In Vitro Activity of Novel Topoisomerase Inhibitors against Francisella tularensis and Burkholderia pseudomallei
Abstract
1. Introduction
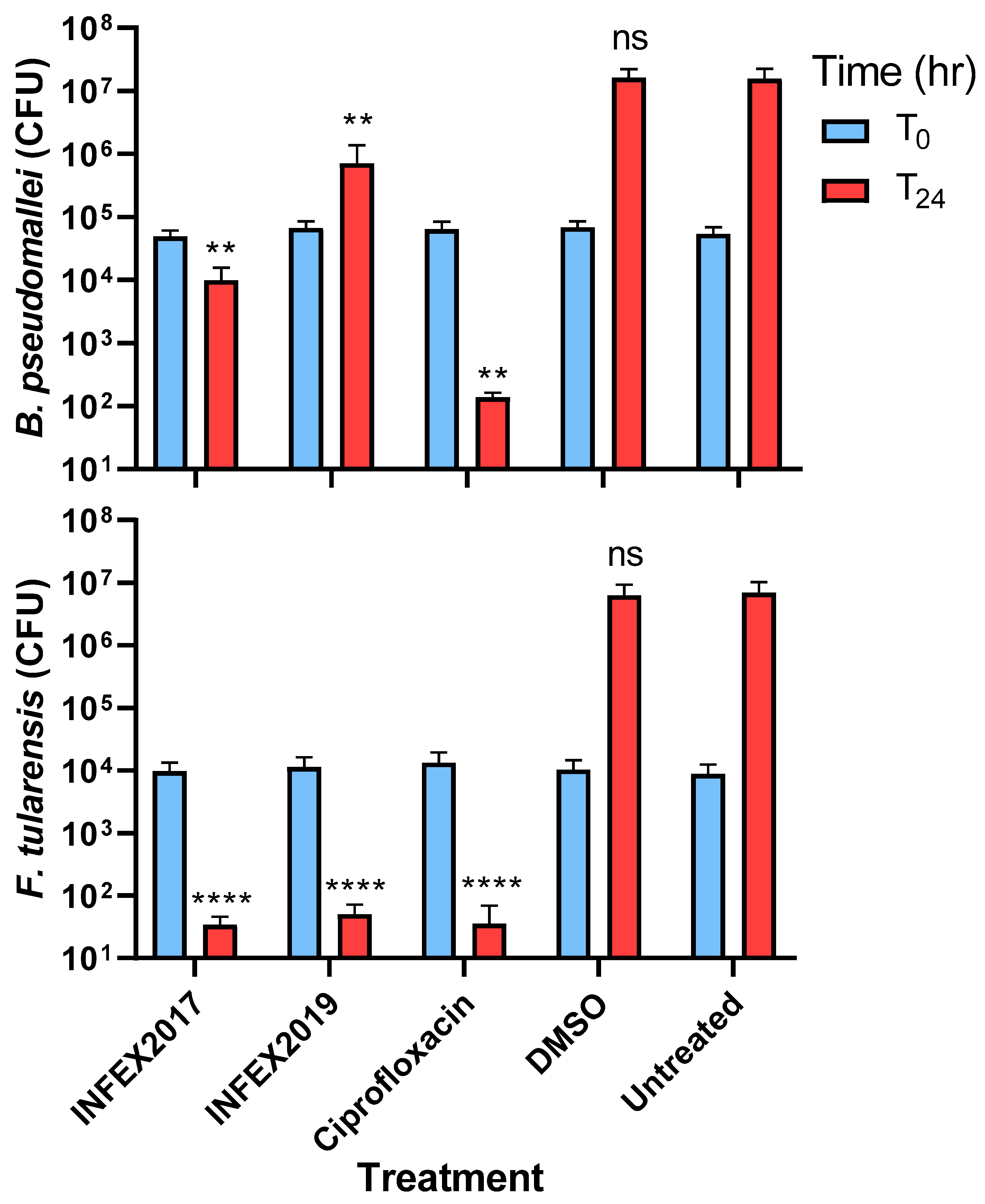
2. Results
3. Materials and Methods
3.1. Reagents
3.2. Bacterial Strains and Cultures
3.3. MIC Assays
3.4. Time-Kill Assays
3.5. Intracellular Assays
3.6. Statistical Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheng, A.C.; Currie, B.J. Melioidosis: Epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 2005, 18, 383–416. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.; Oyston, P.C.; Green, M.; Titball, R.W. Tularemia. Clin. Microbiol. Rev. 2002, 15, 631–646. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.; Hu, Z.; Chen, J.; Tang, M.; Chen, H.; Lu, X.; Cao, L.; Deng, L.; Mao, X.; Li, Q. Molecular epidemiology and antibiotic resistance of Burkholderia pseudomallei isolates from Hainan. China Med. 2019, 98, 14461. [Google Scholar] [CrossRef]
- Narayanan, N.; Lacy, C.R.; Cruz, J.E.; Nahass, M.; Karp, J.; Barone, J.A.; Hermes-DeSantis, E.R. Disaster preparedness: Biological threats and treatment options. Pharmacotherapy 2018, 38, 217–234. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, R.P.; Ward, L.; Currie, B.J. Oral eradication therapy for melioidosis; important but not without risks. Int. J. Infect. Dis. 2019, 80, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Hooper, D.C. Emerging mechanisms of fluoroquinolone resistance. Emerg Infect Dis. 2001, 7, 337–341. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Redgrave, L.S.; Sutton, S.B.; Webber, M.A.; Piddock, L.J.V. Fluoroquinolone resistance: Mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014, 22, 438–445. [Google Scholar] [CrossRef]
- Wright, G.D. Molecular mechanisms of antibiotic resistance. Chem. Commun. 2011, 47, 4055–4061. [Google Scholar] [CrossRef]
- Gibson, E.G.; Bax, B.; Chan, P.F.; Osheroff, N. Mechanistic and structural basis for the actions of the antibacterial Gepotidacin against Staphylococcus aureus gyrase. ACS Infect. Dis. 2019, 5, 570–581. [Google Scholar] [CrossRef]
- Kolarič, A.; Anderluh, M.; Minovski, N. Two decades of successful SAR-grounded stories of the novel bacterial topoisomerase inhibitors (NBTIs). J. Med. Chem. 2020, 63, 5664–5674. [Google Scholar] [CrossRef] [PubMed]
- Jaswal, S.; Nehra, B.; Kumar, S.; Monge, V. Recent advancements in the medicinal chemistry of bacterial type II topoisomerase inhibitors. Bioorg. Chem. 2020, 104, 104266. [Google Scholar] [CrossRef] [PubMed]
- Reck, F.; Alm, R.A.; Brassil, P.; Newman, J.V.; Ciaccio, P.; McNulty, J.; Barthlow, H.; Goteti, K.; Breen, J.; Comita-Prevoir, J.; et al. Novel N-linked aminopiperidine inhibitors of bacterial topoisomerase type II with reduced p K a: Antibacterial agents with an improved safety profile. J. Med. Chem. 2012, 55, 6916–6933. [Google Scholar] [CrossRef]
