CRISPR-Cas9 System: A Prospective Pathway toward Combatting Antibiotic Resistance
Abstract
1. Introduction
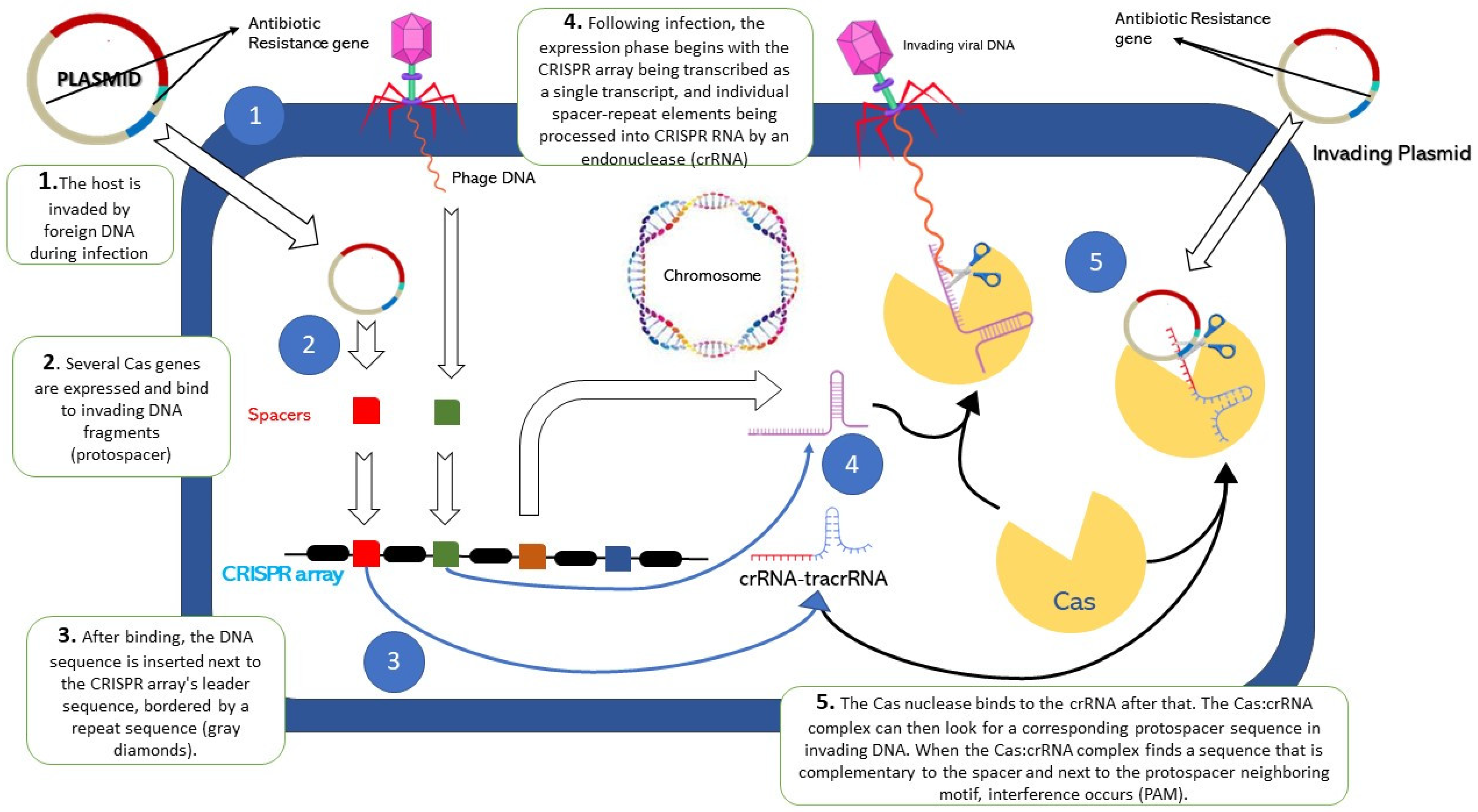
2. Overview of CRISPR Cas9
3. The Perilous Persistence of Drug Resistance
4. Roles of CRISPR-Cas in Antibiotic-Resistant Bacteria
5. Neutralization of Antibiotic-Resistant Genes by CRISPR-Cas System
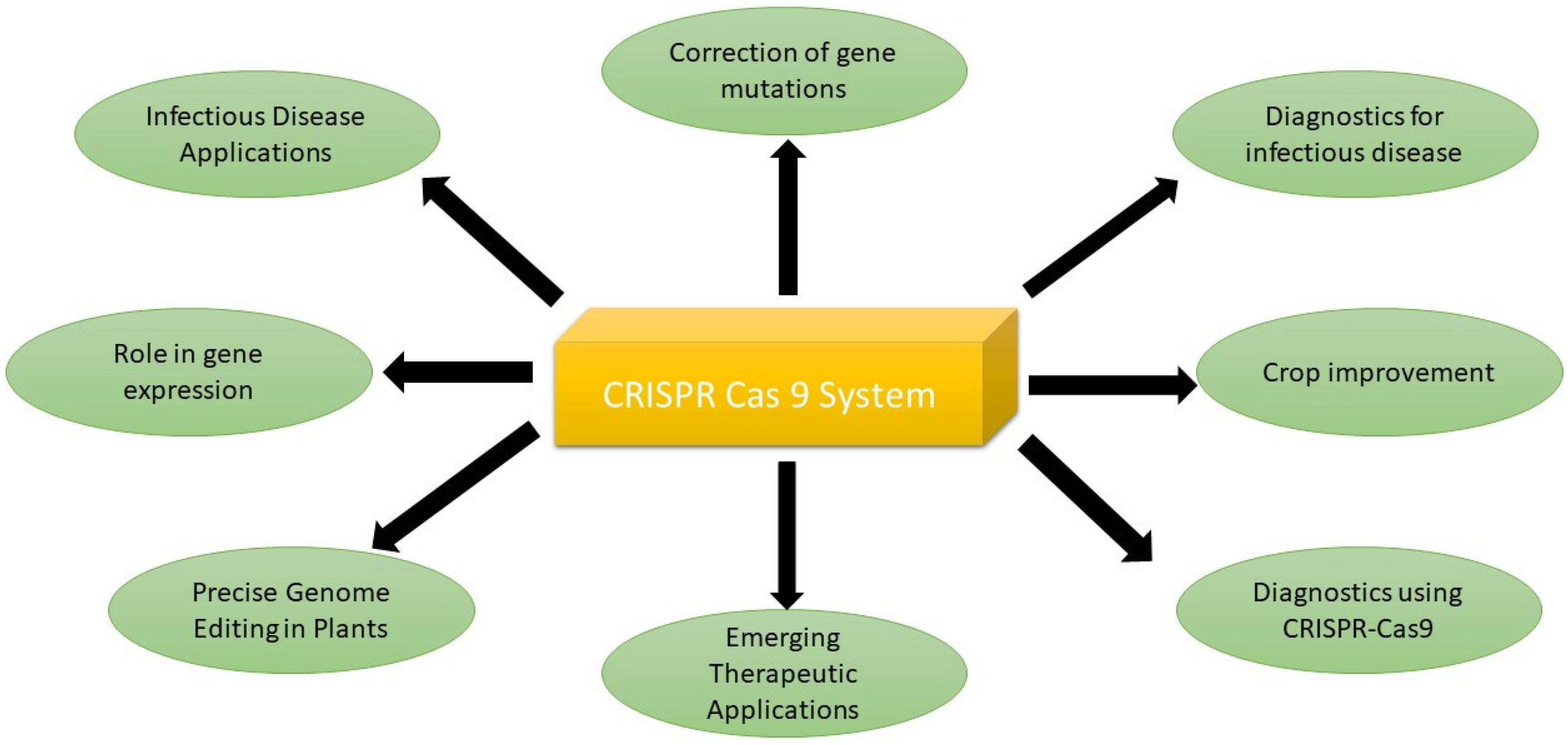
6. Applications of CRISPR-Cas9 System
6.1. Correction of Gene Mutations
6.2. Infectious Disease Applications
6.3. Revolutionizing Fungal Disease Control with CRISPR-Cas9
6.4. Emerging Therapeutic Applications
6.5. Role in Gene Expression
7. Challenges
8. Future of CRISPR
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kumar, M.; Sarma, D.K.; Shubham, S.; Kumawat, M.; Verma, V.; Nina, P.B.; JP, D.; Kumar, S.; Singh, B.; Tiwari, R.R. Futuristic Non-Antibiotic Therapies to Combat Antibiotic Resistance: A Review. Front. Microbiol. 2021, 12, 609459. [Google Scholar] [CrossRef] [PubMed]
- Bilal, H.; Khan, M.N.; Rehman, T.; Hameed, M.F.; Yang, X. Antibiotic Resistance in Pakistan: A Systematic Review of Past Decade. BMC Infect. Dis. 2021, 21, 244. [Google Scholar] [CrossRef] [PubMed]
- Pereira, H.S.; Tagliaferri, T.L.; Mendes, T.A.D.O. Enlarging the Toolbox Against Antimicrobial Resistance: Aptamers and CRISPR-Cas. Front. Microbiol. 2021, 12, 606360. [Google Scholar] [CrossRef] [PubMed]
- Jebri, S.; Rahmani, F.; Hmaied, F. Bacteriophages as Antibiotic Resistance Genes Carriers in Agro-Food Systems. J. Appl. Microbiol. 2021, 130, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Santajit, S.; Indrawattana, N. Mechanisms of Antimicrobial Resistance in Pasteurellaceae. BioMed Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef]
- Gholizadeh, P.; Köse, Ş.; Dao, S.; Ganbarov, K.; Tanomand, A.; Dal, T.; Aghazadeh, M.; Ghotaslou, R.; Rezaee, M.A.; Yousefi, B.; et al. How CRISPR-Cas System Could Be Used to Combat Antimicrobial Resistance. Infect. Drug Resist. 2020, 13, 1111–1121. [Google Scholar] [CrossRef]
- Wojciechowska, M.; Równicki, M.; Mieczkowski, A.; Miszkiewicz, J.; Trylska, J. Antibacterial Peptide Nucleic Acids-Facts and Perspectives. Molecules 2020, 25, 559. [Google Scholar] [CrossRef]
- Hatfull, G.F.; Dedrick, R.M.; Schooley, R.T. Phage Therapy for Antibiotic-Resistant Bacterial Infections. Annu. Rev. Med. 2022, 73, 197–211. [Google Scholar] [CrossRef]
- Shahbazi Dastjerdeh, M.; Kouhpayeh, S.; Sabzehei, F.; Khanahmad, H.; Salehi, M.; Mohammadi, Z.; Shariati, L.; Hejazi, Z.; Rabiei, P.; Manian, M. Zinc Finger Nuclease: A New Approach to Overcome Beta-Lactam Antibiotic Resistance. Jundishapur J. Microbiol. 2016, 9, e29384. [Google Scholar] [CrossRef]
- Aslam, B.; Rasool, M.; Idris, A.; Muzammil, S.; Alvi, R.F.; Khurshid, M.; Rasool, M.H.; Zhang, D.; Ma, Z.; Baloch, Z. CRISPR-Cas System: A Potential Alternative Tool to Cope Antibiotic Resistance. Antimicrob. Resist. Infect. Control 2020, 9, 6–8. [Google Scholar] [CrossRef]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide Sequence of the Iap Gene, Responsible for Alkaline Phosphatase Isozyme Conversion in Escherichia Coli, and Identification of the Gene Product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance against Viruses in Prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Asmamaw, M.; Zawdie, B. Mechanism and Applications of CRISPR/Cas-9-Mediated Genome Editing. Biol. Targets Ther. 2021, 15, 353–361. [Google Scholar]
