Vibrio Phage VMJ710 Can Prevent and Treat Disease Caused by Pathogenic MDR V. cholerae O1 in an Infant Mouse Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Bacterial Strains and Growth Conditions Used
2.3. Phage Isolation
2.4. Phage Purification
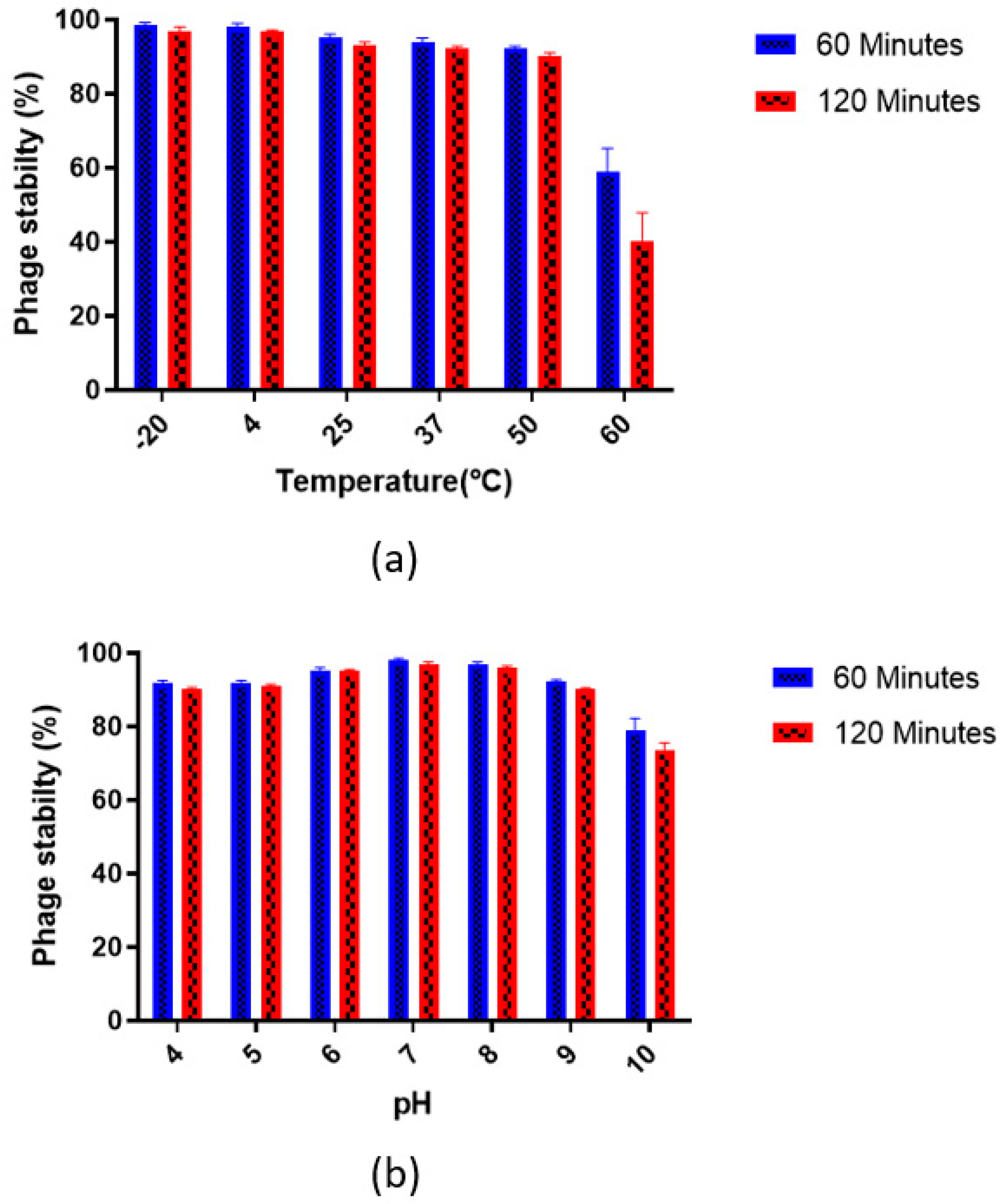
2.5. Phage Stability
2.5.1. pH Stability
2.5.2. Thermal/Temperature Stability
2.6. Phage Morphology
2.7. Genomic DNA Isolation
2.8. Library Preparation, Sequencing, Assembly, and Annotations
2.9. Virulence Factors and Antibiotic Resistance Genes
2.10. Phylogenetic Tree and Comparative Genomics
2.11. Host Range Testing
2.12. Time-Kill Assay
2.13. Antibiofilm Activity of Phage VMJ710 against V. cholerae
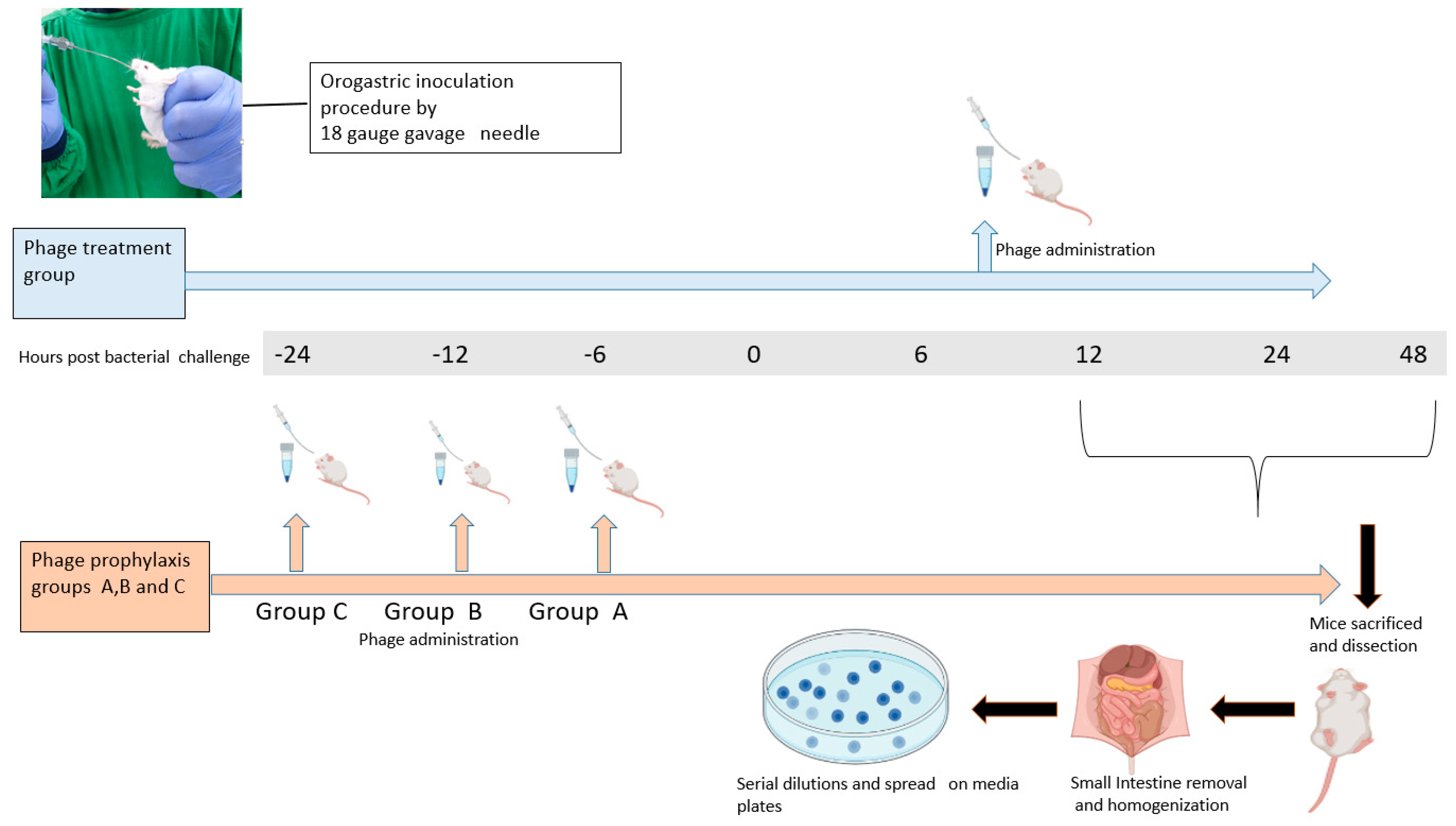
2.14. Phage Efficacy Testing against V. cholerae O1 Using an Infant BALB/c Mouse Model
2.14.1. Animals and Maintenance
2.14.2. Mouse Cholera Infection Model
2.14.3. Prophylactic and Treatment Efficacy Testing against V. cholerae
2.15. Nucleotide Sequence Accession Number
2.16. Statistical Analysis
3. Results
3.1. Sample Collection and Phage Isolation
3.2. Phage Morphology
3.3. Phage Stability
3.4. Genome Features of Vibrio Phage VMJ710
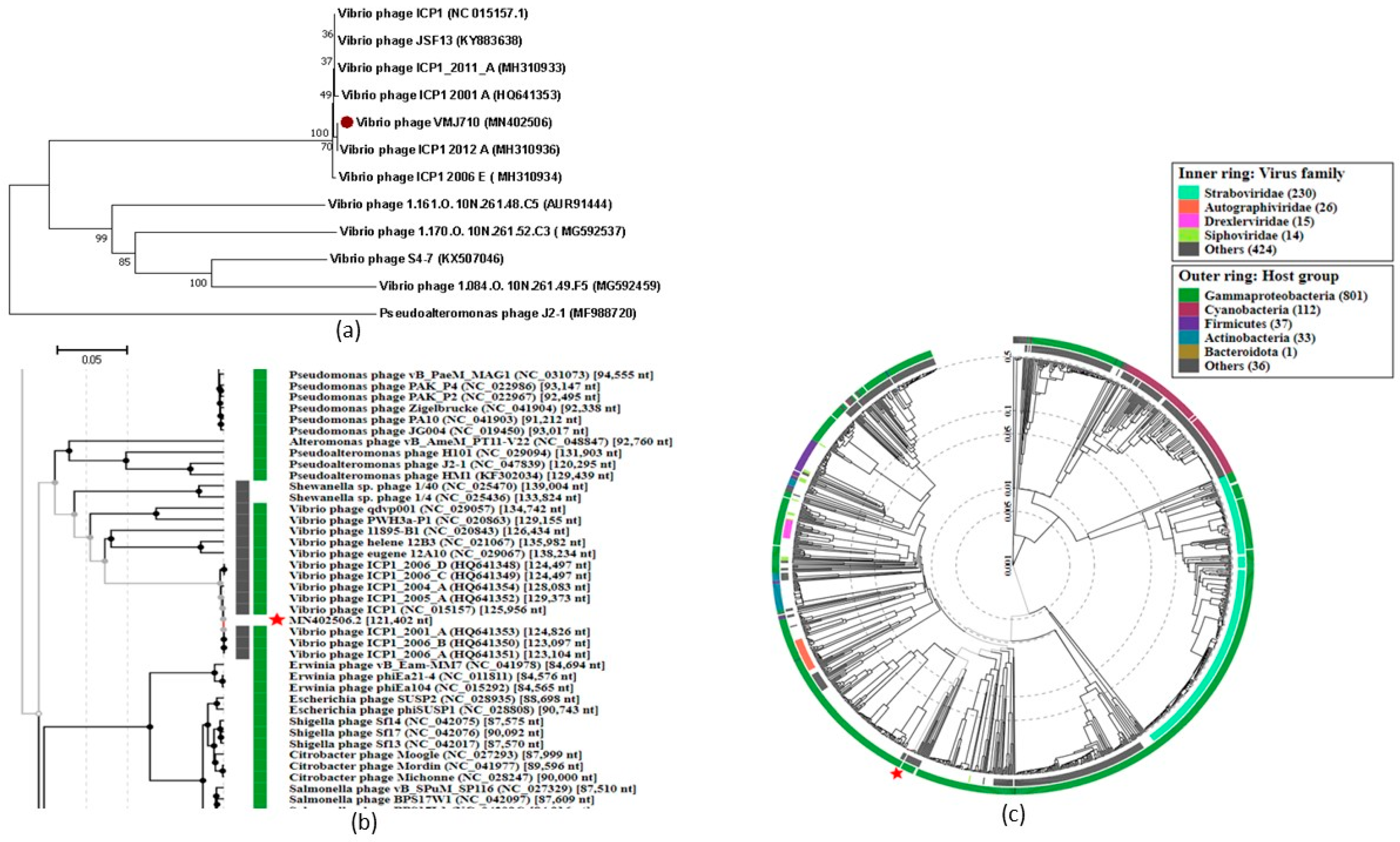
3.5. Phylogenetic Analysis
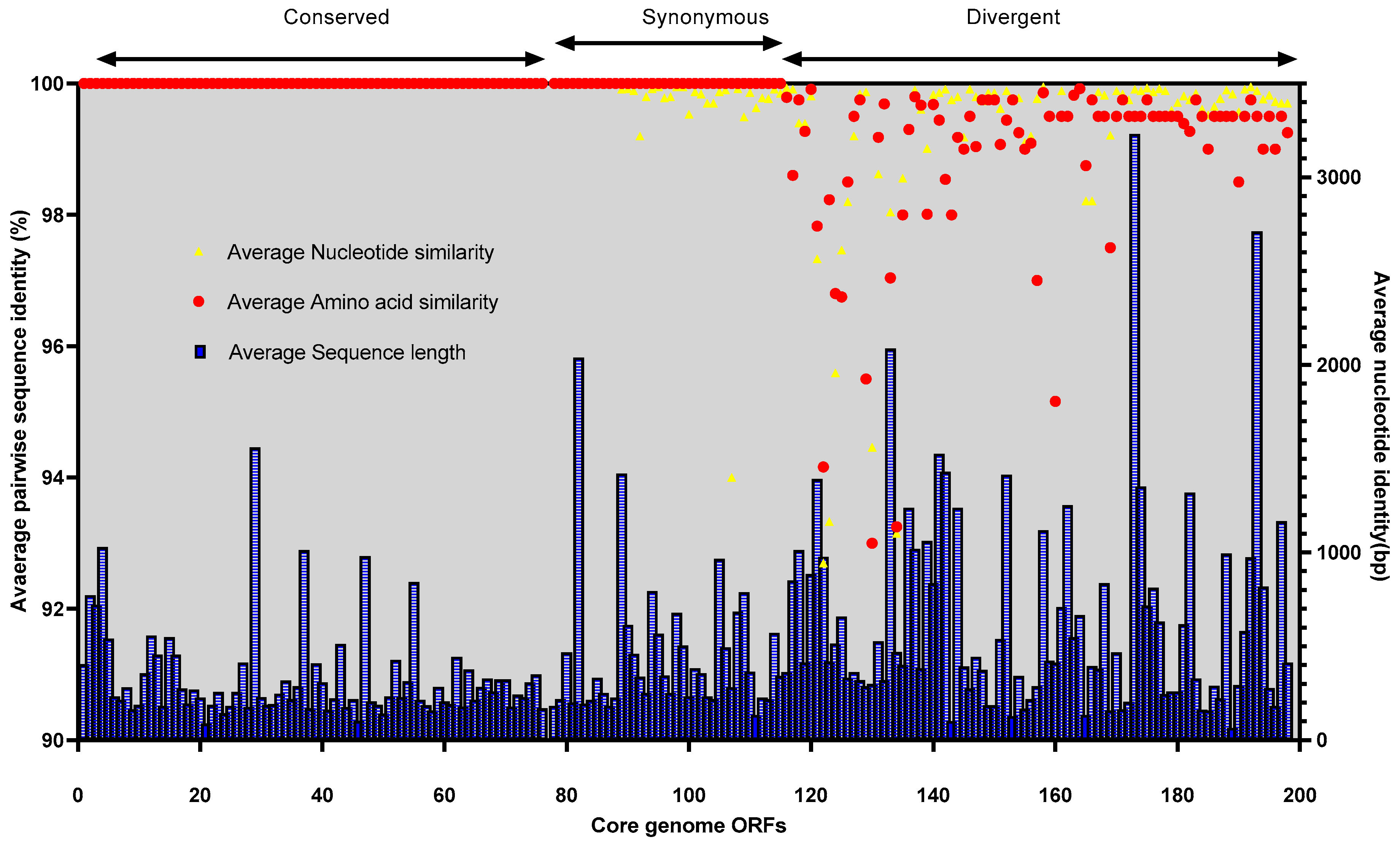
3.6. Comparative Genomics and Core Genome Analysis
3.7. Conserved Functional Domains
3.8. Host Range Testing
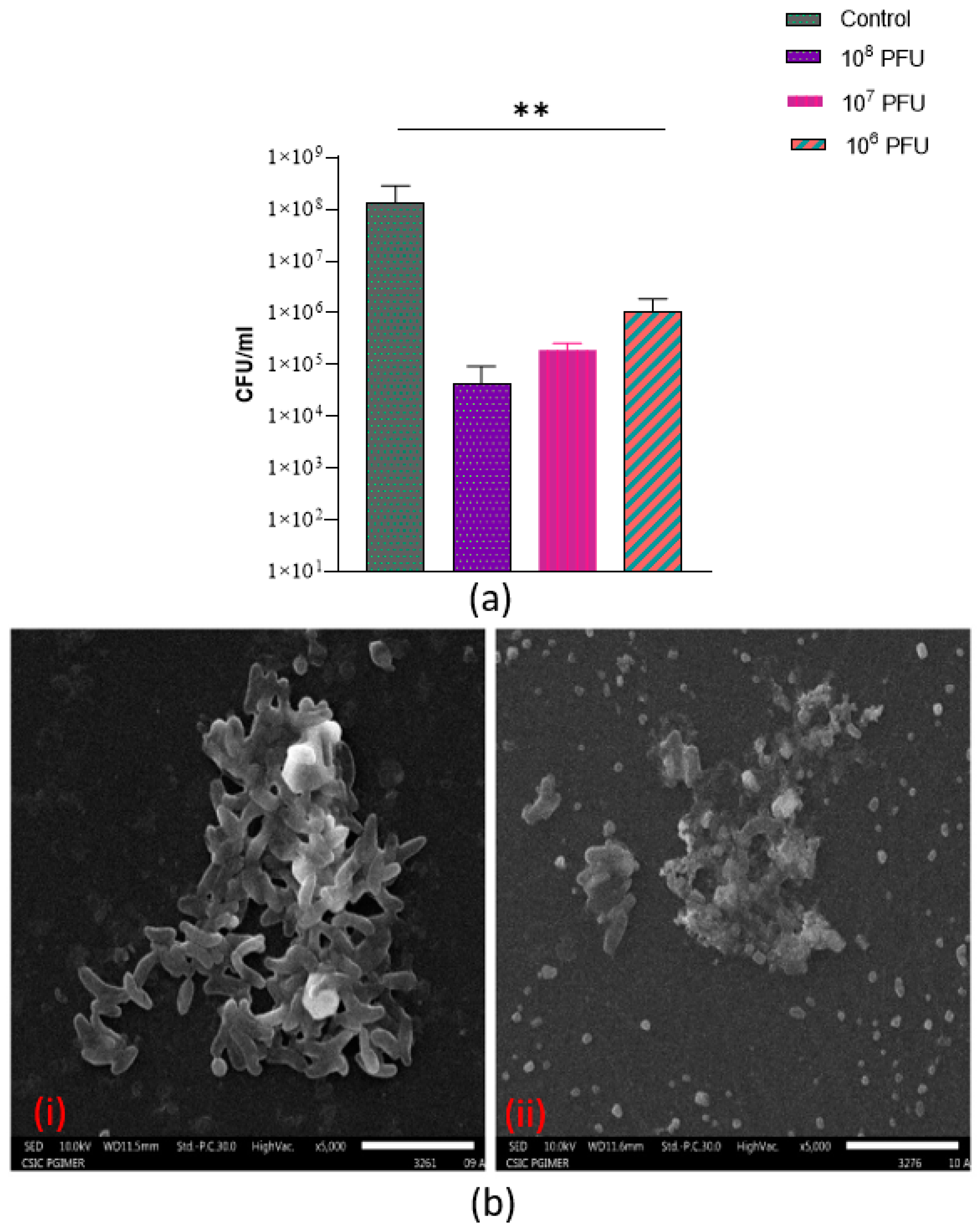
3.9. Anti-Biofilm Activity of Phage VMJ710 against Multidrug-Resistant V. cholerae
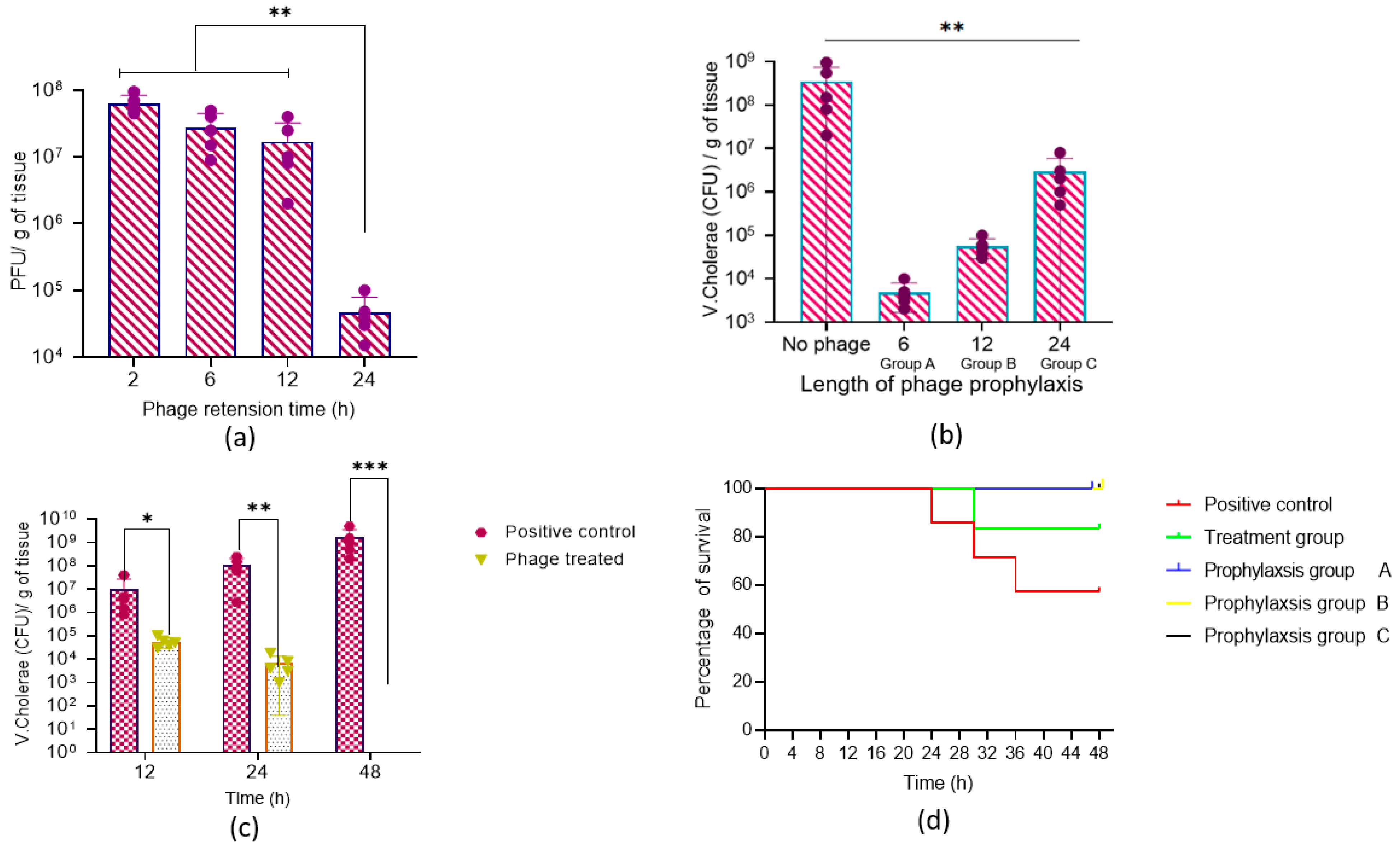
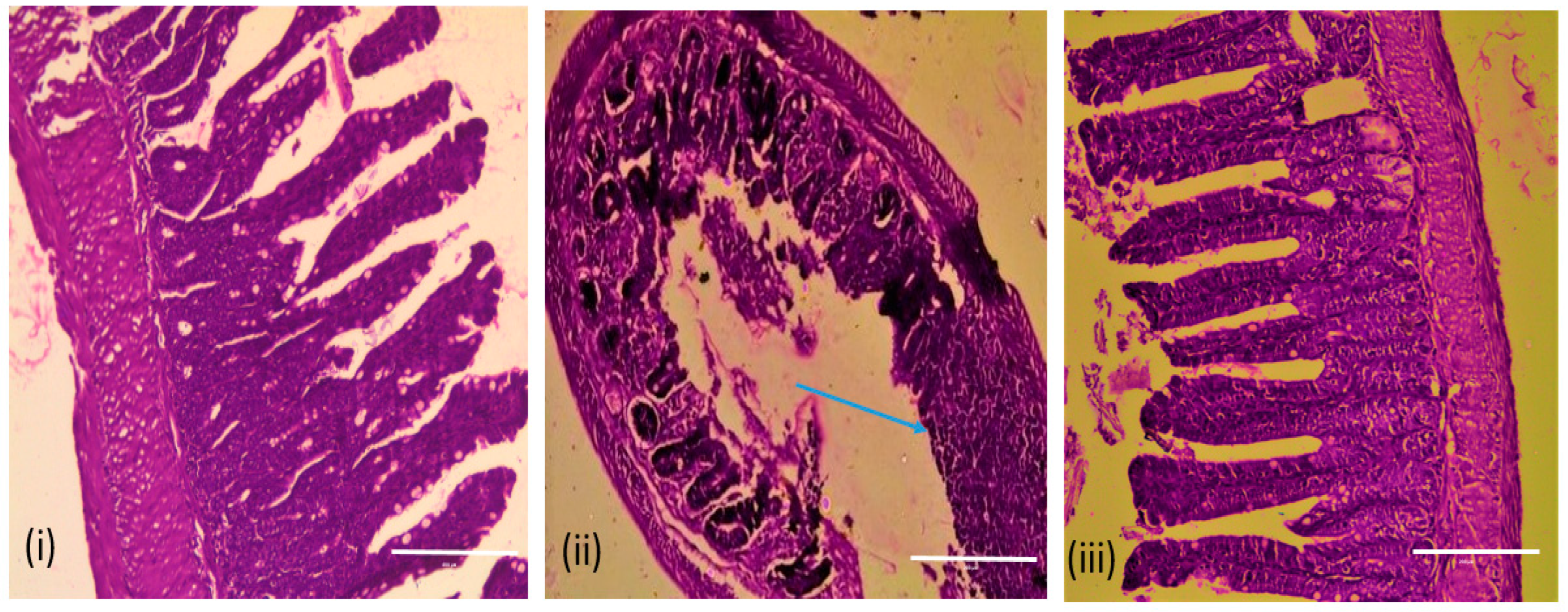
3.10. Efficacy of Vibrio Phage VMJ710 against MDR V. cholerae ELPGI212 in Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ramamurthy, T.; Mutreja, A.; Weill, F.; Das, B.; Ghosh, A.; Nair, G.B. Revisiting the Global Epidemiology of Cholera in Conjunction With the Genomics of Vibrio Cholerae. Front. Public Health 2019, 7, 203. [Google Scholar] [CrossRef] [PubMed]
