Conditions Necessary for the Transfer of Antimicrobial Resistance in Poultry Litter
Abstract
1. Introduction
2. Results
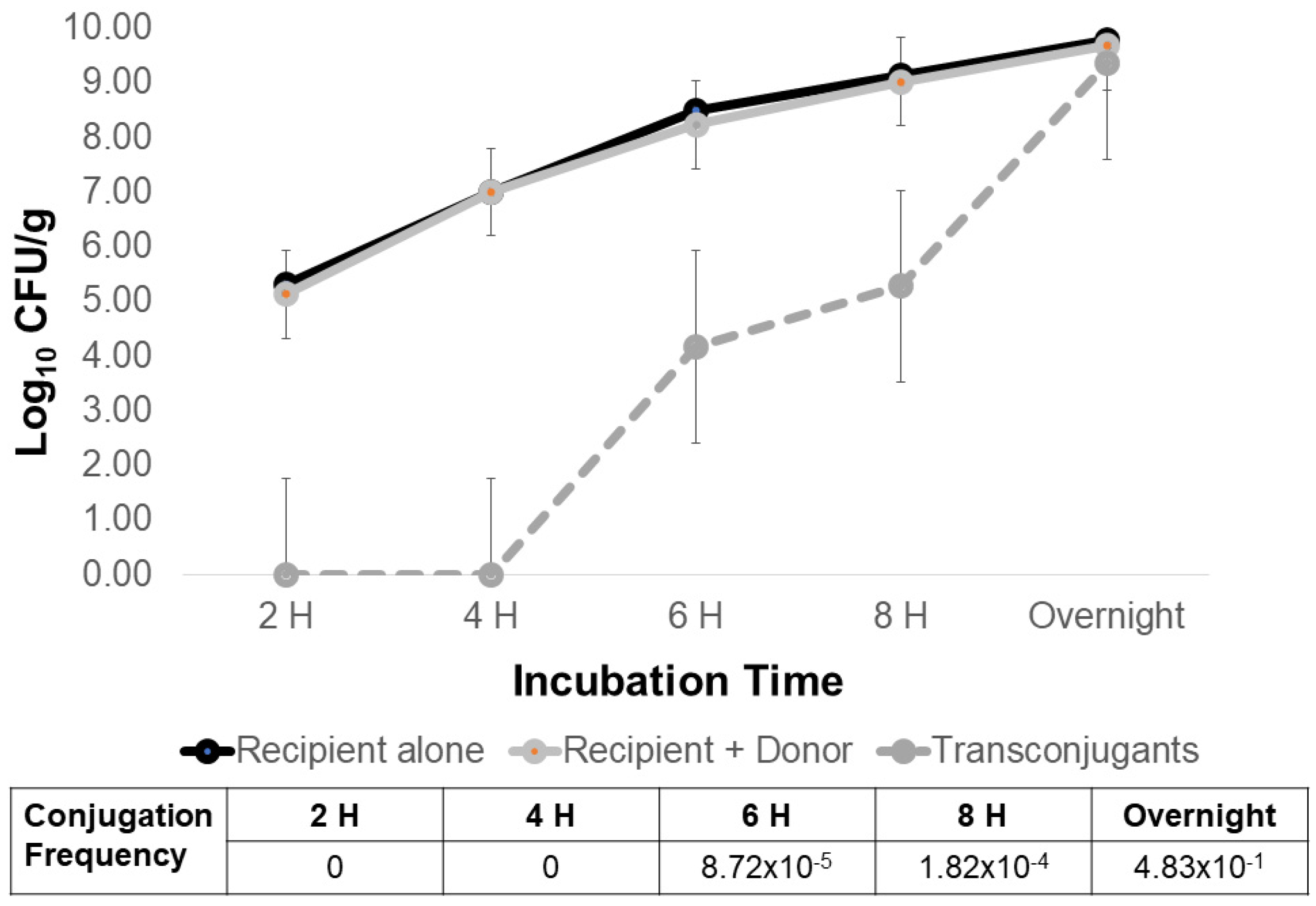
2.1. A Sizable Litter Resistome Did Not