Occurrence of Carbapenemases, Extended-Spectrum Beta-Lactamases and AmpCs among Beta-Lactamase-Producing Gram-Negative Bacteria from Clinical Sources in Accra, Ghana
Abstract
1. Introduction
2. Results
2.1. Spectrum of Gram-Negative Bacteria
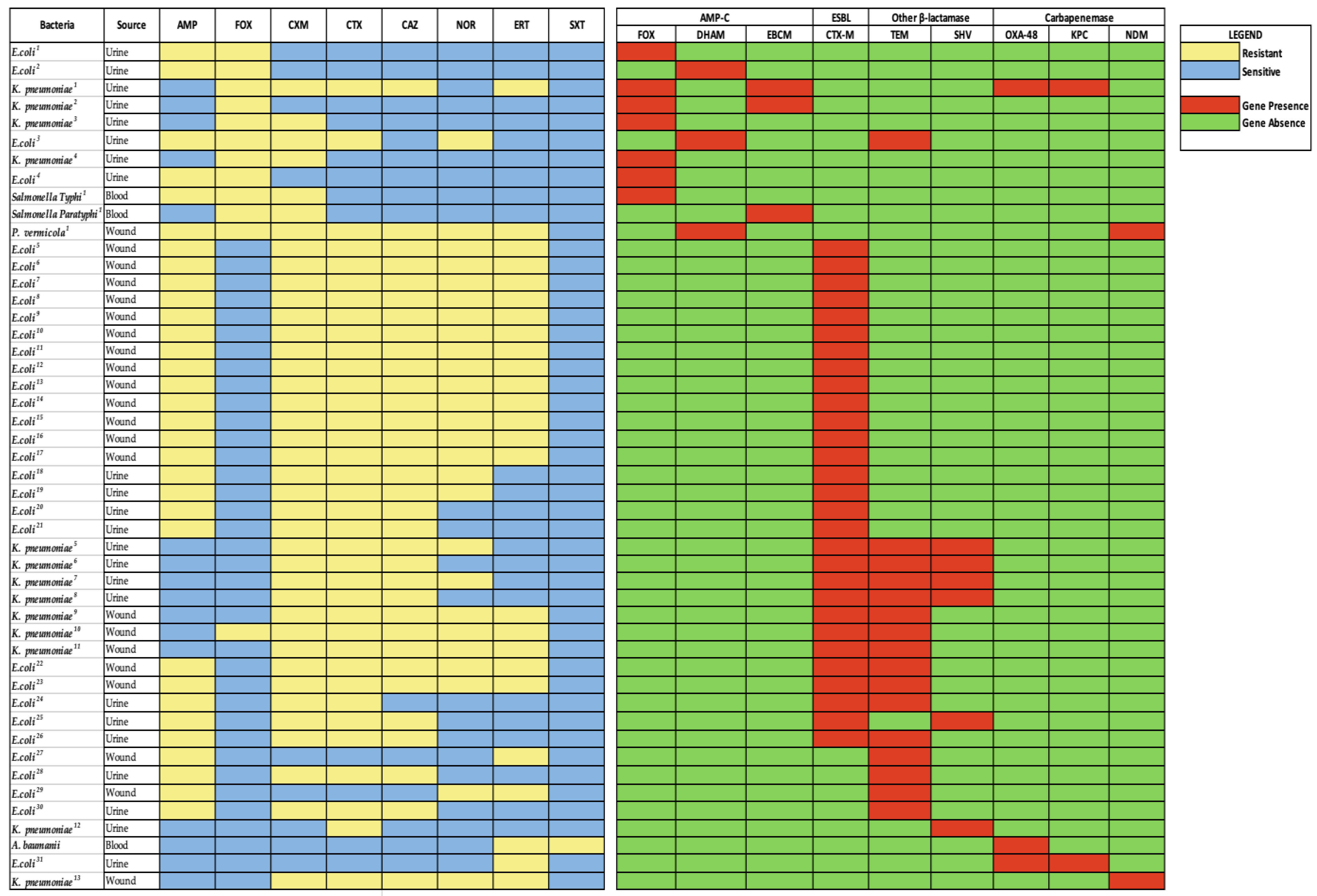
2.2. Antimicrobial Resistance Pattern
2.3. Phenotype and Resistance Gene Markers for AmpC, ESBL and Carbapenemase
2.4. Resistance Gene Distribution among Isolates
3. Discussion
4. Materials and Methods
4.1. Identification of Bacteria
4.2. Antimicrobial Susceptibility Testing
4.3. Phenotypic Screening for AmpC, ESBL and Carbapenem Resistance
4.4. Molecular Detection of Antimicrobial-Resistant Gene Markers of Beta-Lactamases
4.5. Data Analysis
5. Conclusions
6. Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Oliveira DM, P.; Forde, B.M.; Kidd, T.J.; Harris PN, A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- Habeeb, M.A.; Sarwar, Y.; Ali, A.; Salman, M.; Haque, A. Rapid emergence of ESBL producers in E. coli causing urinary and wound infections in Pakistan. Pak. J. Med. Sci. 2013, 29, 540. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.E.; Abbas, M.; Al-Shahrai, A.M.; Elamin, B.K. Phenotypic Characterization and Antibiotic Resistance Patterns of Extended-Spectrum β-Lactamase- and AmpC β-Lactamase-Producing Gram-Negative Bacteria in a Referral Hospital, Saudi Arabia. Can. J. Infect. Dis. Med. Microbiol. 2019, 2019, 6054694. [Google Scholar] [CrossRef]
- Obeng-Nkrumah, N.; Twum-Danso, K.; Krogfelt, K.A.; Newman, M.J. High Levels of Extended-Spectrum Beta-Lactamases in a Major Teaching Hospital in Ghana: The Need for Regular Monitoring and Evaluation of Antibiotic Resistance. Am. J. Trop. Med. Hyg. 2013, 89, 960–964. [Google Scholar] [CrossRef]
- Bush, K. Proliferation and significance of clinically relevant β-lactamases. Ann. N.Y. Acad. Sci. 2013, 1277, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Abdalhamid, B.; Albunayan, S.; Shaikh, A.; Elhadi, N.; Aljindan, R. Prevalence study of plasmid-mediated AmpC β-lactamases in Enterobacteriaceae lacking inducible ampC from Saudi hospitals. J. Med. Microbiol. 2017, 66, 1286–1290. [Google Scholar] [CrossRef]
- Mata, C.; Miró, E.; Rivera, A.; Mirelis, B.; Coll, P.; Navarro, F. Prevalence of acquired AmpC β-lactamases in Enterobacteriaceae lacking inducible chromosomal ampC genes at a Spanish hospital from 1999 to 2007. Clin. Microbiol. Infect. 2010, 16, 472–476. [Google Scholar] [CrossRef]
- Codjoe, F.; Donkor, E. Carbapenem Resistance: A Review. Med. Sci. 2017, 6, 1. [Google Scholar] [CrossRef]