- Faruque, S.M.; Kamruzzaman, M.; Meraj, I.M.; Chowdhury, N.; Nair, G.B.; Sack, R.B.; Colwell, R.R.; Sack, D.A. Pathogenic Potential of Environmental Vibrio Cholerae Strains Carrying Genetic Variants of the Toxin-Coregulated Pilus Pathogenicity Island. Infect. Immun. 2003, 71, 1020–1025. [Google Scholar] [CrossRef] [PubMed]
- Thapa Shrestha, U.; Adhikari, N.; Maharjan, R.; Banjara, M.R.; Rijal, K.R.; Basnyat, S.R.; Agrawal, V.P. Multidrug Resistant Vibrio Cholerae O1 from Clinical and Environmental Samples in Kathmandu City. BMC Infect. Dis. 2015, 15, 104. [Google Scholar] [CrossRef]
- Nelson, E.J.; Nelson, D.S.; Salam, M.A.; Sack, D.A. Antibiotics for Both Moderate and Severe Cholera. N. Engl. J. Med. 2011, 364, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Taneja, N.; Mishra, A.; Batra, N.; Gupta, P.; Mahindroo, J.; Mohan, B. Inland Cholera in Freshwater Environs of North India. Vaccine 2020, 38, A63–A72. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Gupta, S.S.; Arora, N.; Khasnobis, P.; Venkatesh, S.; Sur, D.; Nair, G.B.; Sack, D.A.; Ganguly, N.K. Identification of Burden Hotspots and Risk Factors for Cholera in India: An Observational Study. PLoS ONE 2017, 12, e0183100. [Google Scholar] [CrossRef]
- Naser, I.B.; Hoque, M.M.; Abdullah, A.; Bari, S.M.N.; Ghosh, A.N.; Faruque, S.M. Environmental Bacteriophages Active on Biofilms and Planktonic Forms of Toxigenic Vibrio Cholerae: Potential Relevance in Cholera Epidemiology. PLoS ONE 2017, 12, e0180838. [Google Scholar] [CrossRef]
- Khatoon, Z.; McTiernan, C.D.; Suuronen, E.J.; Mah, T.F.; Alarcon, E.I. Bacterial Biofilm Formation on Implantable Devices and Approaches to Its Treatment and Prevention. Heliyon 2018, 4, e01067. [Google Scholar] [CrossRef]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage Therapy: An Alternative to Antibiotics in the Age of Multi-Drug Resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162–173. [Google Scholar] [CrossRef]
- Chan, B.K.; Abedon, S.T.; Loc-carrillo, C. Phage Cocktails and the Future of Phage Therapy. Futur. Microbiol. 2013, 8, 769–783. [Google Scholar] [CrossRef]
