Approaches Used to Construct Antibiograms for Dogs in a Veterinary Teaching Hospital in the United States
Abstract
1. Introduction
2. Results
2.1. The Isolates
2.2. General Antibiograms
2.3. Antibiograms Stratified by Hospital Section (Medical Service), Sample Type/Anatomic Region of Sample Collection
3. Discussion
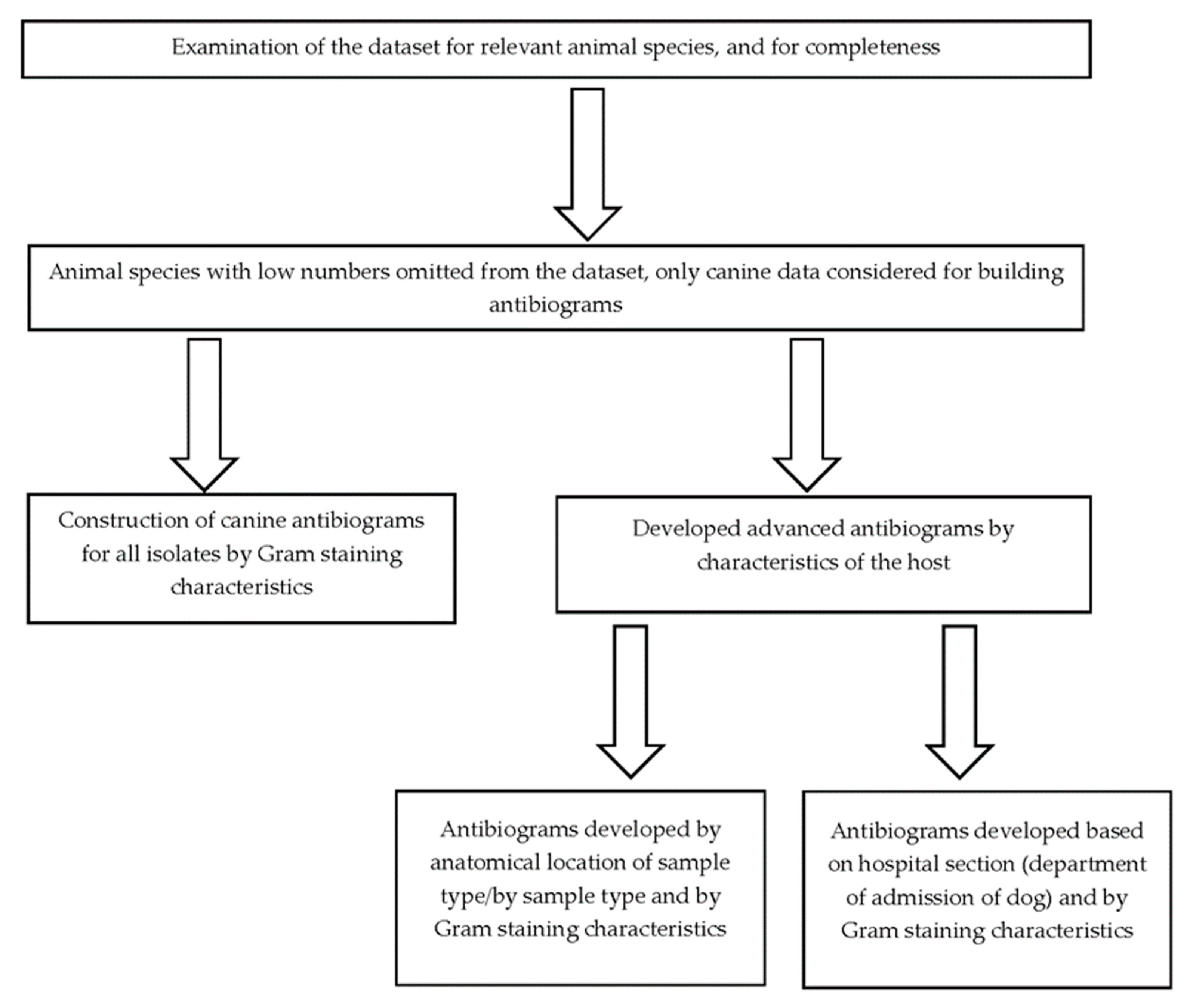
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed]
- Baekkeskov, E.; Rubin, O.; Munkholm, L.; Zaman, W. Antimicrobial Resistance as a Global Health Crisis. Available online: https://oxfordre.com/politics/display/10.1093/acrefore/9780190228637.001.0001/acrefore-9780190228637-e-1626 (accessed on 21 April 2023).