- Watson, B.N.J.; Staals, R.H.J.; Fineran, P.C. CRISPR-Cas-Mediated Phage Resistance Enhances Horizontal Gene Transfer by Transduction. MBio 2018, 9, e02406-17. [Google Scholar] [CrossRef]
- Xu, Y.; Li, Z. CRISPR-Cas Systems: Overview, Innovations and Applications in Human Disease Research and Gene Therapy. Comput. Struct. Biotechnol. J. 2020, 18, 2401–2415. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Haft, D.H.; et al. An Updated Evolutionary Classification of CRISPR-Cas Systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef]
- Medina-Aparicio, L.; Rebollar-Flores, J.E.; Gallego-Hernández, A.L.; Vázquez, A.; Olvera, L.; Gutiérrez-Ríos, R.M.; Calva, E.; Hernandez-Lucas, I. The CRISPR/Cas Immune System Is an Operon Regulated by LeuO, H-NS, and Leucine-Responsive Regulatory Protein in Salmonella Enterica Serovar Typhi. J. Bacteriol. 2011, 193, 2396–2407. [Google Scholar] [CrossRef]
- Jiang, C.; Lv, G.; Tu, Y.; Cheng, X.; Duan, Y.; Zeng, B.; He, B. Applications of CRISPR/Cas9 in the Synthesis of Secondary Metabolites in Filamentous Fungi. Front. Microbiol. 2021, 12, 638096. [Google Scholar] [CrossRef]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA Maturation by Trans-Encoded Small RNA and Host Factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef]
- Heler, R.; Samai, P.; Modell, J.W.; Weiner, C.; Goldberg, G.W.; Bikard, D.; Marraffini, L.A. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature 2015, 519, 199–202. [Google Scholar] [CrossRef]
- Horvath, P.; Barrangou, R. CRISPR/Cas, the Immune System of Bacteria and Archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, Classification and Evolution of CRISPR-Cas Systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Doudna, J.A. CRISPR–Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef] [PubMed]
- Lundgren, M.; Charpentier, E.; Fineran, P.C. CRISPR: Methods and Protocols; Humana Press: New York, NY, USA, 2015; Volume 1311, pp. 1–366. [Google Scholar] [CrossRef]
- Liu, Z.; Dong, H.; Cui, Y.; Cong, L.; Zhang, D. Application of Different Types of CRISPR/Cas-Based Systems in Bacteria. Microb. Cell Fact. 2020, 19, 172. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.M.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and Classification of the CRISPR-Cas Systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef]
- Tang, Y.; Fu, Y. Class 2 CRISPR/Cas: An Expanding Biotechnology Toolbox for and beyond Genome Editing 06 Biological Sciences 0604 Genetics. Cell Biosci. 2018, 8, 59. [Google Scholar] [CrossRef]
- Shmakov, S.; Smargon, A.; Scott, D.; Cox, D.; Pyzocha, N.; Yan, W.; Abudayyeh, O.O.; Gootenberg, J.S.; Makarova, K.S.; Wolf, Y.I. Diversity and Evolution of Class 2 CRISPR–Cas Systems. Nat. Rev. Microbiol. 2017, 15, 169–182. [Google Scholar] [CrossRef]
- Levy, S.B.; Marshall, B. Antibacterial Resistance Worldwide: Causes, Challenges and Responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef]
- Martínez, J.L.; Coque, T.M.; Lanza, V.F.; de la Cruz, F.; Baquero, F. Genomic and Metagenomic Technologies to Explore the Antibiotic Resistance Mobilome. Ann. N. Y. Acad. Sci. 2017, 1388, 26–41. [Google Scholar] [CrossRef]
- Wright, G.D. Q&A: Antibiotic Resistance: Where Does It Come from and What Can We Do about It? BMC Biol. 2010, 8, 123. [Google Scholar]