- Reuland, E.A.; al Naiemi, N.; Raadsen, S.A.; Savelkoul, P.H.M.; Kluytmans, J.A.J.W.; Vandenbroucke-Grauls, C.M.J.E. Prevalence of ESBL-producing Enterobacteriaceae in raw vegetables. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1843–1846. [Google Scholar] [CrossRef]
- Ghafourian, S.; Sadeghifard, N.; Soheili, S.; Sekawi, Z. Extended Spectrum Beta-lactamases: Definition, Classification and Epidemiology. Curr. Issues Mol. Biol. 2015, 17, 11–21. [Google Scholar] [CrossRef]
- Hijazi, S.M.; Fawzi, M.A.; Ali, F.M.; Abd El Galil, K.H. Multidrug-resistant ESBL-producing Enterobacteriaceae and associated risk factors in community infants in Lebanon. J. Infect. Dev. Ctries. 2016, 10, 947–955. [Google Scholar] [CrossRef] [PubMed]
- WHO. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis; WHO: Geneva, Switzerland, 2017; pp. 1–88. [Google Scholar]
- Bradford, P.A. Extended-Spectrum β-Lactamases in the 21st Century: Characterization, Epidemiology, and Detection of This Important Resistance Threat. Clin. Microbiol. Rev. 2001, 14, 933–951. [Google Scholar] [CrossRef] [PubMed]
- Lukac, P.J.; Bonomo, R.A.; Logan, L.K. Extended-Spectrum-Lactamase-Producing Enterobacteriaceae in Children: Old Foe, Emerging Threat. Clin. Infect. Dis. 2015, 60, 1389–1397. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.L.; Ko, W.-C.; von Gottberg, A.; Mohapatra, S.; Casellas, J.M.; Goossens, H.; Mulazimoglu, L.; Trenholme, G.; Klugman, K.P.; Bonomo, R.A.; et al. Antibiotic Therapy for Klebsiella pneumoniae Bacteremia: Implications of Production of Extended-Spectrum-Lactamases. Clin. Infect. Dis. 2004, 39, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Meletis, G. Carbapenem resistance: Overview of the problem and future perspectives. Ther. Adv. Infect. Dis. 2016, 3, 15–21. [Google Scholar] [CrossRef]
- Papp-Wallace, K.M.; Endimiani, A.; Taracila, M.A.; Bonomo, R.A. Carbapenems: Past, Present, and Future. Antimicrob. Agents Chemother. 2011, 55, 4943–4960. [Google Scholar] [CrossRef]
- Livermore, D.M.; Woodford, N. The β-lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. Trends Microbiol. 2006, 14, 413–420. [Google Scholar] [CrossRef]
- Halat, D.H.; Moubareck, C.A. The current burden of Carbapenemases: Review of significant properties and dissemination among gram-negative bacteria. Antibiotics 2020, 9, 186. [Google Scholar] [CrossRef]
- Schultsz, C.; Geerlings, S. Plasmid-Mediated Resistance in Enterobacteriaceae. Drugs 2012, 72, 1–16. [Google Scholar] [CrossRef]
- Bedenić, B.; Plečko, V.; Sardelić, S.; Uzunović, S.; Godič Torkar, K. Carbapenemases in Gram-Negative Bacteria: Laboratory Detection and Clinical Significance. BioMed Res. Int. 2014, 2014, 841951. [Google Scholar] [CrossRef]