- Weese, J.S.; Giguère, S.; Guardabassi, L.; Morley, P.S.; Papich, M.; Ricciuto, D.R.; Sykes, J.E. ACVIM Consensus Statement on Therapeutic Antimicrobial Use in Animals and Antimicrobial Resistance. J. Vet. Intern. Med. 2015, 29, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.H.; Moore, L.S.P.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J.V. Understanding the Mechanisms and Drivers of Antimicrobial Resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef] [PubMed]
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. Trans. R. Soc. Trop. Med. Hyg. 2017, 111, 255–260. [Google Scholar] [CrossRef]
- Hildreth, C.J.; Burke, A.E.; Glass, R.M. Inappropriate Use of Antibiotics. JAMA 2009, 302, 816. [Google Scholar] [CrossRef] [PubMed]
- Leekha, S.; Terrell, C.L.; Edson, R.S. General Principles of Antimicrobial Therapy. Mayo Clin. Proc. 2011, 86, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Guardabassi, L.; Lars Bøgø, J.; Hilde, K. Guide to Antimicrobial Use in Animals, 1st ed.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2008. [Google Scholar]
- De Briyne, N.; Atkinson, J.; Pokludová, L.; Borriello, S.P.; Price, S. Factors Influencing Antibiotic Prescribing Habits and Use of Sensitivity Testing amongst Veterinarians in Europe. Vet. Rec. 2013, 173, 475. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.L.; Sweeney, M.T.; Lubbers, B.V. Antimicrobial Susceptibility Testing of Bacteria of Veterinary Origin. Microbiol. Spectr. 2018, 6, 6.2.08. [Google Scholar] [CrossRef] [PubMed]
- Hindler, J.F.; Stelling, J. Analysis and Presentation of Cumulative Antibiograms: A New Consensus Guideline from the Clinical and Laboratory Standards Institute. Clin. Infect. Dis. 2007, 44, 867–873. [Google Scholar] [CrossRef] [PubMed]
- Fernández, J.; Vazquez, F. The Importance of Cumulative Antibiograms in Diagnostic Stewardship. Clin. Infect. Dis. 2019, 69, 1086–1087. [Google Scholar] [CrossRef] [PubMed]
- Lautenbach, E.; Nachamkin, I. Analysis and Presentation of Cumulative Antimicrobial Susceptibility Data (Antibiograms): Substantial Variability across Medical Centers in the United States. Infect. Control. Hosp. Epidemiol. 2006, 27, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S. Hospital Antibiogram: A Necessity. Indian J. Med. Microbiol. 2010, 28, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Guarascio, A.J.; Brickett, L.M.; Porter, T.J.; Lee, N.D.; Gorse, E.E.; Covvey, J.R. Development of a Statewide Antibiogram to Assess Regional Trends in Antibiotic-Resistant ESKAPE Organisms. J. Pharm. Pract. 2019, 32, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Frey, E.; Jacob, M. Commentary: Using Antibiograms to Promote Antimicrobial Stewardship during Treatment of Bacterial Cystitis and Superficial Bacterial Folliculitis in Companion Animal Practice. J. Am. Vet. Med. Assoc. 2020, 257, 900–903. [Google Scholar] [CrossRef] [PubMed]