- Wright, R.C.T.; Friman, V.-P.; Smith, M.C.M.; Brockhurst, M.A. Resistance Evolution against Phage Combinations Depends on the Timing and Order of Exposure. MBio 2019, 10, e01652-19. [Google Scholar] [CrossRef]
- Isozumi, R.; Yoshimatsu, K.; Yamashiro, T.; Hasebe, F.; Nguyen, B.M.; Ngo, T.C.; Yasuda, S.P.; Koma, T.; Shimizu, K.; Arikawa, J. BlaNDM-1-Positive Klebsiella Pneumoniae from Environment, Vietnam. Emerg. Infect. Dis. 2012, 18, 1383–1385. [Google Scholar] [CrossRef]
- Baquero, F.; Martínez, J.L.; Cantón, R. Antibiotics and Antibiotic Resistance in Water Environments. Curr. Opin. Biotechnol. 2008, 19, 260–265. [Google Scholar] [CrossRef]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic Resistance: A Rundown of a Global Crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef]
- Burley, K.M.; Sedgley, C.M. CRISPR-Cas, a Prokaryotic Adaptive Immune System, in Endodontic, Oral, and Multidrug-Resistant Hospital-Acquired Enterococcus Faecalis. J. Endod. 2012, 38, 1511–1515. [Google Scholar] [CrossRef]
- Yosef, I.; Manor, M.; Kiro, R.; Qimron, U. Temperate and Lytic Bacteriophages Programmed to Sensitize and Kill Antibiotic-Resistant Bacteria. Proc. Natl. Acad. Sci. USA 2015, 112, 7267–7272. [Google Scholar] [CrossRef]
- Kim, J.-S.; Cho, D.-H.; Park, M.; Chung, W.-J.; Shin, D.; Ko, K.S.; Kweon, D.-H. CRISPR/Cas9-Mediated Re-Sensitization of Antibiotic-Resistant Escherichia Coli Harboring Extended-Spectrum β-Lactamases. J. Microbiol. Biotechnol. 2016, 26, 394–401. [Google Scholar] [CrossRef]
- Bikard, D.; Euler, C.W.; Jiang, W.; Nussenzweig, P.M.; Goldberg, G.W.; Duportet, X.; Fischetti, V.A.; Marraffini, L.A. Exploiting CRISPR-Cas Nucleases to Produce Sequence-Specific Antimicrobials. Nat. Biotechnol. 2014, 32, 1146–1150. [Google Scholar] [CrossRef]
- Hu, W.; Kaminski, R.; Yang, F.; Zhang, Y.; Cosentino, L.; Li, F.; Luo, B.; Alvarez-Carbonell, D.; Garcia-Mesa, Y.; Karn, J.; et al. RNA-Directed Gene Editing Specifically Eradicates Latent and Prevents New HIV-1 Infection. Proc. Natl. Acad. Sci. USA 2014, 111, 11461–11466. [Google Scholar] [CrossRef]
- Yin, C.; Zhang, T.; Qu, X.; Zhang, Y.; Putatunda, R.; Xiao, X.; Li, F.; Xiao, W.; Zhao, H.; Dai, S.; et al. In Vivo Excision of HIV-1 Provirus by SaCas9 and Multiplex Single-Guide RNAs in Animal Models. Mol. Ther. 2017, 25, 1168–1186. [Google Scholar] [CrossRef]
- Van Diemen, F.R.; Kruse, E.M.; Hooykaas, M.J.G.; Bruggeling, C.E.; Schürch, A.C.; van Ham, P.M.; Imhof, S.M.; Nijhuis, M.; Wiertz, E.J.H.J.; Lebbink, R.J. CRISPR/Cas9-Mediated Genome Editing of Herpesviruses Limits Productive and Latent Infections. PLoS Pathog. 2016, 12, e1005701. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sheng, C.; Wang, S.; Yang, L.; Liang, Y.; Huang, Y.; Liu, H.; Li, P.; Yang, C.; Yang, X.; et al. Removal of Integrated Hepatitis B Virus DNA Using CRISPR-Cas9. Front. Cell. Infect. Microbiol. 2017, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, E.M.; Kornepati, A.V.R.; Goldstein, M.; Bogerd, H.P.; Poling, B.C.; Whisnant, A.W.; Kastan, M.B.; Cullen, B.R. Inactivation of the Human Papillomavirus E6 or E7 Gene in Cervical Carcinoma Cells by Using a Bacterial CRISPR/Cas RNA-Guided Endonuclease. J. Virol. 2014, 88, 11965–11972. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, F.; van Schaik, W.; Manson McGuire, A.; Godfrey, P.; Griggs, A.; Mazumdar, V.; Corander, J.; Cheng, L.; Saif, S.; Young, S. Emergence of Epidemic Multidrug-Resistant Enterococcus Faecium from Animal and Commensal Strains. MBio 2013, 4, e00534-13. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, M.S.; Clewell, D.B.; Courvalin, P.; Dunny, G.M.; Murray, B.E.; Rice, L.B. The Enterococci: Pathogenesis, Molecular Biology, and Antibiotic Resistance; ASM Press: Washington, DC, USA, 2002; Volume 10, ISBN 1-55581-234-1. [Google Scholar]