- Chaudhary, N.; Mohan, B.; Mavuduru, R.S.; Kumar, Y.; Taneja, N. Characterization, Genome Analysis and in Vitro Activity of a Novel Phage VB_EcoA_RDN8.1 Active against Multi-Drug Resistant and Extensively Drug-Resistant Biofilm-Forming Uropathogenic Escherichia Coli Isolates, India. J. Appl. Microbiol. 2022, 132, 3387–3404. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, N.; Narayan, C.; Mohan, B.; Taneja, N. Indian Journal of Medical Microbiology Characterization and in Vitro Activity of a Lytic Phage RDN37 Isolated from Community Sewage Water Active against MDR Uropathogenic E. Coli. Indian J. Med. Microbiol. 2021, 39, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, N.; Singh, D.; Maurya, R.K.; Mohan, B.; Tanejaa, N. Complete Genome Sequence of Salmonella Phage VB_SenA_SM5, Active against Multidrug-Resistant Salmonella Enterica Serovar Typhi Isolates. Microbiol. Resour. Announc. 2022, 44, e00309-22. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, N.; Singh, D.; Maurya, R.K.; Mohan, B.; Mavuduru, R.S.; Taneja, N. Whole Genome Sequencing and in Vitro Activity Data of Escherichia Phage NTEC3 against Multidrug-Resistant Uropathogenic and Extensively Drug-Resistant Uropathogenic E. coli Isolates. Data Br. 2022, 43, 108479. [Google Scholar] [CrossRef]
- Gu, Y.; Xu, Y.; Xu, J.; Yu, X.; Huang, X.; Liu, G.; Liu, X. Identification of Novel Bacteriophage vB _ EcoP-EG1 with Lytic Activity against Planktonic and Biofilm Forms of Uropathogenic Escherichia coli. Appl. Microbiol. Biotechnol. 2018, 103, 315–326. [Google Scholar] [CrossRef]
- Surekhamol, I.S.; Deepa, G.D.; Somnath Pai, S.; Sreelakshmi, B.; Varghese, S.; Bright Singh, I.S. Isolation and Characterization of Broad Spectrum Bacteriophages Lytic to Vibrio Harveyi from Shrimp Farms of Kerala, India. Lett. Appl. Microbiol. 2014, 58, 197–204. [Google Scholar] [CrossRef]
- Chaudhary, N.; Singh, D.; Narayan, C.; Samui, B.; Mohan, B.; Mavuduru, R.S.; Taneja, N. Complete Genome Sequence of Escherichia Phage 590B, Active against an Extensively Drug-Resistant Uropathogenic Escherichia coli Isolate. Microbiol. Resour. Announc. 2021, 10, 9–10. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Chaudhary, N.; Mohan, B.; Taneja, N. Draft Genome Sequence of Escherichia Phage PGN829.1, Active against Highly Drug-Resistant Uropathogenic Escherichia coli. Microbiol. Resour. Announc. 2018, 7, 20. [Google Scholar] [CrossRef]