- Moquet, O.; Bouchiat, C.; Kinana, A.; Seck, A.; Arouna, O.; Bercion, R.; Breurec, S.; Garin, B. Class D OXA-48 Carbapenemase in Multidrug-Resistant Enterobacteria, Senegal. Emerg. Infect. Dis. 2011, 17, 143–144. [Google Scholar] [CrossRef] [PubMed]
- Osei, M.-M.; Dayie, N.T.K.D.; Azaglo, G.S.K.; Tettey, E.Y.; Nartey, E.T.; Fenny, A.P.; Manzi, M.; Kumar, A.M.v.; Labi, A.-K.; Opintan, J.A.; et al. Alarming Levels of Multidrug Resistance in Aerobic Gram-Negative Bacilli Isolated from the Nasopharynx of Healthy Under-Five Children in Accra, Ghana. Int. J. Environ. Res. Public Health 2022, 19, 10927. [Google Scholar] [CrossRef] [PubMed]
- Donkor, E.S.; Horlortu, P.Z.; Dayie, N.T.; Obeng-Nkrumah, N.; Labi, A.-K. Community acquired urinary tract infections among adults in Accra, Ghana. Infect. Drug Resist. 2019, 12, 2059–2067. [Google Scholar] [CrossRef] [PubMed]
- Mambie, A.; Vuotto, F.; Poitrenaud, D.; Weyrich, P.; Cannesson, O.; Dessein, R.; Faure, K.; Guery, B.; Galpérine, T. Cefoxitin: An alternative to carbapenems in urinary tract infections due to extended-spectrum beta-lactamase-producing Enterobacteriaceae. Med. Et Mal. Infect. 2016, 46, 215–219. [Google Scholar] [CrossRef]
- Aldrich, C.; Hartman, H.; Feasey, N.; Chattaway, M.A.; Dekker, D.; Al-Emran, H.M.; Larkin, L.; McCormick, J.; Sarpong, N.; le Hello, S.; et al. Emergence of phylogenetically diverse and fluoroquinolone resistant Salmonella Enteritidis as a cause of invasive nontyphoidal Salmonella disease in Ghana. PLoS Negl. Trop. Dis. 2019, 13, e0007485. [Google Scholar] [CrossRef]
- Leopold, S.J.; van Leth, F.; Tarekegn, H.; Schultsz, C. Antimicrobial drug resistance among clinically relevant bacterial isolates in sub-Saharan Africa: A systematic review. J. Antimicrob. Chemother. 2014, 69, 2337–2353. [Google Scholar] [CrossRef]
- Kumburu, H.H.; Sonda, T.; Mmbaga, B.T.; Alifrangis, M.; Lund, O.; Kibiki, G.; Aarestrup, F.M. Patterns of infections, aetiological agents and antimicrobial resistance at a tertiary care hospital in northern Tanzania. Trop. Med. Int. Health 2017, 22, 454–464. [Google Scholar] [CrossRef]
- Ntirenganya, C.; Muvunyi, C.M.; Manzi, O.; Ogbuagu, O. High Prevalence of Antimicrobial Resistance Among Common Bacterial Isolates in a Tertiary Healthcare Facility in Rwanda. Am. J. Trop. Med. Hyg. 2015, 92, 865–870. [Google Scholar] [CrossRef]
- Dela, H.; Egyir, B.; Majekodunmi, A.O.; Behene, E.; Yeboah, C.; Ackah, D.; Bongo, R.N.A.; Bonfoh, B.; Zinsstag, J.; Bimi, L.; et al. Diarrhoeagenic E. coli occurrence and antimicrobial resistance of Extended Spectrum Beta-Lactamases isolated from diarrhoea patients attending health facilities in Accra, Ghana. PLoS ONE 2022, 17, e0268991. [Google Scholar] [CrossRef]