- Frey, E.; Jacob, M. Development of a Method for Creating Antibiograms for Use in Companion Animal Private Practices. J. Am. Vet. Med. Assoc. 2020, 257, 950–960. [Google Scholar] [CrossRef] [PubMed]
- Iyori, K.; Shishikura, T.; Shimoike, K.; Minoshima, K.; Imanishi, I.; Toyoda, Y. Influence of Hospital Size on Antimicrobial Resistance and Advantages of Restricting Antimicrobial Use Based on Cumulative Antibiograms in Dogs with Staphylococcus Pseudintermedius Infections in Japan. Vet. Dermatol. 2021, 32, 668-e178. [Google Scholar] [CrossRef] [PubMed]
- Bourély, C.; Cazeau, G.; Jarrige, N.; Leblond, A.; Madec, J.Y.; Haenni, M.; Gay, E. Antimicrobial Resistance Patterns of Bacteria Isolated from Dogs with Otitis. Epidemiol. Infect. 2019, 147, e121. [Google Scholar] [CrossRef] [PubMed]
- Scarborough, R.; Bailey, K.; Galgut, B.; Williamson, A.; Hardefeldt, L.; Gilkerson, J.; Browning, G. Use of Local Antibiogram Data and Antimicrobial Importance Ratings to Select Optimal Empirical Therapies for Urinary Tract Infections in Dogs and Cats. Antibiotics 2020, 9, 924. [Google Scholar] [CrossRef] [PubMed]
- Marshfieldslabs. Canine Prevalent Pathogens & Antimicrobial Susceptibility Patterns: January 1, 2017 to December 31, 2018. Available online: https://wwwmarshfieldlabsorg/Sites/Ltrm/Vet/Documents/Annual_Cumulative_Antibiogram_Caninepdf (accessed on 27 January 2023).
- Papich, M.G. Antibiotic Treatment of Resistant Infections in Small Animals. Vet. Clin. N. Am. Small Anim. Pract. 2013, 43, 1091–1107. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Analysis and Presentation of Cumulative Antimicrobial Susceptibility Test Data; Approved Guideline—Fourth Edition. CLSI document M39-A4; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2014. [Google Scholar]
| Escherichia coli | Pseudomonas spp. | Proteus mirabilis | |
|---|---|---|---|
| Number of Isolates | 100 | 27 | 26 |
| Antimicrobial | |||
| Amikacin | 99% | 100% | 92% |
| Amoxicillin/clavulanic acid | 78% | 4% | 100% |
| Ampicillin | 64% | 4% | 84% |
| Cefazolin | 79% | 4% | 88% |
| Cefovecin | 82% | 7% | 96% |
| Cefoxitin | 83% | 4% | 96% |
| Cefpodoxime | 82% | 4% | 96% |
| Ceftiofur | 79% | 7% | 96% |
| Cephalothin ** | 100% | 100% | |
| Chloramphenicol | 84% | 4% | 69% |
| Clindamycin ¥ | 2% | 4% | 0% |
| Doxycycline | 78% | 7% | 0% |
| Enrofloxacin | 85% | 70% | 92% |
| Erythromycin ¥ | 10% | 4% | 0% |
| Gentamycin | 94% | 85% | 85% |
| Imipenem | 100% | 100% | 96% |
| Marbofloxacin | 83% | 92% | |
| Oxacillin + 2%NACL | 0% | 7% | 0% |
| Penicillin | 0% | 4% | 0% |