- Hunt, M.; Gall, A.; Ong, S.H.; Brener, J.; Ferns, B.; Goulder, P.; Nastouli, E.; Keane, J.A.; Kellam, P.; Otto, T.D. IVA: Accurate de Novo Assembly of RNA Virus Genomes. Bioinformatics 2015, 31, 2374–2376. [Google Scholar] [CrossRef]
- Besemer, J.; Borodovsky, M. GeneMark: Web Software for Gene Finding in Prokaryotes, Eukaryotes and Viruses. Nucleic Acids Res. 2005, 33, 451–454. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying Bacterial Genes and Endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D.; Canback, B. ARAGORN, a Program to Detect TRNA Genes and TmRNA Genes in Nucleotide Sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an Update of CRISRFinder, Includes a Portable Version, Enhanced Performance and Integrates Search for Cas Proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A Comparative Genomics Tool for Circular Genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A Comparative Pathogenomic Platform with an Interactive Web Interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Chaudhary, N.; Maurya, R.K.; Singh, D.; Mohan, B.; Taneja, N. Genome Analysis and Antibiofilm Activity of Phage 590B. Pathogen 2022, 11, 1448. [Google Scholar] [CrossRef]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The Viral Proteomic Tree Server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple Alignment of Conserved Genomic Sequence with Rearrangements. Genom. Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple Prokaryote Genome Comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed]
- Kutter, E. Phage Host Range and Efficiency of Plating. In Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2009; Volume 501, pp. 141–149. ISBN 9781588296825. [Google Scholar]
- Kaur, H.; Kalia, M.; Chaudhary, N.; Singh, V.; Yadav, V.K.; Modgil, V.; Kant, V.; Mohan, B.; Bhatia, A.; Taneja, N. Repurposing of FDA Approved Drugs against Uropathogenic Escherichia coli: In Silico, in Vitro, and in Vivo Analysis. Microb. Pathog. 2022, 169, 105665. [Google Scholar] [CrossRef]