- Roberts, A.P.; Mullany, P. Tn 916-like Genetic Elements: A Diverse Group of Modular Mobile Elements Conferring Antibiotic Resistance. FEMS Microbiol. Rev. 2011, 35, 856–871. [Google Scholar] [CrossRef] [PubMed]
- Brouns, S.J.J.; Jore, M.M.; Lundgren, M.; Westra, E.R.; Slijkhuis, R.J.H.; Snijders, A.P.L.; Dickman, M.J.; Makarova, K.S.; Koonin, E.V.; Van Der Oost, J. Small CRISPR RNAs Guide Antiviral Defense in Prokaryotes. Science 2008, 321, 960–964. [Google Scholar] [CrossRef]
- Marraffini, L.A.; Sontheimer, E.J. CRISPR Interference: RNA-Directed Adaptive Immunity in Bacteria and Archaea. Nat. Rev. Genet. 2010, 11, 181–190. [Google Scholar] [CrossRef]
- Touchon, M.; Charpentier, S.; Pognard, D.; Picard, B.; Arlet, G.; Rocha, E.P.C.; Denamur, E.; Branger, C. Antibiotic Resistance Plasmids Spread among Natural Isolates of Escherichia Coli in Spite of CRISPR Elements. Microbiology 2012, 158, 2997–3004. [Google Scholar] [CrossRef]
- Díez-Villaseñor, C.; Guzmán, N.M.; Almendros, C.; García-Martínez, J.; Mojica, F.J.M. CRISPR-Spacer Integration Reporter Plasmids Reveal Distinct Genuine Acquisition Specificities among CRISPR-Cas IE Variants of Escherichia Coli. RNA Biol. 2013, 10, 792–802. [Google Scholar] [CrossRef]
- Long, J.; Zhang, J.; Xi, Y.; Zhao, J.; Jin, Y.; Yang, H.; Chen, S.; Duan, G. Genomic Insights into CRISPR-Harboring Plasmids in the Klebsiella Genus: Distribution, Backbone Structures, Antibiotic Resistance, and Virulence Determinant Profiles. Antimicrob. Agents Chemother. 2023, 67, e01189-22. [Google Scholar] [CrossRef]
- Pawluk, A.; Shah, M.; Mejdani, M.; Calmettes, C.; Moraes, T.F.; Davidson, A.R.; Maxwell, K.L. Disabling a Type IE CRISPR-Cas Nuclease with a Bacteriophage-Encoded Anti-CRISPR Protein. MBio 2017, 8, e01751-17. [Google Scholar] [CrossRef]
- Touchon, M.; Rocha, E.P.C. The Small, Slow and Specialized CRISPR and Anti-CRISPR of Escherichia and Salmonella. PLoS ONE 2010, 5, e11126. [Google Scholar] [CrossRef]
- Smith, R.A.; M’ikanatha, N.M.; Read, A.F. Antibiotic Resistance: A Primer and Call to Action. Health Commun. 2015, 30, 309–314. [Google Scholar] [CrossRef]
- Marraffini, L.A.; Sontheimer, E.J. Self versus Non-Self Discrimination during CRISPR RNA-Directed Immunity. Nature 2010, 463, 568–571. [Google Scholar] [CrossRef]
- Citorik, R.J.; Mimee, M.; Lu, T.K. Sequence-Specific Antimicrobials Using Efficiently Delivered RNA-Guided Nucleases. Nat. Biotechnol. 2014, 32, 1141–1145. [Google Scholar] [CrossRef]
- Vercoe, R.B.; Chang, J.T.; Dy, R.L.; Taylor, C.; Gristwood, T.; Clulow, J.S.; Richter, C.; Przybilski, R.; Pitman, A.R.; Fineran, P.C. Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands. PLoS Genet. 2013, 9, e1003454. [Google Scholar] [CrossRef]
- Esvelt, K.M.; Smidler, A.L.; Catteruccia, F.; Church, G.M. Emerging Technology: Concerning RNA-Guided Gene Drives for the Alteration of Wild Populations. eLife 2014, 3, e03401. [Google Scholar] [CrossRef]