| Ticarcillin | 64% | 100% | 85% |
| Ticarcillin/clavulanic acid | 75% | 100% | 96% |
| Trimethoprim/sulfamethoxazole | 85% | 11% | 85% |
| Staphylococcus spp. | Streptococcus spp. | Corynebacterium spp. | Enterococcus spp. | |
|---|---|---|---|---|
| Number of Isolates | 170 | 64 | 30 | 65 |
| Antimicrobial | ||||
| Amikacin | 89% | 84% | 93% | 0% |
| Amoxicillin/clavulanic acid | 84% | 100% | 59% | 72% |
| Ampicillin | 16% | 95% | 38% | 72% |
| Cefazolin | 85% | 100% | 50% | 0% |
| Cefovecin | 80% | 95% | 48% | 0% |
| Cefoxitin | 84% | 95% | 41% | 0% |
| Cefpodoxime | 80% | 95% | 38% | 0% |
| Ceftiofur | 81% | 97% | 53% | 0% |
| Cephalothin ** | 100% | 100% | ||
| Chloramphenicol | 86% | 92% | 80% | 88% |
| Clindamycin ¥ | 68% | 90% | 69% | 0% |
| Doxycycline | 67% | 78% | 90% | 51% |
| Enrofloxacin | 72% | 34% | 38% | 6% |
| Erythromycin ¥ | 68% | 3% | 63% | 31% |
| Gentamycin | 76% | 92% | 87% | 2% |
| Imipenem | 85% | 100% | 59% | 74% |
| Marbofloxacin | 79% | 60% | ||
| Oxacillin + 2%NACL | 85% | 98% | 21% | 6% |
| Penicillin | 0% | 92% | 0 | 69% |
| Ticarcillin | 75% | 97% | 60% | 15% |
| Ticarcillin/clavulanic acid | 84% | 97% | 52% | 18% |
| Trimethoprim/sulfamethoxazole | 75% | 94% | 87% | 3% |
| Dermatology | Surgery | Oncology | ||||||
|---|---|---|---|---|---|---|---|---|
| Staphylococcus spp. | Streptococcus spp. | Staphylococcus spp. | Staphylococcus spp. | Streptococcus spp. | Enterococcus spp. | |||
| Number of Isolates | 41 | 12 | Number of Isolates | 20 | Number of Isolates | 33 | 20 | 17 |
| Antimicrobial | ||||||||
| Amikacin | 93% | 75% | Amikacin | 85% | Amikacin | 91% | 85% | 0% |
| Amoxicillin/clavulanic acid | 85% | 100% | Amoxicillin/clavulanic acid | 65% | Amoxicillin/clavulanic acid | 85% | 100% | 65% |
| Ampicillin | ⱡ 10% | 100% | Ampicillin * | 13% | Ampicillin | 24% | 95% | 65% |
| Cefazolin | 85% | 100% | Cefazolin | 65% | Cefazolin | 85% | 100% | 0% |
| Cefovecin | 78% | 100% | Cefovecin | 60% | Cefovecin | 85% | 100% | 0% |
| Cefoxitin | 85% | 100% | Cefoxitin | 65% | Cefoxitin | 85% | 95% | 0% |
| Cefpodoxime | 78% | 100% | Cefpodoxime | 60% | Cefpodoxime | 85% | 100% | 0% |
| Ceftiofur | 85% | 100% | Ceftiofur | 60% | Ceftiofur | 79% | 100% | 0% |
| Cephalothin ** | 100% | 100% | Cephalothin ** | 100% | Cephalothin ** | 100% | 100% | |
| Chloramphenicol | 88% | 100% | Chloramphenicol | 85% | Chloramphenicol | 76% | 95% | 88% |
| Clindamycin | 68% | 92% | Clindamycin | 55% | Clindamycin | 67% | 95% | 0% |
| Doxycycline | 71% | 92% | Doxycycline | 90% | Doxycycline | 61% | 75% | 47% |
| Enrofloxacin | 68% | 42% | Enrofloxacin | 60% | Enrofloxacin | 76% | 25% | 0% |
| Erythromycin | 71% | 0% | Erythromycin | 55% | Erythromycin | 67% | 5% | 35% |
