, Y. Cloning, sequencing and heterologous expression of the medermycin biosynthetic gene cluster of Streptomyces sp. AM-7161: Towards comparative analysis of the benzoisochromanequinone gene clusters. Microbiology 2003, 149, 1633–1645. [Google Scholar] [CrossRef]
- Ichinose, K.; Bedford, D.J.; Tornus, D.; Bechthold, A.; Bibb, M.J.; Peter Revill, W.; Floss, H.G.; Hopwood, D.A. The granaticin biosynthetic gene cluster of Streptomyces violaceoruber Tü22: Sequence analysis and expression in a heterologous host. Chem. Biol. 1998, 5, 647–659. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Moreno, M.A.; Martínez, E.; Caballero, J.L.; Ichinose, K.; Hopwood, D.A.; Malpartida, F. DNA sequence and functions of the actVI region of the actinorhodin biosynthetic gene cluster of Streptomyces coelicolor A3(2). J. Biol. Chem. 1994, 269, 24854–24863. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Moreno, M.A.; Martínez, E.; Boto, L.; Hopwood, D.A.; Malpartida, F. Nucleotide sequence and deduced functions of a set of cotranscribed genes of Streptomyces coelicolor A3(2) including the polyketide synthase for the antibiotic actinorhodin. J. Biol. Chem. 1992, 267, 19278–19290. [Google Scholar] [CrossRef]
- Fernández-Moreno, M.A.; Caballero, J.L.; Hopwood, D.A.; Malpartida, F. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell 1991, 66, 769–780. [Google Scholar] [CrossRef]
- Oja, T.; Palmu, K.; Lehmussola, H.; Leppäranta, O.; Hännikäinen, K.; Niemi, J.; Mäntsälä, P.; Metsä-Ketelä, M. Characterization of the alnumycin gene cluster reveals unusual gene products for pyran ring formation and dioxan biosynthesis. Chem. Biol. 2008, 15, 1046–1057. [Google Scholar] [CrossRef]
- Oja, T.; Niiranen, L.; Sandalova, T.; Klika, K.D.; Niemi, J.; Mäntsälä, P.; Schneider, G.; Metsä-Ketelä, M. Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway. Proc. Natl. Acad. Sci. USA 2013, 110, 1291–1296. [Google Scholar] [CrossRef]
- Bieber, B.; Nüske, J. Alnumycin Useful as Antibiotic. German Patent Application DE19745914A, 17 October 1997. [Google Scholar]
- Oja, T.; Galindo, P.S.M.; Taguchi, T.; Manner, S.; Vuorela, P.M.; Ichinose, K.; Metsä-Ketelä, M.; Fallarero, A. Effective antibiofilm polyketides against Staphylococcus aureus from the pyranonaphthoquinone biosynthetic pathways of Streptomyces species. Antimicrob. Agents Chemother. 2015, 59, 6046–6052. [Google Scholar] [CrossRef] [PubMed]
- Grocholski, T.; Oja, T.; Humphrey, L.; Mäntsälä, P.; Niemi, J.; Metsä-Ketelä, M. Characterization of the two-component monooxygenase system AlnT/AlnH reveals early timing of quinone formation in alnumycin biosynthesis. J. Bacteriol. 2012, 194, 2829–2836. [Google Scholar] [CrossRef]
- Xue, C.-M.; Tian, L.; Lin, W.-H.; Deng, Z.-W. Anthraquinone derivatives from Micromonospora rhodorangea. Nat. Prod. Res. 2009, 23, 533–538. [Google Scholar] [CrossRef]
- Poumale, H.M.P.; Ngadjui, B.T.; Helmke, E.; Laatsch, H. New anthraquinones from a marine Streptomyces sp.—Isolation, structure determination and biological activities. Z. Naturforsch. 2006, 61, 1450–1454. [Google Scholar] [CrossRef]
- Huang, Y.-F.; Tian, L.; Sun, Y.; Pei, Y.-H. Two new compounds from marine Streptomyces sp. FX-58. J. Asian. Nat. Prod. Res. 2006, 8, 495–498. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.M.; Zhang, J.L.; Zhou, K.; Yue, L.M.; Zhang, Y.; Wang, C.Y.; Wang, K.L.; Xu, Y. Anthraquinones as potential antibiofilm agents against methicillin-resistant Staphylococcus aureus. Front. Microbiol. 2021, 12, 709826. [Google Scholar] [CrossRef] [PubMed]
- Deitz, W.H.; Bailey, J.H.; Froehlich, E.J. In vitro antibacterial properties of nalidixic acid, a new drug active against gram-negative organisms. Antimicrob. Agents Chemother. 1963, 161, 583–587. [Google Scholar]
- Béjar, S.; Bouché, J.P. The spacing of Escherichia coli DNA gyrase sites cleaved in vivo by treatment with oxolinic acid and sodium dodecyl sulfate. Biochemie 1984, 66, 693–700. [Google Scholar] [CrossRef]