- Bulter, A.; Helliwell, M.V.; Zhange, Y.; Hancox, J.C.; Dempsey, C.E. An update on the structure of hERG. Front. Pharmacol. 2020, 10, 1572. [Google Scholar]
- Biedenbach, D.J.; Bouchillon, S.K.; Hackel, M.; Miller, L.A.; Scangarella-Oman, N.E.; Jakielaszek, C.; Sahm, D.F. In vitro activity of gepotidacin, a novel triazaacenaphthylene bacterial topoisomerase inhibitor, against a broad spectrum of bacterial pathogens. Antimicrob. Agents. Chemother. 2016, 60, 1918–1923. [Google Scholar] [CrossRef] [PubMed]
- Jacobsson, S.; Golparian, D.; Scangarella-Oman, N.; Unemo, M. In vitro activity of the novel triazaacenaphthylene gepotidacin (GSK2140944) against MDR Neisseria gonorrhoeae. J. Antimicrob. Chemother. 2018, 73, 2072–2077. [Google Scholar] [CrossRef] [PubMed]
- Jakielaszek, C.; Hossain, M.; Qian, L.; Fishman, C.; Widdowson, K.; Hilliard, J.J.; Mannino, F.; Raychaudhuri, A.; Carniel, E.; Demons, S.; et al. Gepotidacin is efficacious in a nonhuman primate model of pneumonic plague. Sci. Transl. Med. 2022, 14, 1787. [Google Scholar] [CrossRef] [PubMed]
- Lyons, A.; Kirkham, J.; Blades, K.; Orr, D.; Dauncey, E.; Smith, O.; Dick, E.; Walker, R.; Matthews, T.; Bunt, A.; et al. Discovery and structure-activity relationships of a novel oxazolidinone class of bacterial type II topoisomerase inhibitors. Bioorg. Med. Chem. Lett. 2022, 65, 128648. [Google Scholar] [CrossRef]
- Larsson, P.; Oyston, P.C.F.; Chain, P.; Chu, M.C.; Duffield, M.; Fuxelius, H.-H.; Garcia, E.; Hälltorp, G.; Johansson, D.; E Isherwood, K.; et al. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Gen. 2005, 37, 153–159. [Google Scholar] [CrossRef]
- Holden, M.T.G.; Titball, R.W.; Peacock, S.J.; Cerdeño-Tárraga, A.M.; Atkins, T.; Crossman, L.C.; Pitt, T.; Churcher, C.; Mungall, K.; Bentley, S.D.; et al. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. USA 2004, 101, 14240–14245. [Google Scholar] [CrossRef]
- Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard. CSLI Document M07-A10; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015.
- Eyles, J.E.; Hartley, M.G.; Laws, T.R.; Oyston, P.C.F.; Griffin, K.F.; Titball, R.W. Protection afforded against aerosol challenge by systemic immunisation with inactivated Francisella tularensis live vaccine strain (LVS). Microb. Pathog. 2008, 44, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Methods for Determining Bactericidal Activity of Antimicrobial Agents: Approved Guideline; CLSI Document M26-A; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 1999.