Result in Transfer of Antimicrobial Resistance from Poultry Litter to Salmonella
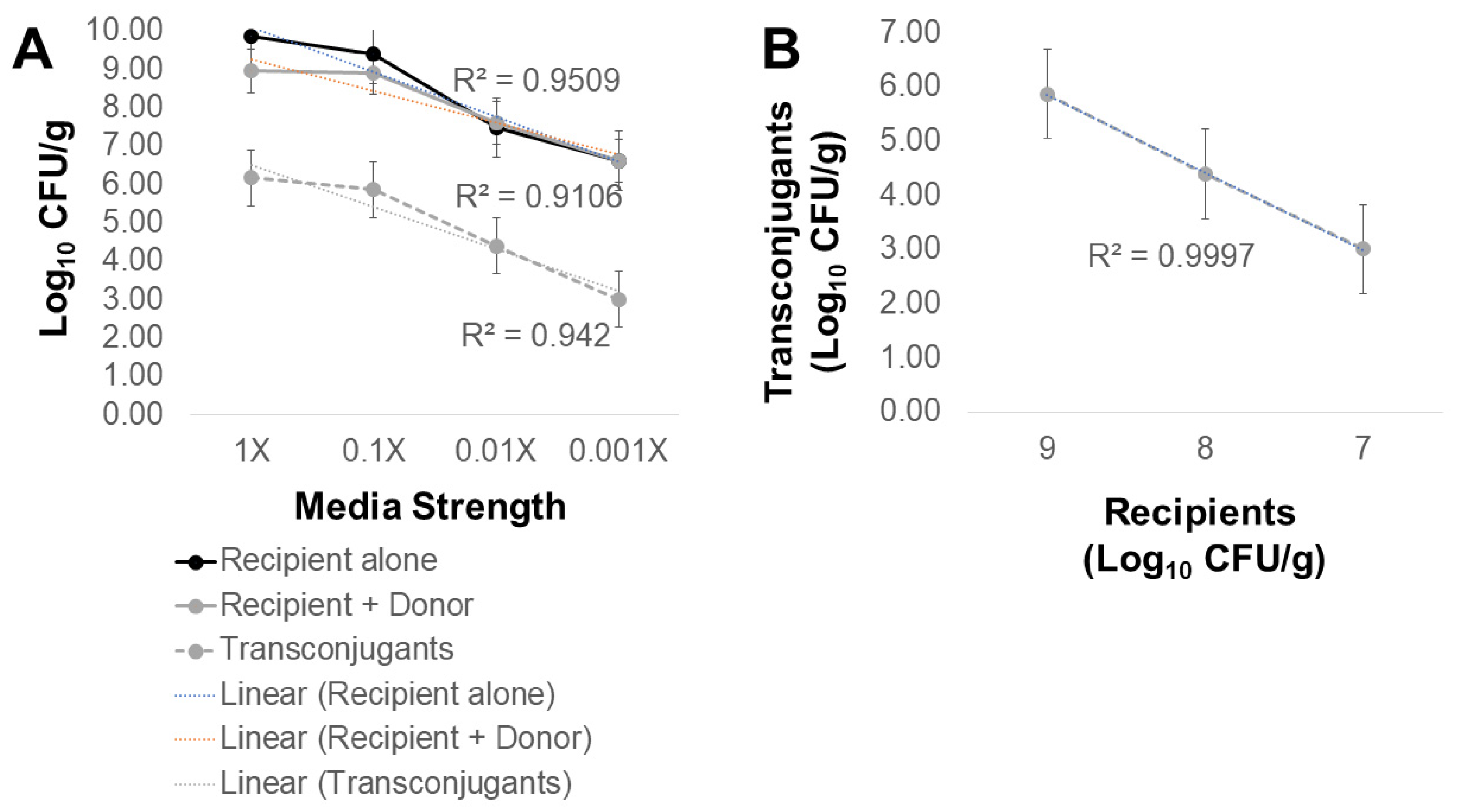
2.2. What Conditions Are Optimal for Transfer of AMR from Poultry Litter Resistome to Salmonella?