- Ouedraogo, A.-S.; Sanou, M.; Kissou, A.; Sanou, S.; Solaré, H.; Kaboré, F.; Poda, A.; Aberkane, S.; Bouzinbi, N.; Sano, I.; et al. High prevalence of extended-spectrum ß-lactamase producing enterobacteriaceae among clinical isolates in Burkina Faso. BMC Infect. Dis. 2016, 16, 326. [Google Scholar] [CrossRef]
- Nakayama, T.; Ueda, S.; Huong BT, M.; Tuyen, L.D.; Komalamisra, C.; Kusolsuk, T.; Hirai, I.; Yamamoto, Y. Wide dissemination of extended-spectrumβ-lactamase-producing Escherichia coli in community residents in the Indochinese peninsula. Infect. Drug Resist. 2015, 12, 3317–3325. [Google Scholar] [CrossRef]
- Duval, V.; Maiga, I.; Maiga, A.; Guillard, T.; Brasme, L.; Forte, D.; Madoux, J.; Vernet-Garnier, V.; de Champs, C. High prevalence of CTX-M-type beta-lactamases among clinical isolates of enterobacteriaceae in Bamako, Mali. Antimicrob Agents Chemother 2009, 53, 4957–4958. [Google Scholar] [CrossRef] [PubMed]
- Doi, Y.; Park, Y.S.; Rivera, J.I.; Adams-Haduch, J.M.; Hingwe, A.; Sordillo, E.M.; Lewis, J.S.; Howard, W.J.; Johnson, L.E.; Polsky, B.; et al. Community-Associated Extended-Spectrum β-Lactamase–Producing Escherichia coli Infection in the United States. Clin. Infect. Dis. 2013, 56, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Peirano, G.; van Greune, C.H.J.; Pitout, J.D.D. Characteristics of infections caused by extended-spectrum β-lactamase–producing Escherichia coli from community hospitals in South Africa. Diagn. Microbiol. Infect. Dis. 2011, 69, 449–453. [Google Scholar] [CrossRef] [PubMed]
- Chukwu, E.E.; Awoderu, O.B.; Enwuru, C.A.; Afocha, E.E.; Lawal, R.G.; Ahmed, R.A.; Olanrewaju, I.; Onwuamah, C.K.; Audu, R.A.; Ogunsola, F.T. High prevalence of resistance to third-generation cephalosporins detected among clinical isolates from sentinel healthcare facilities in Lagos, Nigeria. Antimicrob. Resist. Infect. Control 2022, 11, 134. [Google Scholar] [CrossRef] [PubMed]
- Agyekum, A.; Fajardo-Lubián, A.; Ansong, D.; Partridge, S.R.; Agbenyega, T.; Iredell, J.R. blaCTX-M-15 carried by IncF-type plasmids is the dominant ESBL gene in Escherichia coli and Klebsiella pneumoniae at a hospital in Ghana. Diagn. Microbiol. Infect. Dis. 2016, 84, 328–333. [Google Scholar] [CrossRef]
- Shaaban, M.; Elshaer, S.L.; Abd El-Rahman, O.A. Prevalence of extended-spectrum β-lactamases, AmpC, and carbapenemases in Proteus mirabilis clinical isolates. BMC Microbiol. 2022, 22, 247. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, I.; Haruna, M.; Yahaya, H. Prevalence and antibiotic susceptibility of AmpC and ESBLs producing clinical isolates at a tertiary health care center in Kano, north-west Nigeria. Afr. J. Clin. Exp. Microbiol. 2013, 14, 109–119. [Google Scholar] [CrossRef]