- Gomaa, A.A.; Klumpe, H.E.; Luo, M.L.; Selle, K.; Barrangou, R.; Beisel, C.L. Programmable Removal of Bacterial Strains by Use of Genome- Targeting CRISPR-Cas Systems. MBio 2014, 5, e00928-13. [Google Scholar] [CrossRef]
- Melnikov, A.A.; Tchernov, A.P.; Fodor, I.; Bayev, A.A. Lambda Phagemids and Their Transducing Properties. Gene 1984, 28, 29–35. [Google Scholar] [CrossRef]
- Rasheed, J.K.; Anderson, G.J.; Yigit, H.; Queenan, A.M.; Doménech-Sánchez, A.; Swenson, J.M.; Biddle, J.W.; Ferraro, M.J.; Jacoby, G.A.; Tenover, F.C. Characterization of the Extended-Spectrum β-Lactamase Reference Strain, Klebsiella Pneumoniae K6 (ATCC 700603), Which Produces the Novel Enzyme SHV-18. Antimicrob. Agents Chemother. 2000, 44, 2382–2388. [Google Scholar] [CrossRef]
- Chung, Y.; Tanaka, S.; Chu, F.; Nurieva, R.I.; Martinez, G.J.; Rawal, S.; Wang, Y.-H.; Lim, H.; Reynolds, J.M.; Zhou, X.; et al. Follicular Regulatory T Cells Expressing Foxp3 and Bcl-6 Suppress Germinal Center Reactions. Nat. Med. 2011, 17, 983–988. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, S.; Gupta, K.C. Novel Polyethylenimine-Derived Nanoparticles for in Vivo Gene Delivery. Expert Opin. Drug Deliv. 2013, 10, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Zuris, J.A.; Thompson, D.B.; Shu, Y.; Guilinger, J.P.; Bessen, J.L.; Hu, J.H.; Maeder, M.L.; Joung, J.K.; Chen, Z.-Y.; Liu, D.R. Cationic Lipid-Mediated Delivery of Proteins Enables Efficient Protein-Based Genome Editing in Vitro and in Vivo. Nat. Biotechnol. 2015, 33, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, B.; Xie, H.; Ren, Q.; Liu, X.; Li, F.; Lv, X.; He, X.; Cheng, C.; Deng, R.; et al. Efficient Human Genome Editing Using SaCas9 Ribonucleoprotein Complexes. Biotechnol. J. 2019, 14, 1800689. [Google Scholar] [CrossRef]
- Kang, Y.K.; Kwon, K.; Ryu, J.S.; Lee, H.N.; Park, C.; Chung, H.J. Nonviral Genome Editing Based on a Polymer-Derivatized CRISPR Nanocomplex for Targeting Bacterial Pathogens and Antibiotic Resistance. Bioconjug. Chem. 2017, 28, 957–967. [Google Scholar] [CrossRef]
- Pérez-Mendoza, D.; de la Cruz, F. Escherichia Coli Genes Affecting Recipient Ability in Plasmid Conjugation: Are There Any? BMC Genom. 2009, 10, 71. [Google Scholar] [CrossRef]
- Jani, K.; Srivastava, V.; Sharma, P.; Vir, A.; Sharma, A. Easy Access to Antibiotics; Spread of Antimicrobial Resistance and Implementation of One Health Approach in India. J. Epidemiol. Glob. Health 2021, 11, 444–452. [Google Scholar] [CrossRef]
- Hausner, M.; Wuertz, S. High Rates of Conjugation in Bacterial Biofilms as Determined by Quantitative in Situ Analysis. Appl. Environ. Microbiol. 1999, 65, 3710–3713. [Google Scholar] [CrossRef]
- Peters, J.E. Targeted Transposition with Tn7 Elements: Safe Sites, Mobile Plasmids, CRISPR/Cas and Beyond. Mol. Microbiol. 2019, 112, 1635–1644. [Google Scholar] [CrossRef]
- Hamilton, T.A.; Pellegrino, G.M.; Therrien, J.A.; Ham, D.T.; Bartlett, P.C.; Karas, B.J.; Gloor, G.B.; Edgell, D.R. Efficient Inter-Species Conjugative Transfer of a CRISPR Nuclease for Targeted Bacterial Killing. Nat. Commun. 2019, 10, 4544. [Google Scholar] [CrossRef]
- Gray, G.S.; Fitch, W.M. Evolution of Antibiotic Resistance Genes: The DNA Sequence of a Kanamycin Resistance Gene from Staphylococcus Aureus. Mol. Biol. Evol. 1983, 1, 57–66. [Google Scholar] [CrossRef]
- Kiga, K.; Tan, X.-E.; Ibarra-Chávez, R.; Watanabe, S.; Aiba, Y.; Sato’o, Y.; Li, F.-Y.; Sasahara, T.; Cui, B.; Kawauchi, M. Development of CRISPR-Cas13a-Based Antimicrobials Capable of Sequence-Specific Killing of Target Bacteria. Nat. Commun. 2020, 11, 2934. [Google Scholar] [CrossRef]