- Dua, P.; Karmakar, A.; Ghosh, C. Virulence Gene Profiles, Biofilm Formation, and Antimicrobial Resistance of Vibrio Cholerae Non-O1/Non-O139 Bacteria Isolated from West Bengal, India. Heliyon 2018, 4, e01040. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Sharma, P.; Kalia, N.; Singh, J.; Kaur, S. Anti-Biofilm Properties of the Fecal Probiotic Lactobacilli against Vibrio Spp. Front. Cell. Infect. Microbiol. 2018, 8, 120. [Google Scholar] [CrossRef] [PubMed]
- Yen, M.; Cairns, L.S.; Camilli, A. A Cocktail of Three Virulent Bacteriophages Prevents Vibrio Cholerae Infection in Animal Models. Nat. Commun. 2017, 8, 14187. [Google Scholar] [CrossRef]
- Turner, D.; Shkoporov, A.N.; Lood, C.; Millard, A.D.; Dutilh, B.E.; Alfenas-Zerbini, P.; van Zyl, L.J.; Aziz, R.K.; Oksanen, H.M.; Poranen, M.M.; et al. Abolishment of Morphology-Based Taxa and Change to Binomial Species Names: 2022 Taxonomy Update of the ICTV Bacterial Viruses Subcommittee. Arch. Virol. 2023, 168, 74. [Google Scholar] [CrossRef]
- Seed, K.D.; Bodi, K.L.; Kropinski, A.M.; Ackermann, H.W.; Calderwood, S.B.; Qadri, F.; Camilli, A. Evidence of a Dominant Lineage of Vibrio Cholerae-Specific Lytic Bacteriophages Shed by Cholera Patients over a 10-Year Period in Dhaka, Bangladesh. MBio 2011, 2, e00334-10. [Google Scholar] [CrossRef]
- Angermeyer, A.; Das, M.M.; Singh, D.V.; Seed, K.D. Analysis of 19 Highly Conserved Vibrio Cholerae Bacteriophages Isolated from Environmental and Patient Sources over a Twelve-Year Period. Viruses 2018, 10, 299. [Google Scholar] [CrossRef]
- Naser, I.B.; Hoque, M.M.; Nahid, M.A.; Tareq, T.M.; Rocky, M.K.; Faruque, S.M. Analysis of the CRISPR-Cas System in Bacteriophages Active on Epidemic Strains of Vibrio cholerae in Bangladesh. Sci. Rep. 2017, 7, 14880. [Google Scholar] [CrossRef]
- Seed, K.D.; Lazinski, D.W.; Calderwood, S.B.; Camilli, A. A Bacteriophage Encodes Its Own CRISPR/Cas Adaptive Response to Evade Host Innate Immunity. Nature 2013, 494, 489–491. [Google Scholar] [CrossRef]