- Arnold, R.S.; Thom, K.A.; Sharma, S.; Phillips, M.; Kristie Johnson, J.; Morgan, D.J. Emergence of Klebsiella pneumoniae Carbapenemase-Producing Bacteria. South. Med. J. 2011, 104, 40–45. [Google Scholar] [CrossRef]
- Codjoe, F.S. Detection and Characterisation of Carbapenem-Resistant Gram-Negative Bacilli Infections in Ghana. Ph.D. Thesis, Sheffield Hallam University, Sheffield, UK, 2016. [Google Scholar]
- Quansah, E.; Amoah Barnie, P.; Omane Acheampong, D.; Obiri-Yeboah, D.; Odarkor Mills, R.; Asmah, E.; Cudjoe, O.; Dadzie, I. Geographical Distribution of β-Lactam Resistance among Klebsiella spp. from Selected Health Facilities in Ghana. Trop. Med. Infect. Dis. 2019, 4, 117. [Google Scholar] [CrossRef]
- Monnheimer, M.; Cooper, P.; Amegbletor, H.K.; Pellio, T.; Groß, U.; Pfeifer, Y.; Schulze, M.H. High Prevalence of Carbapenemase-Producing Acinetobacter baumannii in Wound Infections, Ghana, 2017/2018. Microorganisms 2021, 9, 537. [Google Scholar] [CrossRef] [PubMed]
- Codjoe, F.S.; Brown, C.A.; Smith, T.J.; Miller, K.; Donkor, E.S. Genetic relatedness in carbapenem-resistant isolates from clinical specimens in Ghana using ERIC-PCR technique. PLoS ONE 2019, 14, e0222168. [Google Scholar] [CrossRef] [PubMed]
- Kazemian, H.; Heidari, H.; Ghanavati, R.; Ghafourian, S.; Yazdani, F.; Sadeghifard, N.; Valadbeigi, H.; Maleki, A.; Pakzad, I. Phenotypic and Genotypic Characterization of ESBL-, AmpC-, and Carbapenemase-Producing Klebsiella pneumoniae and Escherichia coli Isolates. Med. Princ. Pract. 2019, 28, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Obeng-Nkrumah, N.; Labi, A.K.; Blankson, H.; Awuah-Mensah, G.; Oduro-Mensah, D.; Anum, J.; Teye, J.; Kwashie, S.D.; Bako, E.; Ayeh-Kumi, P.F.; et al. Household cockroaches carry CTX-M-15-, OXA-48- And NDM-1-producing enterobacteria, and share beta-lactam resistance determinants with humans. BMC Microbiol. 2019, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M. Prevalence and antibiogram of Extended Spectrum β-Lactamase (ESBL) producing Gram negative bacilli and further molecular characterization of ESBL producing Escherichia coli and Klebsiella spp. J. Clin. Diagn. Res. 2013, 7, 2173–2177. [Google Scholar] [CrossRef]
- Khurana, S.; Mathur, P.; Kapil, A.; Valsan, C.; Behera, B. Molecular epidemiology of beta-lactamase producing nosocomial Gram-negative pathogens from North and South Indian hospitals. J. Med. Microbiol. 2017, 66, 999–1004. [Google Scholar] [CrossRef] [PubMed]