- Barnard, F.M.; Maxwell, A. Interaction between DNA gyrase and quinolones: Effects of alanine mutations at GyrA subunit residues Ser83 and Asp87. Antimicrob. Agents Chemother. 2001, 45, 1994–2000. [Google Scholar] [CrossRef]
- Anderson, V.E.; Zaniewski, R.P.; Kaczmarek, F.S.; Gootz, T.D.; Osheroff, N. Quinolones inhibit DNA religation mediated by Staphylococcus aureus topoisomerase IV. J. Biol. Chem. 1999, 274, 35927–35932. [Google Scholar] [CrossRef]
- Chen, C.-R.; Malik, M.; Snyder, M.; Drlica, K. DNA gyrase and topoisomerase IV on the bacterial chromosome: Quinolone-induced DNA cleavage. J. Mol. Biol. 1996, 258, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Kampranis, S.C.; Maxwell, A. The DNA gyrase-quinolone complex. J. Biol. Chem. 1998, 273, 22615–22626. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.-S.; Cui, Y.; Liao, X.-M.; Tan, X.-B.; Cao, X. Effect of emodin on the cariogenic properties of Streptococcus mutans and the development of caries in rats. Exp. Ther. Med. 2014, 8, 1308–1312. [Google Scholar] [CrossRef] [PubMed]
- Friedman, M.; Xu, A.; Lee, R.; Nguyen, D.N.; Phan, T.A.; Hamada, S.M.; Panchel, R.; Tam, C.C.; Kim, J.H.; Cheng, L.W.; et al. The inhibitory activity of anthraquinones against pathogenic protozoa, bacteria, and fungi and the relationship to structure. Molecules 2020, 25, 3101–3113. [Google Scholar] [CrossRef]
- Lenta, B.N.; Weniger, B.; Antheaume, C.; Noungoue, D.T.; Ngouela, S.; Assob, J.C.N.; Vonthron-Sénécheau, C.; Fokou, P.A.; Devkota, K.P.; Tsamo, E. Anthraquinones from the stem bark of Stereospermum zenkeri with antimicrobial activity. Phytochemistry 2007, 68, 1595–1599. [Google Scholar] [CrossRef]
- Lu, C.; Wang, H.; Lv, W.; Xu, P.; Zhu, J.; Xie, J.; Liu, B.; Lou, Z. Antibacterial properties of anthraquinones extracted from rhubarb against Aeromonas hydrophila. Fish. Sci. 2011, 77, 375–384. [Google Scholar] [CrossRef]
- Amorim, J.C.; Cabrera Bermeo, A.E.; Vásquez Urgilés, V.E.; Martínez León, M.R.; Carpio Arévalo, J.M. An in-silico evaluation of anthraquinones as potential inhibitors of DNA gyrase B of Mycobacterium tuberculosis. Microorganisms 2022, 10, 2434–2448. [Google Scholar] [CrossRef]
- Liu, M.; Peng, W.; Qin, R.; Yan, Z.; Cen, Y.; Zheng, X.; Pan, X.; Jiang, W.; Li, B.; Li, X.; et al. The direct anti-MRSA effect of emodin via damaging cell membrane. Appl. Microbiol. Biotechnol. 2015, 99, 7699–7709. [Google Scholar] [CrossRef]
- Doughari, J.H.; Ndakidemi, P.A.; Human, I.S.; Benade, S. Antioxidant, antimicrobial and antiverotoxic potentials of extracts of Curtisia dentata. J. Ethnopharmacol. 2012, 141, 1041–1050. [Google Scholar] [CrossRef]
- Kuo, I.-P.; Lee, P.-T.; Nan, F.-H. Rheum officinale extract promotes the innate immunity of orange-spotted grouper (Epinephelus coioides) and exerts strong bactericidal activity against six aquatic pathogens. Fish. Shellfish. Immunol. 2020, 102, 117–124. [Google Scholar] [CrossRef]
- Omosa, L.K.; Midiwo, J.O.; Mbaveng, A.T.; Tankeo, S.B.; Seukep, J.A.; Voukeng, I.K.; Dzotam, J.K.; Isemeki, J.; Derese, S.; Omolle, R.A.; et al. Antibacterial activities and structure–activity relationships of a panel of 48 compounds from Kenyan plants against multidrug resistant phenotypes. Springerplus 2016, 5, 901–916. [Google Scholar] [CrossRef]
- Comini, L.R.; Núñez Montoya, S.C.; Páez, P.L.; Argüello, G.A.; Albesa, I.; Cabrera, J.L. Antibacterial activity of anthraquinone derivatives from Heterophyllaea pustulata (Rubiaceae). J. Photochem. Photobiol. B 2011, 102, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.; Peng, W.; Li, X.; Liu, M.; Li, B.; Qin, R.; Jiang, W.; Cen, Y.; Pan, X.; Yan, Z.; et al. Emodin is identified as the active component of ether extracts from Rhizoma Polygoni Cuspidati, for anti-MRSA activity. Can. J. Physiol. Pharmacol. 2015, 93, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Liu, X.; Peng, Y.; Zhan, R.; Xu, H.; Ge, X. Comparative analysis of methicillin-sensitive and resistant Staphylococcus aureus exposed to emodin based on proteomic profiling. Biochem. Biophys. Res. Commun. 2017, 494, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Küster, E.; Williams, S.T. Selection of media for isolation of streptomycetes. Nature 1964, 202, 928–929. [Google Scholar] [CrossRef]