- Nakagawa, H.; Arisaka, F.; Ishii, S. Isolation and Characterization of the Bacteriophage T4 Tail-Associated Lysozyme. J. Virol. 1985, 54, 460–466. [Google Scholar] [CrossRef]
- Kala, S.; Cumby, N.; Sadowski, P.D.; Hyder, B.Z.; Kanelis, V.; Davidson, A.R.; Maxwell, K.L. HNH Proteins Are a Widespread Component of Phage DNA Packaging Machines. Proc. Natl. Acad. Sci. USA 2014, 111, 6022–6027. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S.R.; Gilcrease, E.B. Determining DNA Packaging Strategy by Analysis of the Termini of the Chromosomes in Tailed-Bacteriophage Virions. Methods Mol. Biol. 2009, 502, 91–111. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, P.; Das, J.; Foley, I. Biofilm Susceptibility to Antimicrobials. Adv. Dent. Res. 1997, 11, 160–167. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial Biofilms: A Common Cause of Persistent Infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Davies, D. Understanding Biofilm Resistance to Antibacterial Agents. Nat. Rev. Drug Discov. 2003, 11, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Grenier, D.; Grignon, L.; Gottschalk, M. Characterisation of Biofilm Formation by a Streptococcus Suis Meningitis Isolate. Vet. J. 2009, 179, 292–295. [Google Scholar] [CrossRef]
- Chan, B.; Abedon, S. Bacteriophages and Their Enzymes in Biofilm Control. Curr. Pharm. Des. 2014, 21, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Landini, P. Cross-Talk Mechanisms in Biofilm Formation and Responses to Environmental and Physiological Stress in Escherichia coli. Res. Microbiol. 2009, 160, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Curtin, J.J.; Donlan, R.M. Using Bacteriophages to Reduce Formation of Catheter-Associated Biofilms by Staphylococcus epidermidis. Antimicrob. Agents Chemother. 2006, 50, 1268–1275. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival Mechanisms of Clinically Relevant Microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Ryu, J.H.; Beuchat, L.R. Inactivation of Escherichia Coli O157:H7 in Biofilm on Stainless Steel by Treatment with an Alkaline Cleaner and a Bacteriophage. J. Appl. Microbiol. 