| Isolates | Urine | Wound | Blood | Throat | Stool | Ear |
|---|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |
| Escherichia coli (n = 83) | 61 (73) | 18 (22) | 3 (4) | _ | _ | 1 (1.2) |
| Klebsiella pneumoniae (n = 30) | 25 (83) | 3 (10) | 2 (7) | _ | _ | _ |
| Proteus mirabilis (n = 18) | 8 (44) | 2 (11) | 7 (39) | _ | 1 (6) | _ |
| Pseudomonas aeruginosa (n = 8) | _ | 8 (100) | _ | _ | _ | _ |
| Pseudomonas stutzeri (n = 1) | _ | _ | 1 (100) | _ | _ | _ |
| Enterobacter cloacae (n = 7) | 5 (71) | _ | 2 (29) | _ | _ | _ |
| Enterobacter asburiae (n = 4) | 4 (100) | _ | _ | _ | _ | _ |
| Enterobacter kobei (n = 4) | 3 (75) | 1 (25) | _ | _ | _ | _ |
| Enterobacter aerogenes (n = 1) | 1 (100) | _ | _ | _ | _ | _ |
| Salmonella paratyphi (n = 3) | 1 (33) | _ | 1 (33) | 1 (33) | _ | _ |
| Salmonella typhi (n = 3) | _ | _ | 3 (100) | _ | _ | _ |
| Salmonella enterica (n = 2) | _ | _ | 1 (50) | _ | 1 (50) | _ |
| Acinetobacter baumannii (n = 2) | 1 (50) | _ | 1 (50) | _ | _ | _ |
| Acinetobacter nosocomialis (n = 2) | 2 (100) | _ | _ | _ | _ | _ |
| Neisseria subflava (n = 2) | _ | _ | _ | 2 (100) | _ | _ |
| Klebsiella oxytoca (n = 1) | _ | 1 (100) | _ | _ | _ | _ |
| Providencia stuartii (n = 1) | _ | 1 (100) | _ | _ | _ | _ |
| Providencia vermicola (n = 1) | _ | 1 (100) | _ | _ | _ | _ |
| Citrobacter koseri (n = 1) | 1 (100) | _ | _ | _ | _ | _ |
| Citrobacter youngae (n = 1) | _ | _ | _ | _ | 1 (100) | _ |
| Citrobacter freundi (n = 1) | 1 (100) | _ | _ | _ | _ | _ |
| Neisseria meningiditis (n = 1) | _ | _ | _ | 1 (100) | _ | _ |
| Kerstersia gyiorum (n = 1) | _ | 1 (100) | _ | _ | _ | _ |
| Achromobacter xylososidans (n = 1) | _ | 1 (100) | _ | _ | _ | _ |
| Alcaligenes faecalis (n = 1) | _ | 1 (100) | _ | _ | _ | _ |
| Cupriavidus gilardii (n = 1) | 1 (100) | _ | _ | _ | _ | _ |
| Total (181) | 114 (63) | 38 (21) | 21 (12) | 4 (2.2) | 3 (1.6) | 1 (0.5) |
| Bacterial Isolates | AMP N (%) | CTX N (%) | NOR N (%) | CAZ N (%) | ERT N (%) | FOX N (%) | CXM N (%) | MDR N (%) |
|---|---|---|---|---|---|---|---|---|
| E. coli (n = 83) | 76 (92) | 27 (33) | 23 (28) | 23 (28) | 18 (22) | 5 (6) | 29 (35) | 40 (48) |
| K. pneumoniae (n = 30) | #_ | 12 (40) | 5 (17) | 9 (30) | 6 (20) | 5 (17) | 10 (33) | 11 (37) |
| P. mirabilis (n = 18) | 4 (22) | 1 (6) | 0 | 6 (33) | 2 (11) | 0 | 1 (6) | 1 (6) |
| Enterobacter spp. (n = 16) | #_ | 3 (19) | 2 (13) | 4 (25) | 3 (19) | 8 (50) | 3 (19) | 11 (69) |
| Citrobacter spp. (n = 3) | #_ | 1 (33) | 0 | 0 | 0 | 2 (67) | 0 | 2 (67) |
| Salmonella Typhi (n = 3) | 2 (67) | 0 | 0 | 0 | 0 | 1 (33) | $_ | 1 (33) |