| Gentamycin | 66% | 92% | Gentamycin | 70% | Gentamycin | 82% | 95% | 0% |
| Imipenem | 85% | 100% | Imipenem | 70% | Imipenem | 85% | 100% | 71% |
| Marbofloxacin | 71% | 67% | Marbofloxacin | 75% | Marbofloxacin | 85% | 50% | |
| Oxacillin + 2%NACL | 85% | 100% | Oxacillin + 2%NACL | 70% | Oxacillin + 2%NACL | 85% | 100% | 6% |
| Penicillin * | 0% | 100% | Penicillin * | 0% | Penicillin * | 0% | 95% | 65% |
| Ticarcillin * | 79% | 100% | Ticarcillin * | 44% | Ticarcillin * | 80% | 100% | 24% |
| Ticarcillin/clavulanic acid | 85% | 100% | Ticarcillin/clavulanic acid | 65% | Ticarcillin/clavulanic acid | 85% | 100% | 24% |
| Trimethoprim/sulfamethoxazole | 68% | 100% | Trimethoprim/sulfamethoxazole | 80% | Trimethoprim/sulfamethoxazole | 79% | 95% | 0% |
| Medicine | Emergency | ||||
|---|---|---|---|---|---|
| Staphylococcus spp. | Enterococcus spp. | Staphylococcus spp. | Enterococcus spp. | ||
| Number of Isolates | 28 | 15 | Number of Isolates | 16 | 13 |
| Antimicrobial | |||||
| Amikacin | 86% | 0% | Amikacin | 88% | 0% |
| Amoxicillin/clavulanic acid | 82% | 80% | Amoxicillin/clavulanic acid | 94% | 69% |
| Ampicillin | * 6% | 80% | Ampicillin * | 13% | 69% |
| Cefazolin | 82% | 0% | Cefazolin | 94% | 0% |
| Cefovecin | 79% | 0% | Cefovecin | 88% | 0% |
| Cefoxitin | 82% | 0% | Cefoxitin | 94% | 0% |
| Cefpodoxime | 79% | 0% | Cefpodoxime | 88% | 0% |
| Ceftiofur | 79% | 0% | Ceftiofur | 88% | 0% |
| Cephalothin ** | 100% | Cephalothin ** | 100% | ||
| Chloramphenicol | 86% | 100% | Chloramphenicol | 94% | 62% |
| Clindamycin | 61% | 0% | Clindamycin | 69% | 0% |
| Doxycycline | 54% | 60% | Doxycycline | 75% | 31% |
| Enrofloxacin | 71% | 7% | Enrofloxacin | 69% | 8% |
| Erythromycin | 61% | 27% | Erythromycin | 69% | 8% |
| Gentamycin | 75% | 7% | Gentamycin | 81% | 0% |
| Imipenem | 82% | 80% | Imipenem | 94% | 695 |
| Marbofloxacin | 71% | Marbofloxacin | 88% | ||
| Oxacillin + 2%NACL | 82% | 7% | Oxacillin + 2%NACL | 94% | 0% |
| Penicillin | * 0% | 73% | Penicillin | * 0% | 69% |
| Ticarcillin | * 72% | 13% | Ticarcillin | * 88% | 0% |
| Ticarcillin/clavulanic acid | 82% | 20% | Ticarcillin/clavulanic acid | 94% | 0% |
| Trimethoprim/sulfamethoxazole | 61% | 7% | Trimethoprim/sulfamethoxazole | 81% | 0% |
| Oncology | Medicine | Emergency | |||
|---|---|---|---|---|---|
| Escherichia coli | Escherichia coli | Escherichia coli | |||
| Number of Isolates | 28 | Number of Isolates | 26 | Number of Isolates | 15 |
| Antimicrobial | |||||
| Amikacin | 100% | Amikacin | 100% | Amikacin | 100% |
| Amoxicillin/clavulanic acid | 71% | Amoxicillin/clavulanic acid | 81% | Amoxicillin/clavulanic acid | 67% |
| Ampicillin | 57% | Ampicillin | 69% | Ampicillin | 73% |