2005, 99, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Drilling, A.; Hons, B.; Morales, S.; Boase, S.; Jervis-bardy, J. Safety and Efficacy of Topical Bacteriophage and Ethylenediaminetetraacetic Acid Treatment Of. Int. Forum Allergy Rhinol. 2014, 4, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Abel, S.; Waldor, M.K. Infant Rabbit Model for Diarrheal Diseases. Curr. Protoc. Microbiol. 2015, 38, 6. [Google Scholar] [CrossRef]
- Matson, J.S. Infant mouse model of Vibrio cholerae infection and colonization. In Vibrio Cholerae: Methods and Protocols; Humana Press: Totowa, NJ, USA, 2018; pp. 147–152. [Google Scholar]




| Vibrio Phage Name | Taxonomic Class | Isolation Year | Isolation Source | Genomic Size (bp) | G+C Content | Genbank Accession Number |
|---|---|---|---|---|---|---|
| ICP1 | Caudoviricetes | 2001 | Stool | 125,956 | 37.1 | HQ641347 |
| ICP1_2012_A | Caudoviricetes | 2012 | Stool | 121,418 | 37.2 | MH310936 |
| VMJ710 | Caudoviricetes | 2015 | Water | 121,402 | 37.2 | MN402506 |
| JSF13 | Caudoviricetes | 2017 | Water | 128,814 | 37.2 | KY883638 |
| ICP1_2011_A | Caudoviricetes | 2011 | Stool | 126,861 | 37.1 | MH310933 |
| Date of Collection, Antibiotic Sensitivity, Source of Isolation, and Phage Killing of V. cholerae Strains. | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V. cholerae Isolates | Date of Collection | Source | Antibiotic Sensitivity | |||||||||||
| Amikacin | Cefotaxime | Ciprofloxacin | Gentamycin | Norfloxacin | Nalidixic acid | Ampicillin | Furoxan | Chloramphenicol | Tetracycline | Cotrimoxazole | Phage Killing (EOP) | |||
| 219 | 10 August 2015 | ST | ||||||||||||
| 235 | 12 August 2015 | ST | ||||||||||||
| 66 | 23 August 2015 | ST | ||||||||||||
| ELPGI212 | 14 September 2015 | ST | ||||||||||||
| 231 | 14 September 2015 | ST | ||||||||||||
| 223 | 20 September 2015 | ST | ||||||||||||
| 220 | 20 September 2015 | ST | ||||||||||||
| 15-238 | 25 October 2015 | ST | ||||||||||||
| 1672 | 30 October 2015 | ST | ||||||||||||
| VMJ3 | 8 November 2015 | SW | ||||||||||||
| CHD5 | 15 November 2015 | ST | ||||||||||||
| LDH4 | 5 August 2016 | SW | ||||||||||||