| Salmonella Paratyphi (n = 3) | 0 | 0 | 0 | 0 | 0 | 1 (33) | $_ | 1 (33) |
| Neisseria spp. (n = 3) | NA | 1 (33) | NA | NA | NA | 0 | NA | 0 |
| Salmonella enterica. (n = 2) | 0 | 0 | 0 | 1 (50) | 0 | 0 | $_ | 0 |
| K. oxytoca (n = 1) | #_ | 0 | 0 | 0 | 1 (100) | 0 | 1 (100) | 1 (100) |
| Providencia vermicola (n = 1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) |
| Providencia stuartii (n = 1) | 1 (100) | 0 | 1 (100) | 0 | 1 (100) | 0 | 0 | 1 (100) |
| Kerstersia gyiorum (n = 1) | 0 | 0 | 1 (100) | 0 | 0 | 0 | 1 (100) | 1 (100) |
| Achromobacter xylososidans (n = 1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) |
| Alcaligenes faecalis (n = 1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 | 1 (100) | 1 (100) |
| Cupriavidus gilardii (n = 1) | NA | 0 | 0 | 1 (100) | 1 (100) | 0 | NA | 1 (100) |
| Total (n = 168) | 86 (51) | 48 (29) | 35 (21) | 47 (28) | 35 (21) | 24 (14) | 48 (29) | 74 (44) |
| Isolate | AmpC | ESBL | Carbapenemase | |||
|---|---|---|---|---|---|---|
| Phenotypic (n = 28) | Genotypic (n = 11) | Phenotypic (n = 36) | Genotypic (n = 29) | Phenotypic (n = 35) | Genotypic (n = 5) | |
| E. coli (n = 83) | 5 | 4 (80%) | 27 | 22 (81%) | 18 | 1 (6%) |
| K. pneumoniae (n = 30) | 5 | 4 (80%) | 8 | 7 (88%) | 6 | 2 (33%) |
| Proteus mirabilis (n = 18) | - | - | 1 | - | 2 | - |
| Enterobacter spp. (n = 16) | 8 | - | - | - | 3 | - |
| Salmonella spp. (n = 8) | 2 | 2 (100%) | - | - | - | - |
| Acinetobacter spp. (n = 4) | 4 | - | - | - | 3 | 1 (33%) |
| Citrobacter spp. (n = 3) | 2 | - | - | - | - | - |
| K. oxytoca (n = 1) | - | - | - | - | 1 | - |
| Providencia vermicola (n = 1) | 1 | 1 (100%) | - | - | 1 | 1 (100%) |
| Achromobacter xylososidans (n = 1) | 1 | - | - | - | - | - |
| Cupriavidus gilardii (n = 1) | - | - | - | - | 1 | - |
| Gene | Primers (5′-3′) | Size (Bp) | Cycling Conditions | References |
|---|---|---|---|---|
| ESBL | ||||
| CTX-M | FP: GAAGGTCATCAAGAAGGTGCG RP: GCATTGCCACGCTTTTCATAG | 560 | Initial denaturation at 95 °C for 5 min, followed by 30 cycles of denaturation at 95 °C for 30 s, primer annealing at 60 °C for 30 s, extension at 72 °C for 2 min and a final elongation temperature at 72 °C for 10 min. | [48] |
| OTHER BETA-LACTAMASE | ||||
| SHV | FP: GTCAGCGAAAAACACCTTGCC RP: GTCTTATCGGCGATAAACCAG | 383 | ||
| TEM | FP: GAGACAATAACCCTGGTAAAT RP: AGAAGTAAGTTGGCAGCAGTG | 420 | ||
| CARBAPENEMASE | ||||
| KPC | FP: ATGTCACTGTATCGCCGTC RP: AATCCCTCCGAGCGCGAG | 863 | Amplification was carried out at 94 °C for 3 min as the initial step for denaturation, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 61.6 °C for 30 s and extension at 72 °C for 1 min. Final elongation was at 72 °C for 7 min. | [49] |