| Cefazolin | 79% | Cefazolin | 85% | Cefazolin | 73% |
| Cefovecin | 82% | Cefovecin | 88% | Cefovecin | 73% |
| Cefoxitin | 86% | Cefoxitin | 85% | Cefoxitin | 73% |
| Cefpodoxime | 82% | Cefpodoxime | 88% | Cefpodoxime | 73% |
| Ceftiofur | 82% | Ceftiofur | 81% | Ceftiofur | 67% |
| Cephalothin ** | 100% | Cephalothin ** | 100% | Cephalothin ** | 9% |
| Chloramphenicol | 86% | Chloramphenicol | 85% | Chloramphenicol | 93% |
| Clindamycin ¥ | 0% | Clindamycin | 4% | Clindamycin | 7% |
| Doxycycline | 75% | Doxycycline | 81% | Doxycycline | 87% |
| Enrofloxacin | 82% | Enrofloxacin | 85% | Enrofloxacin | 93% |
| Erythromycin ¥ | 11% | Erythromycin | 12% | Erythromycin | 7% |
| Gentamycin | 93% | Gentamycin | 96% | Gentamycin | 100% |
| Imipenem | 100% | Imipenem | 100% | Imipenem | 100% |
| Marbofloxacin | 82% | Marbofloxacin | 85% | Marbofloxacin | 93% |
| Oxacillin + 2%NACL | 0% | Oxacillin + 2%NACL | 0% | Oxacillin + 2%NACL | 0% |
| Penicillin | 0% | Penicillin | 0% | Penicillin | 0% |
| Ticarcillin | 57% | Ticarcillin | 69% | Ticarcillin | 73% |
| Ticarcillin/clavulanic acid | 71% | Ticarcillin/clavulanic acid | 80% | Ticarcillin/clavulanic acid | 73% |
| Trimethoprim/sulfamethoxazole | 79% | Trimethoprim/sulfamethoxazole | 88% | Trimethoprim/sulfamethoxazole | 100% |
| Samples from the Ear | Samples from the Skin | ||
|---|---|---|---|
| Staphylococcus spp. | Staphylococcus spp. | ||
| Number of Isolates | 21 | Number of Isolates | 43 |
| Antimicrobial | |||
| Amikacin | 90% | Amikacin | 93% |
| Amoxicillin/clavulanic acid | 95% | Amoxicillin/clavulanic acid | 88% |
| Ampicillin * | 29% | Ampicillin * | 3% |
| Cefazolin | 95% | Cefazolin | 88% |
| Cefovecin | 90% | Cefovecin | 81% |
| Cefoxitin | 95% | Cefoxitin | 88% |
| Cefpodoxime | 90% | Cefpodoxime | 81% |
| Ceftiofur | 95% | Ceftiofur | 88% |
| Cephalothin ** | 100% | Cephalothin ** | 100% |
| Chloramphenicol | 95% | Chloramphenicol | 84% |
| Clindamycin | 81% | Clindamycin | 63% |
| Doxycycline | 71% | Doxycycline | 65% |
| Enrofloxacin | 76% | Enrofloxacin | 70% |
| Erythromycin | 81% | Erythromycin | 65% |
| Gentamycin | 86% | Gentamycin | 67% |
| Imipenem | 95% | Imipenem | 88% |
| Marbofloxacin | 81% | Marbofloxacin | 72% |
| Oxacillin + 2%NACL | 95% | Oxacillin + 2%NACL | 88% |
| Penicillin * | 0% | Penicillin * | 0% |
| Ticarcillin * | 93% | Ticarcillin * | 81% |
| Ticarcillin/clavulanic acid | 95% | Ticarcillin/clavulanic acid | 88% |
| Trimethoprim/sulfamethoxazole | 71% | Trimethoprim/sulfamethoxazole | 63% |
| Gram-Negative | Gram-Positive | ||||||
|---|---|---|---|---|---|---|---|
| Escherichia coli | Pseudomonas spp. | Proteus mirabilis | Staphylococcus spp. | Streptococcus spp. | Enterococcus spp. | ||