- Roach, D.R.; Leung, C.Y.; Henry, M.; Morello, E.; Singh, D.; Di Santo, J.P.; Weitz, J.S.; Debarbieux, L. Synergy between the Host Immune System and Bacteriophage Is Essential for Successful Phage Therapy against an Acute Respiratory Pathogen. Cell Host Microbe 2017, 22, 38–47. [Google Scholar] [CrossRef]
- Wiersinga, W.J.; Rhodes, A.; Cheng, A.C.; Peacock, S.J.; Prescott, H.C. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA J. Am. Med. Assoc. 2020, 324, 782–793. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, C.; Zheng, Y.; Zhao, Y.; Wang, Y.; Hao, J.; Zhao, X.; Yi, K.; Shi, L.; Kang, C. Virus-like Nanoparticle as a Co-Delivery System to Enhance Efficacy of CRISPR/Cas9-Based Cancer Immunotherapy. Biomaterials 2020, 258, 120275. [Google Scholar] [CrossRef]
- Jacków, J.; Guo, Z.; Hansen, C.; Abaci, H.E.; Doucet, Y.S.; Shin, J.U.; Hayashi, R.; DeLorenzo, D.; Kabata, Y.; Shinkuma, S.; et al. CRISPR/Cas9-Based Targeted Genome Editing for Correction of Recessive Dystrophic Epidermolysis Bullosa Using IPS Cells. Proc. Natl. Acad. Sci. USA 2019, 116, 26846–26852. [Google Scholar] [CrossRef]
- Li, H.; Yang, Y.; Hong, W.; Huang, M.; Wu, M.; Zhao, X. Applications of Genome Editing Technology in the Targeted Therapy of Human Diseases: Mechanisms, Advances and Prospects. Signal Transduct. Target. Ther. 2020, 5, 1. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, L.; Huang, X. Genome Modification by CRISPR/Cas9. FEBS J. 2014, 281, 5186–5193. [Google Scholar] [CrossRef]
- Guk, K.; Keem, J.O.; Hwang, S.G.; Kim, H.; Kang, T.; Lim, E.K.; Jung, J. A Facile, Rapid and Sensitive Detection of MRSA Using a CRISPR-Mediated DNA FISH Method, Antibody-like DCas9/SgRNA Complex. Biosens. Bioelectron. 2017, 95, 67–71. [Google Scholar] [CrossRef]
- Kaboli, S.; Babazada, H. CRISPR Mediated Genome Engineering and Its Application in Industry. Curr. Issues Mol. Biol. 2018, 26, 81–92. [Google Scholar] [CrossRef]
- Liao, B.; Chen, X.; Zhou, X.; Zhou, Y.; Shi, Y.; Ye, X.; Liao, M.; Zhou, Z.; Cheng, L.; Ren, B. Applications of CRISPR/Cas Gene-Editing Technology in Yeast and Fungi. Arch. Microbiol. 2022, 204, 79. [Google Scholar] [CrossRef] [PubMed]
- Doerflinger, M.; Forsyth, W.; Ebert, G.; Pellegrini, M.; Herold, M.J. CRISPR/Cas9—The Ultimate Weapon to Battle Infectious Diseases? Cell. Microbiol. 2017, 19, e12693. [Google Scholar] [CrossRef] [PubMed]
- Vyas, V.K.; Barrasa, M.I.; Fink, G.R. A Candida Albicans CRISPR System Permits Genetic Engineering of Essential Genes and Gene Families. Sci. Adv. 2015, 1, e1500248. [Google Scholar] [CrossRef] [PubMed]
- Min, K.; Ichikawa, Y.; Woolford, C.A.; Mitchell, A.P. Candida Albicans Gene Deletion with a Transient CRISPR-Cas9 System. MSphere 2016, 1, e00130-16. [Google Scholar] [CrossRef]
- Fuller, K.K.; Chen, S.; Loros, J.J.; Dunlap, J.C. Development of the CRISPR/Cas9 System for Targeted Gene Disruption in Aspergillus Fumigatus. Eukaryot. Cell 2015, 14, 1073–1080. [Google Scholar] [CrossRef]
- Wang, Y.; Wei, D.; Zhu, X.; Pan, J.; Zhang, P.; Huo, L.; Zhu, X. A ‘Suicide’CRISPR-Cas9 System to Promote Gene Deletion and Restoration by Electroporation in Cryptococcus Neoformans. Sci. Rep. 2016, 6, 31145. [Google Scholar] [CrossRef]