- Bax, B.D.; Chan, P.F.; Eggleston, D.S.; Fosberry, A.; Gentry, D.R.; Gorrec, F.; Giordano, I.; Hann, M.M.; Hennessy, A.; Hibbs, M.; et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature 2010, 466, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Charrier, C.; Salisbury, A.M.; Savage, V.J.; Duffy, T.; Moyo, E.; Chaffer-Malam, N.; Ooi, N.; Newman, R.; Cheung, J.; Metzger, R.; et al. Novel bacterial topoisomerase inhibitors with potent broad-spectrum activity against drug-resistant bacteria. Antimicrob. Agents. Chemother. 2017, 61, e02100-16. [Google Scholar] [CrossRef]
- Ross, B.N.; Myers, J.N.; Muruato, L.A.; Tapia, D.; Torres, A.G. Evaluating new compounds to treat Burkholderia pseudomallei infections. Front. Cell. Infect. Microbiol. 2018, 8, 210. [Google Scholar] [CrossRef]
- Bommineni, G.R.; Kapilashrami, K.; Cummings, J.E.; Lu, L.; Knudson, S.E.; Gu, C.; Walker, S.G.; Slayden, S.A.; Tonge, P.J. Thiolactomycin-based inhibitors of bacterial β-Ketoacyl-ACP synthases with in vivo activity. J. Med. Chem. 2016, 59, 5377–5390. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.E.; Kingry, L.C.; Rholl, D.A.; Schweizer, H.P.; Tonge, P.J.; Slayden, R.A. The Burkholderia pseudomallei enoyl-acyl carrier protein reductase fabI1 is essential for in vivo growth and is the target of a novel chemotherapeutic with efficacy. Antimicrob. Agents. Chemother. 2014, 58, 931–935. [Google Scholar] [CrossRef]
- Cummings, J.E.; Beaupre, A.J.; Knudson, S.E.; Liu, N.; Yu, W.; Neckles, C.; Wang, H.; Khanna, A.; Bommineni, G.R.; Trunck, L.A.; et al. Substituted diphenyl ethers as a novel chemotherapeutic platform against Burkholderia pseudomallei. Antimicrob. Agents. Chemother. 2014, 58, 1646–1651. [Google Scholar] [CrossRef] [PubMed]

| Inhibitor/Antibiotic | MIC (μg/mL) | |
|---|---|---|
| B. pseudomallei | F. tularensis | |
| INFEX993 | 2 | 0.06–0.125 |
| INFEX2017 | 4 | 0.5–1 |
| INFEX2018 | 8 | 8–16 |
| INFEX2019 | 4 | 0.5–4 |
| INFEX2020 | 8 | 16–32 |
| Ciprofloxacin | 0.5–2 | 0.03–0.5 |
| Doxycycline | 0.5–4 | 2–4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© Crown copyright (2023), Dstl. This material is licensed under the terms of the Open Government Licence except where otherwise stated. To view this licence, visit http://www.nationalarchives.gov.uk/doc/open-government-licence/version/3 or write to the Information Policy Team, The National Archives, Kew, London TW9 4DU, or email: psi@nationalarchives.gov.uk.
Share and Cite
Whelan, A.O.; Cooper, I.; Ooi, N.; Orr, D.; Blades, K.; Kirkham, J.; Lyons, A.; Barnes, K.B.; Richards, M.I.; Salisbury, A.-M.; et al. In Vitro Activity of Novel Topoisomerase Inhibitors against Francisella tularensis and Burkholderia pseudomallei. Antibiotics 2023, 12, 983. https://doi.org/10.3390/antibiotics12060983
Whelan AO, Cooper I, Ooi N, Orr D, Blades K, Kirkham J, Lyons A, Barnes KB, Richards MI, Salisbury A-M, et al. In Vitro Activity of Novel Topoisomerase Inhibitors against Francisella tularensis and Burkholderia pseudomallei. Antibiotics. 2023; 12(6):983. https://doi.org/10.3390/antibiotics12060983
Chicago/Turabian StyleWhelan, Adam O., Ian Cooper, Nicola Ooi, David Orr, Kevin Blades, James Kirkham, Amanda Lyons, Kay B. Barnes, Mark I. Richards, Anne-Marie Salisbury, and et al. 2023. "In Vitro Activity of Novel Topoisomerase Inhibitors against Francisella tularensis and Burkholderia pseudomallei" Antibiotics 12, no. 6: 983. https://doi.org/10.3390/antibiotics12060983
APA StyleWhelan, A. O., Cooper, I., Ooi, N., Orr, D., Blades, K., Kirkham, J., Lyons, A., Barnes, K. B., Richards, M. I., Salisbury, A.-M., Craighead, M., & Harding, S. V. (2023). In Vitro Activity of Novel Topoisomerase Inhibitors against Francisella tularensis and Burkholderia pseudomallei. Antibiotics, 12(6), 983. https://doi.org/10.3390/antibiotics12060983