| Number of Isolates | 71 | 13 | 14 | Number of Isolates | 49 | 27 | 45 |
| Antimicrobial | |||||||
| Amikacin | 99% | 100% | 93% | Amikacin | 90% | 89% | 0% |
| Amoxicillin/clavulanic acid | 80% | 8% | 100% | Amoxicillin/clavulanic acid | 86% | 100% | 73% |
| Ampicillin | 69% | 8% | 86% | Ampicillin | 23% | 93% | 73% |
| Cefazolin | 82% | 8% | 93% | Cefazolin | 86% | 100% | 0% |
| Cefovecin | 86% | 15% | 93% | Cefovecin | 82% | 96% | 0% |
| Cefoxitin | 84% | 8% | 93% | Cefoxitin | 86% | 93% | 0% |
| Cefpodoxime | 84% | 8% | 93% | Cefpodoxime | 82% | 93% | 0% |
| Ceftiofur | 80% | 15% | 93% | Ceftiofur | 76% | 96% | 0% |
| Cephalothin ** | 100% | 100% | Cephalothin ** | 100% | 100% | ||
| Chloramphenicol | 83% | 8% | 71% | Chloramphenicol | 78% | 89% | 89% |
| Clindamycin ¥ | 1% | 8% | 0% | Clindamycin | 69% | 96% | 0% |
| Doxycycline | 82% | 15% | 0% | Doxycycline | 59% | 78% | 49% |
| Enrofloxacin | 87% | 77% | 93% | Enrofloxacin | 76% | 26% | 4% |
| Erythromycin ¥ | 11% | 8% | 0% | Erythromycin | 69% | 7% | 31% |
| Gentamycin | 94% | 100% | 86% | Gentamycin | 80% | 92% | 2% |
| Imipenem | 100% | 100% | 100% | Imipenem | 86% | 100% | 76% |
| Marbofloxacin | 86% | 93% | Marbofloxacin | 82% | 48% | ||
| Oxacillin + 2%NACL | 0% | 15% | 0% | Oxacillin + 2%NACL | 86% | 100% | 7% |
| Penicillin | 0% | 8% | 0% | Penicillin * | 0% | 89% | 69% |
| Ticarcillin | 69% | 100% | 86% | Ticarcillin * | 80% | 96% | 13% |
| Ticarcillin/clavulanic acid | 79% | 100% | 93% | Ticarcillin/clavulanic acid | 86% | 96% | 16% |
| Trimethoprim/sulfamethoxazole | 86% | 23% | 93% | Trimethoprim/sulfamethoxazole | 80% | 89% | 4% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ekakoro, J.E.; Guptill, L.; Hendrix, K.; Anderson, M.; Ruple, A. Approaches Used to Construct Antibiograms for Dogs in a Veterinary Teaching Hospital in the United States. Antibiotics 2023, 12, 1034. https://doi.org/10.3390/antibiotics12061034
Ekakoro JE, Guptill L, Hendrix K, Anderson M, Ruple A. Approaches Used to Construct Antibiograms for Dogs in a Veterinary Teaching Hospital in the United States. Antibiotics. 2023; 12(6):1034. https://doi.org/10.3390/antibiotics12061034
Chicago/Turabian StyleEkakoro, John E., Lynn Guptill, Kenitra Hendrix, Melinda Anderson, and Audrey Ruple. 2023. "Approaches Used to Construct Antibiograms for Dogs in a Veterinary Teaching Hospital in the United States" Antibiotics 12, no. 6: 1034. https://doi.org/10.3390/antibiotics12061034
APA StyleEkakoro, J. E., Guptill, L., Hendrix, K., Anderson, M., & Ruple, A. (2023). Approaches Used to Construct Antibiograms for Dogs in a Veterinary Teaching Hospital in the United States. Antibiotics, 12(6), 1034. https://doi.org/10.3390/antibiotics12061034