- Távora, F.T.P.K.; dos Santos Diniz, F.D.A.; de Moraes Rêgo-Machado, C.; Freitas, N.C.; Arraes, F.B.M.; de Andrade, E.C.; Furtado, L.L.; Osiro, K.O.; de Sousa, N.L.; Cardoso, T.B. CRISPR/Cas-and Topical RNAi-Based Technologies for Crop Management and Improvement: Reviewing the Risk Assessment and Challenges towards a More Sustainable Agriculture. Front. Bioeng. Biotechnol. 2022, 10, 913728. [Google Scholar]
- Shanmugam, K.; Ramalingam, S.; Venkataraman, G.; Hariharan, G.N. The CRISPR/Cas9 System for Targeted Genome Engineering in Free-Living Fungi: Advances and Opportunities for Lichenized Fungi. Front. Microbiol. 2019, 10, 62. [Google Scholar] [CrossRef]
- Bagherpour, G.; Ghasemi, H.; Zand, B.; Zarei, N.; Roohvand, F.; Ardakani, E.M.; Azizi, M.; Khalaj, V. Oral Administration of Recombinant Saccharomyces boulardii Expressing Ovalbumin-CPE Fusion Protein Induces Antibody Response in Mice. Front. Microbiol. 2018, 9, 723. [Google Scholar] [CrossRef]
- Manrique, P.; Bolduc, B.; Walk, S.T.; Van der Oost, J.; De Vos, W.M.; Young, M.J. Healthy Human Gut Phageome. Proc. Natl. Acad. Sci. USA 2016, 113, 10400–10405. [Google Scholar] [CrossRef]
- Ramachandran, G.; Bikard, D. Editing the Microbiome the CRISPR Way. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180103. [Google Scholar] [CrossRef]
- Manghwar, H.; Li, B.; Ding, X.; Hussain, A.; Lindsey, K.; Zhang, X.; Jin, S. CRISPR/Cas Systems in Genome Editing: Methodologies and Tools for SgRNA Design, Off-Target Evaluation, and Strategies to Mitigate Off-Target Effects. Adv. Sci. 2020, 7, 1902312. [Google Scholar] [CrossRef]
- Pavlovic, G.; Erbs, V.; Andre, P.; Sylvie, J.; Eisenman, B.; Dreyer, D.; Lorentz, R.; Wattenhofer-Donze, M.; Birling, M.-C.; Herault, Y. Generation of Targeted Overexpressing Models by CRISPR/Cas9 and Need of Careful Validation of Your Knock-in Line Obtained by Nuclease Genome Editing. Transgenic Res. 2016, 25, 254–255. [Google Scholar]
- Li, J.; Shou, J.; Guo, Y.; Tang, Y.; Wu, Y.; Jia, Z.; Zhai, Y.; Chen, Z.; Xu, Q.; Wu, Q. Efficient Inversions and Duplications of Mammalian Regulatory DNA Elements and Gene Clusters by CRISPR/Cas9. J. Mol. Cell Biol. 2015, 7, 284–298. [Google Scholar] [CrossRef]
- van Overbeek, M.; Capurso, D.; Carter, M.M.; Thompson, M.S.; Frias, E.; Russ, C.; Reece-Hoyes, J.S.; Nye, C.; Gradia, S.; Vidal, B.; et al. DNA Repair Profiling Reveals Nonrandom Outcomes at Cas9-Mediated Breaks. Mol. Cell 2016, 63, 633–646. [Google Scholar] [CrossRef]
- Wilson, L.O.W.; O’Brien, A.R.; Bauer, D.C. The Current State and Future of CRISPR-Cas9 GRNA Design Tools. Front. Pharmacol. 2018, 9, 749. [Google Scholar] [CrossRef]

| Class | Type | Subtype | Signature Genes | Target | Effector | Families of Encoded Proteins | CRISPR Protein | Associated Type-Subtype | Function | References |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 (A multi-Cas protein) | I | A, B, C, D, E, F, U | I-A; Cas8a2, Cas5 I-B; Cas8b I-C; Cas8c I-D; Cas10d I-E; Cse1, Cse2 I-F: Csy1, Csy2, Csy3, Cas6f | dsDNA | Cascade | COG1518 | Cas 1 | I, II, III-A, III-B, IV, possibly VI | DNA Nuclease | [15] |
| 1 | III | A, B, C, D | III-A: Csm2, Csx10 all1473 III-B Cmr5, MTH326 (Cas10 or Csx11) | ssRNA | Cascade | COG1203 | Cas 2 | I, II, III-A, III-B, V, some VI | RNA Nuclease | [15] |
| 1 | IV | A, B | DinG (Csf4) | dsDNA | Cascade | COG1468 | Cas 3 | I | DNA nuclease and helicase | [15,22] |
| 1 | II | A | Csn-2 | dsDNA | SpCas9 | COG1343 | Cas 4 | Mostly type I, II & V | DNA nuclease | [15,24] |