| OXA-48 | FP: GCTTGATCGCCCTCGATT RP: GATTTGCTCCGTGGCCGAAA | 281 | ||
| IMP | FP: GGCAGTCGCCCTAAAACAAA RP: TAGTTACTTGGCTGTGATGG | 737 | ||
| VIM | FP: AAAGTTATGCCGCACTCACC RP: TGCAACTTCATGTTATGCCG | 865 | ||
| NDM | FP: GGTGCATGCCCGGTGAAATC RP: ATGCTGGCCTTGGGGAACG | 660 | ||
| AMPC | ||||
| MOXM | FR: GCTGCTCAAGGAGCACAGGAT RP: CACATTGACATAGGTGTGGTGC | 520 | Amplification was carried out at 94 °C for 15 min as the initial step for denaturation, 25 cycles of denaturation at 94 °C for 30 s, annealing at 64 °C for 90 s and extension at 72 °C for 60 s. Final elongation was at 72 °C for 10 min. | [47] |
| CITM | FR: TGGCCAGAACTGACAGGCAAA RP: TTT CTC CTG AAC GTG GCT GGC | 462 | ||
| DHAM | FR: AACTTTCACAGGTGTGCTGGGT RP: CCGTACGCATACTGGCTTTGC | 405 | ||
| FOXM | FP: AACATGGGGTATCAGGGAGATG RP: CAAAGCGCGTAACCGGATTGG | 190 | ||
| ACCM | FP: AACAGCCTCAGCAGCCGGTTA RP: TTCGCCGCAATCATCCCTAGC | 346 | ||
| EBCM | FR: TCGGTAAAGCCGATGTTGCGG RP: CTTCCACTGCGGCTGCCAGTT | 302 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Owusu, F.A.; Obeng-Nkrumah, N.; Gyinae, E.; Kodom, S.; Tagoe, R.; Tabi, B.K.A.; Dayie, N.T.K.D.; Opintan, J.A.; Egyir, B. Occurrence of Carbapenemases, Extended-Spectrum Beta-Lactamases and AmpCs among Beta-Lactamase-Producing Gram-Negative Bacteria from Clinical Sources in Accra, Ghana. Antibiotics 2023, 12, 1016. https://doi.org/10.3390/antibiotics12061016
Owusu FA, Obeng-Nkrumah N, Gyinae E, Kodom S, Tagoe R, Tabi BKA, Dayie NTKD, Opintan JA, Egyir B. Occurrence of Carbapenemases, Extended-Spectrum Beta-Lactamases and AmpCs among Beta-Lactamase-Producing Gram-Negative Bacteria from Clinical Sources in Accra, Ghana. Antibiotics. 2023; 12(6):1016. https://doi.org/10.3390/antibiotics12061016
Chicago/Turabian StyleOwusu, Felicia A., Noah Obeng-Nkrumah, Esther Gyinae, Sarkodie Kodom, Rhodalyn Tagoe, Blessing Kofi Adu Tabi, Nicholas T. K. D. Dayie, Japheth A. Opintan, and Beverly Egyir. 2023. "Occurrence of Carbapenemases, Extended-Spectrum Beta-Lactamases and AmpCs among Beta-Lactamase-Producing Gram-Negative Bacteria from Clinical Sources in Accra, Ghana" Antibiotics 12, no. 6: 1016. https://doi.org/10.3390/antibiotics12061016
APA StyleOwusu, F. A., Obeng-Nkrumah, N., Gyinae, E., Kodom, S., Tagoe, R., Tabi, B. K. A., Dayie, N. T. K. D., Opintan, J. A., & Egyir, B. (2023). Occurrence of Carbapenemases, Extended-Spectrum Beta-Lactamases and AmpCs among Beta-Lactamase-Producing Gram-Negative Bacteria from Clinical Sources in Accra, Ghana. Antibiotics, 12(6), 1016. https://doi.org/10.3390/antibiotics12061016