- Kieser, T.; Bibb, M.; Buttner, M.; Chater, K.; Hopwood, D. Practical Streptomyces Genetics; John Innes Foundation: Norwich, UK, 1985. [Google Scholar]
- Koren, S.; Walzen, B.; Berlin, K.; Miller, J.; Bergmann, N.; Phillippy, A. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Miller, J.; Koren, S.; Sutton, G. Assembly algorithms for next-generation sequencing data. Genomics 2010, 95, 312–327. [Google Scholar] [CrossRef]
- Wick, R.; Judd, L.; Gorrie, C.; Holt, K. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. AntiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, 339–346. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. AntiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, 29–35. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef] [PubMed]
- CLSI M07-A8; Method for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2009.
- Hallett, P.; Grimshaw, A.J.; Wigley, D.B.; Maxwell, A. Cloning of the DNA gyrase genes under tac promoter control: Overproduction of the gyrase A and B proteins. Gene 1990, 93, 139–142. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M.G. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60–74. [Google Scholar] [CrossRef]
- Nolan, R.A. Amino acids and growth factors in vitamin-free casamino acids. Mycologia 1971, 63, 1231–1234. [Google Scholar] [CrossRef]
- Nolan, R.A.; Nolan, W.G. Elemental analysis of vitamin-free casamino acids. Appl. Microbiol. 1972, 24, 290–291. [Google Scholar] [CrossRef]
- Seixas Chaves, D.F. Malt extract as a healthy substitute for refined sugar. Am. J. Biomed. Sci. Res. 2019, 4, 52–53. [Google Scholar] [CrossRef]
- Zhang, J.; Reddy, J.; Buckland, B.; Greasham, R. Toward consistent and productive complex media for industrial fermentations: Studies on yeast extract for a recombinant yeast fermentation process. Biotechnol. Bioeng. 2003, 82, 640–652. [Google Scholar] [CrossRef]
- Grant, C.L.; Pramer, D. Minor element composition of yeast extract. J. Bacteriol. 1962, 84, 869–870. [Google Scholar] [CrossRef] [PubMed]
- Porter, M.A.; Jones, A.M. Variability in soy flour composition. JAOCS 2003, 80, 557–562. [Google Scholar] [CrossRef]
- Davami, F.; Eghbalpour, F.; Nematollahi, L.; Barkhordari, F.; Mahboudi, F. Effects of peptone supplementation in different culture media on growth, metabolic pathway and productivity of CHO DG44 cells; a new insight into amino acid profiles. Iran Biomed. J. 2015, 19, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Perassolo, M.; Quevedo, C.; Busto, V.; Ianone, F.; Giulietti, A.M.; Talou, J.R. Enhance of anthraquinone production by effect of proline and aminoindan-2-phosphonic acid in Rubia tinctorum suspension cultures. Enzyme Microb. Technol. 2007, 41, 181–185. [Google Scholar] [CrossRef]
- Kim, Y.J.; Song, J.Y.; Moon, M.H.; Smith, C.P.; Hong, S.-K.; Chang, Y.K. pH shock induces overexpression of regulatory and biosynthetic genes for actinorhodin productionin Streptomyces coelicolor A3(2). Appl. Microbiol. Biotechnol. 2007, 76, 1119–1130. [Google Scholar] [CrossRef] [PubMed]
- Farooq, U.; Khan, S.; Naz, S.; Khan, A.; Khan, A.; Ahmed, A.; Rauf, A.; Bukhari, S.M.; Khan, S.A.; Kamil, A.; et al. Three new anthraquinone derivatives isolated from Symplocos racemosa and their antibiofilm activity. Chin. J. Nat. Med. 2017, 15, 944–949. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Metsä-Ketelä, M.; Hertweck, C. Ketosynthase III as a gateway to engineering the biosynthesis of antitumoral benastatin derivatives. J. Biotechnol. 2009, 140, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Meadows, E.S.; Khosla, C. In vitro reconstitution and analysis of the chain initiating enzymes of the R1128 polyketide synthase. Biochemistry 2001, 40, 14855–14861. [Google Scholar] [CrossRef]