| 2 | II | A | dsDNA | SpCas9 | COG3512 | Cas 5 | Type I, I | Ribonuclease responsible for converting pre-crRNA to mature crRNA. | [24] | |
| 2 | II | B | Cas9 (Csx12 subfamily) | dsDNA/ssRNA | FnCas9 | COG1343 and COG3512 | Cas 6 | Most type IIIB and type I | Pre-crRNA is converted to mature crRNA by ribonuclease. | [24] |
| 2 | II | C | N/A | dsDNA | NmCas9 | COG1343 and COG3512 | Cas 7 | I, III, IV | It binds with crRNA and comprises of an RNA recognition motif. | [5] |
| 2 | V | A | Csm4, Csx10, Cmr3 | dsDNA | Cas12a (Cpf1) | COG1688 (RAMP) | Cas 8 | Most type I | It forms effector complex large subunit in type I | [25] |
| 2 | V | B | Cas5, Csy2 | dsDNA | Cas12b (C2c1) | COG1688 (RAMP) | Cas 9 | II only | DNA nuclease | [26] |
| 2 | V | C | Csc1, Csf3 | dSDNA | Cas12c (C2c3) | COG1688 (RAMP) | Cas 10 | Some type I, most type III | It forms effector complex large subunit in type III. | [25] |
| 2 | VI | A | - | ssRNA | Cas13a (C2c2) | COG1583 and COG5551 (RAMP) | Cas 12 (cpf1) | V | crRNA sorting, DNA nuclease | [25,27] |
| 2 | VI | B | Cmr6 | ssRNA | Cas13b (C2c4) | (RAMP) | Cas 13 (C2c2) | VI | crRNA sorting, RNA nuclease | [15,28] |
| 2 | VI | C | - | ssRNA | Cas13c (C2c7) | (RAMP) | Csm, Cmr | III | Nucleases for single-stranded DNA and RNA | [15,28] |
| 2 | VI | D | - | ssRNA | Cas13d | (RAMP) | RNase III | II | tracrRNA is processed, and crRNA maturation is aided by this system. | [22] |
| CRISPR System | Organism | Gene Target | Reference |
|---|---|---|---|
| Type I-E (Cas3) | E. coli | blaNDM-1, blaCTX-M-15 | [37] |
| Type II (Cas9) | E. coli | BlaTEM, blaSHV | [38] |
| Type II (Cas9) | S. aureus | aph-3, mecA | [39] |
| Type II (Cas9) | HIV-1 | LTR U3 region | [40] |
| Type II (Cas9) | HIV-1 | LTR, gag, pol | [41] |
| Type II (Cas9) | HSV-1 | EBNA-1, OriP | [42] |
| Type II (Cas9) | HBV | Repeat region of integrated genome | [43] |
| Type II (Cas9) | HPV | E6, E7 | [44] |
| CRISPR-Cas | Mechanism/Function | Delivery Vehicle |
|---|---|---|
| CRISPR-SpCas9 | single-RNA-mediated DNA endonuclease |
|
| CRISPRi | single-RNA-mediated inhibition of mRNA transcription |
|
| CRISPRa | single-RNA-mediated activation of mRNA transcription |
|
| CRISPR-SaCas9 | single-RNA-mediated DNA endonuclease |
|
| FnCas9 | single-RNA-mediated PAM-independent inhibiting of translation of target RNA |
|
| C2c1/3 | dual-RNA-guided DNA endonuclease | No mammalian expression vector |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Javed, M.U.; Hayat, M.T.; Mukhtar, H.; Imre, K. CRISPR-Cas9 System: A Prospective Pathway toward Combatting Antibiotic Resistance. Antibiotics 2023, 12, 1075. https://doi.org/10.3390/antibiotics12061075
Javed MU, Hayat MT, Mukhtar H, Imre K. CRISPR-Cas9 System: A Prospective Pathway toward Combatting Antibiotic Resistance. Antibiotics. 2023; 12(6):1075. https://doi.org/10.3390/antibiotics12061075
Chicago/Turabian StyleJaved, Muhammad Uzair, Muhammad Tahir Hayat, Hamid Mukhtar, and Kalman Imre. 2023. "CRISPR-Cas9 System: A Prospective Pathway toward Combatting Antibiotic Resistance" Antibiotics 12, no. 6: 1075. https://doi.org/10.3390/antibiotics12061075
APA StyleJaved, M. U., Hayat, M. T., Mukhtar, H., & Imre, K. (2023). CRISPR-Cas9 System: A Prospective Pathway toward Combatting Antibiotic Resistance. Antibiotics, 12(6), 1075. https://doi.org/10.3390/antibiotics12061075