| 183 | 19 July 2016 | ST | ||||||||||||
| 100 | 20 August 2016 | ST | ||||||||||||
| 221 | 20 August 2016 | ST | ||||||||||||
| 187 | 11 September 2016 | ST | ||||||||||||
| 163 | 20 September 2015 | ST | ||||||||||||
| 174 | 20 September 2016 | ST | ||||||||||||
| 226 | 16 October 2016 | ST | ||||||||||||
| 222 | 17 October 2016 | ST | ||||||||||||
| 238 | 25 October 2016 | ST | ||||||||||||
| 229 | 25 October 2016 | ST | ||||||||||||
| 236 | 28 October 2016 | ST | ||||||||||||
| 218 | 5 November 2016 | ST | ||||||||||||
| 211 | 10 November 2016 | ST | ||||||||||||
| VMJ1 | 11 November 2016 | ST | ||||||||||||
| Footnotes ST-Stool SW-Sewage water | Color codes of the antibiotic sensitivity profile | Key to EOP | EOP | |||||||||||
| >1.00 | ||||||||||||||
| 0.100–1.00 | ||||||||||||||
| Sensitive | 0.001–0.099 | |||||||||||||
| Intermediate | Reference 1.00 | |||||||||||||
| Resistant | No growth | |||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaudhary, N.; Mohan, B.; Kaur, H.; Modgil, V.; Kant, V.; Bhatia, A.; Taneja, N. Vibrio Phage VMJ710 Can Prevent and Treat Disease Caused by Pathogenic MDR V. cholerae O1 in an Infant Mouse Model. Antibiotics 2023, 12, 1046. https://doi.org/10.3390/antibiotics12061046
Chaudhary N, Mohan B, Kaur H, Modgil V, Kant V, Bhatia A, Taneja N. Vibrio Phage VMJ710 Can Prevent and Treat Disease Caused by Pathogenic MDR V. cholerae O1 in an Infant Mouse Model. Antibiotics. 2023; 12(6):1046. https://doi.org/10.3390/antibiotics12061046
Chicago/Turabian StyleChaudhary, Naveen, Balvinder Mohan, Harpreet Kaur, Vinay Modgil, Vishal Kant, Alka Bhatia, and Neelam Taneja. 2023. "Vibrio Phage VMJ710 Can Prevent and Treat Disease Caused by Pathogenic MDR V. cholerae O1 in an Infant Mouse Model" Antibiotics 12, no. 6: 1046. https://doi.org/10.3390/antibiotics12061046
APA StyleChaudhary, N., Mohan, B., Kaur, H., Modgil, V., Kant, V., Bhatia, A., & Taneja, N. (2023). Vibrio Phage VMJ710 Can Prevent and Treat Disease Caused by Pathogenic MDR V. cholerae O1 in an Infant Mouse Model. Antibiotics, 12(6), 1046. https://doi.org/10.3390/antibiotics12061046