- Tang, Y.; Koppisch, A.T.; Khosla, C. The acyltransferase homologue from the initiation module of the R1128 polyketide synthase is an acyl-ACP thioesterase that edits acetyl primer units. Biochemistry 2004, 43, 9546–9555. [Google Scholar] [CrossRef]
- Taguchi, T.; Kunieda, K.; Takeda-Shitaka, M.; Takaya, D.; Kawano, N.; Kimberley, M.R.; Booker-Milburn, K.I.; Stephenson, G.R.; Umeyama, H.; Ebizuka, Y.; et al. Remarkably different structures and reaction mechanisms of ketoreductases for the opposite stereochemical control in the biosynthesis of BIQ antibiotics. Bioorg. Med. Chem. 2004, 12, 5917–5927. [Google Scholar] [CrossRef] [PubMed]
- Gartman, J.A.; Tambar, U.K. Recent total syntheses of anthraquinone-based natural products. Tetrahedron 2022, 105, 132501. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Redman, N.; Litke, A.P.; Zeng, J.; Zhan, J.; Chan, K.Y.; Chang, C.-W.T. Synthesis and antibacterial activity study of a novel class of cationic anthraquinone analogs. Bioorg. Med. Chem. 2011, 19, 498–503. [Google Scholar] [CrossRef]
- Degtyareva, N.N.; Gong, C.; Story, S.; Levinson, N.S.; Oyelere, A.K.; Green, K.D.; Garneau-Tsodikova, S.; Arya, D.P. Antimicrobial activity, AME resistance, and A-Site binding studies of anthraquinone–neomycin conjugates. ACS Infect. Dis. 2017, 3, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Nicolaou, K.C.; Das, D.; Lu, Y.; Rout, S.; Pitsinos, E.N.; Lyssikatos, J.; Schammel, A.; Sandoval, J.; Hammond, M.; Aujay, M.; et al. Total synthesis and biological evaluation of tiancimycins A and B, yangpumicin A, and related anthraquinone-fused enediyne antitumor antibiotics. J. Am. Chem. Soc. 2020, 142, 2549–2561. [Google Scholar] [CrossRef] [PubMed]
| Compound | E. coli ΔtolC | E. coli DSM 30083 | A. baumannii DSM 30007 | B. subtilis 168 | S. aureus DSM 20231 | S. aureus Mu50 |
|---|---|---|---|---|---|---|
| Alnumycin | 100 µg/mL | >100 µg/mL | >100 µg/mL | 8 µg/mL | 10 µg/mL | 8 µg/mL |
| 1,6-dihydro 8-propylanthraquinone | 10 µg/mL | >100 µg/mL | >100 µg/mL | 10 µg/mL | 10 µg/mL | 8 µg/mL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sagurna, L.; Heinrich, S.; Kaufmann, L.-S.; Rückert-Reed, C.; Busche, T.; Wolf, A.; Eickhoff, J.; Klebl, B.; Kalinowski, J.; Bandow, J.E. Characterization of the Antibacterial Activity of Quinone-Based Compounds Originating from the Alnumycin Biosynthetic Gene Cluster of a Streptomyces Isolate. Antibiotics 2023, 12, 1116. https://doi.org/10.3390/antibiotics12071116
Sagurna L, Heinrich S, Kaufmann L-S, Rückert-Reed C, Busche T, Wolf A, Eickhoff J, Klebl B, Kalinowski J, Bandow JE. Characterization of the Antibacterial Activity of Quinone-Based Compounds Originating from the Alnumycin Biosynthetic Gene Cluster of a Streptomyces Isolate. Antibiotics. 2023; 12(7):1116. https://doi.org/10.3390/antibiotics12071116
Chicago/Turabian StyleSagurna, Leonie, Sascha Heinrich, Lara-Sophie Kaufmann, Christian Rückert-Reed, Tobias Busche, Alexander Wolf, Jan Eickhoff, Bert Klebl, Jörn Kalinowski, and Julia E. Bandow. 2023. "Characterization of the Antibacterial Activity of Quinone-Based Compounds Originating from the Alnumycin Biosynthetic Gene Cluster of a Streptomyces Isolate" Antibiotics 12, no. 7: 1116. https://doi.org/10.3390/antibiotics12071116
APA StyleSagurna, L., Heinrich, S., Kaufmann, L.-S., Rückert-Reed, C., Busche, T., Wolf, A., Eickhoff, J., Klebl, B., Kalinowski, J., & Bandow, J. E. (2023). Characterization of the Antibacterial Activity of Quinone-Based Compounds Originating from the Alnumycin Biosynthetic Gene Cluster of a Streptomyces Isolate. Antibiotics, 12(7), 1116. https://doi.org/10.3390/antibiotics12071116