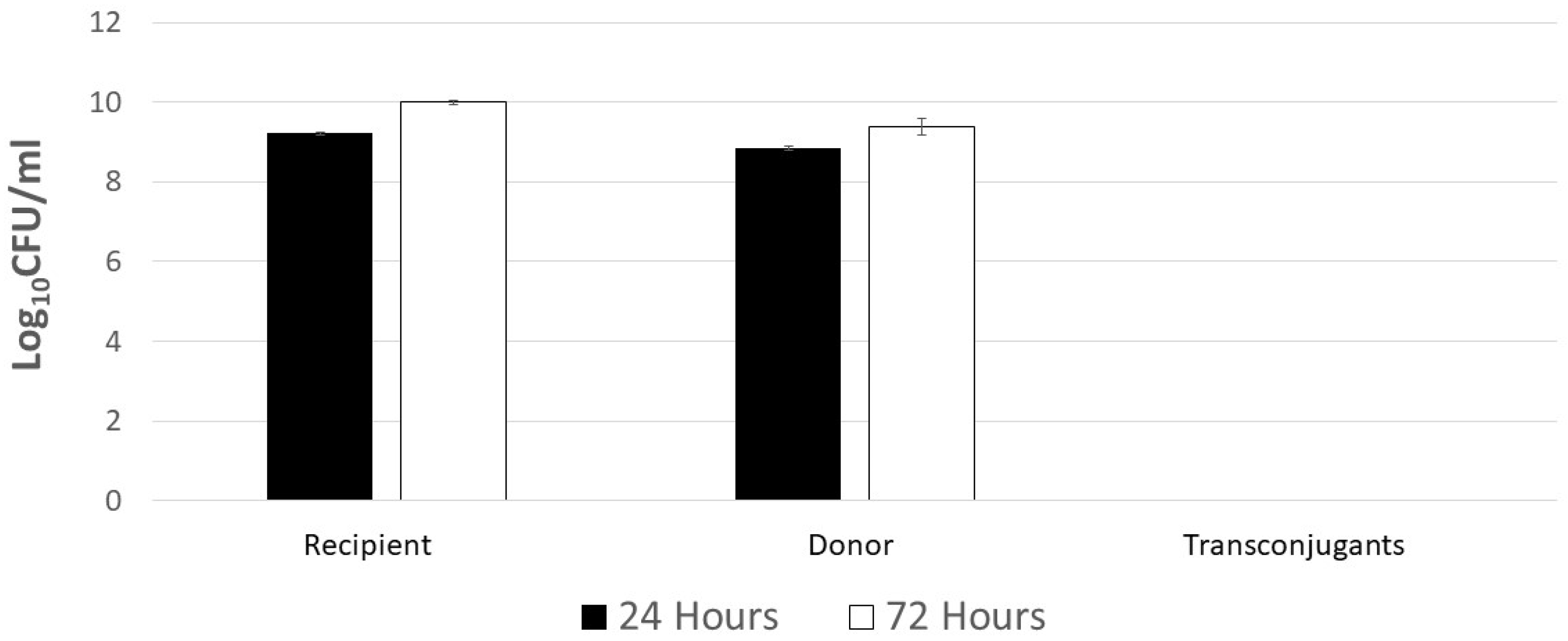
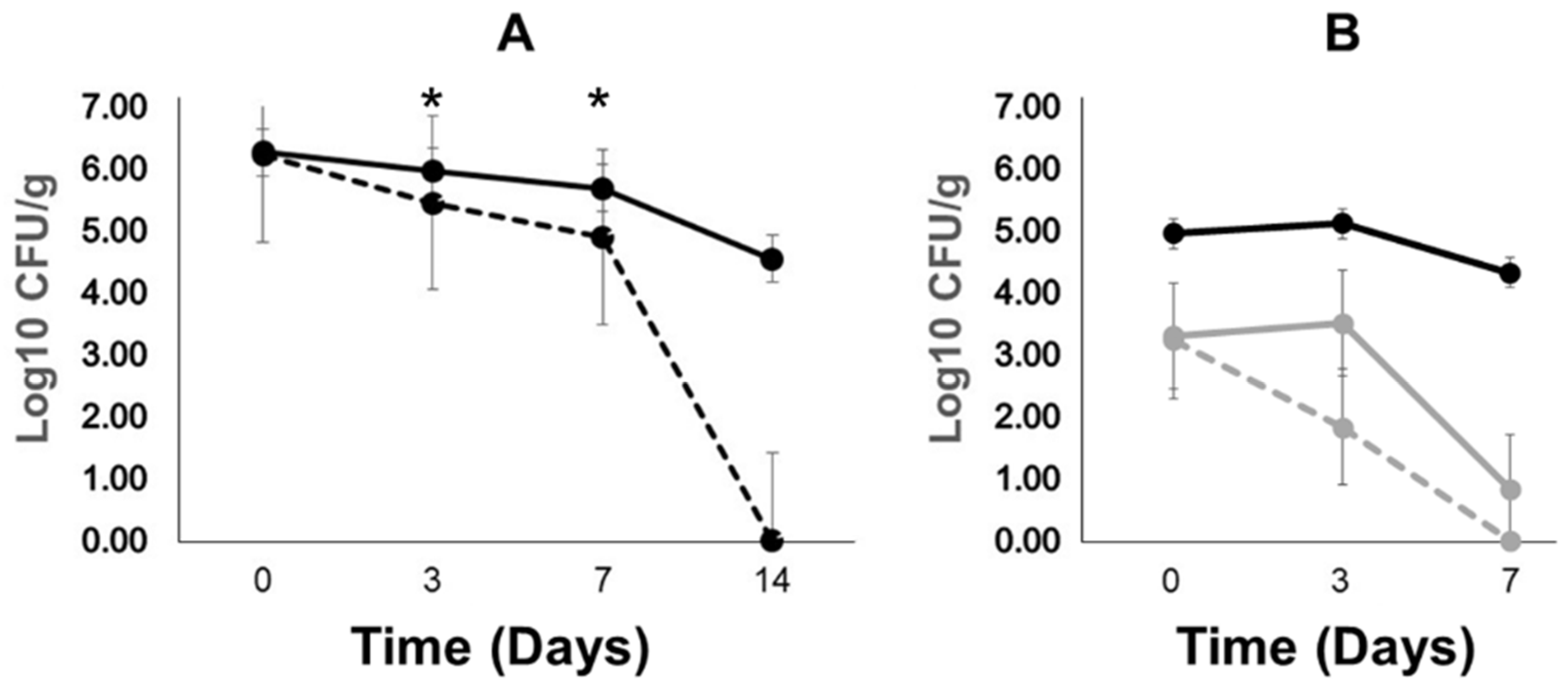
2.3. Poultry Litter Bacteria Are Not Inhibiting Plasmid Transfer between Escherichia coli and Salmonella
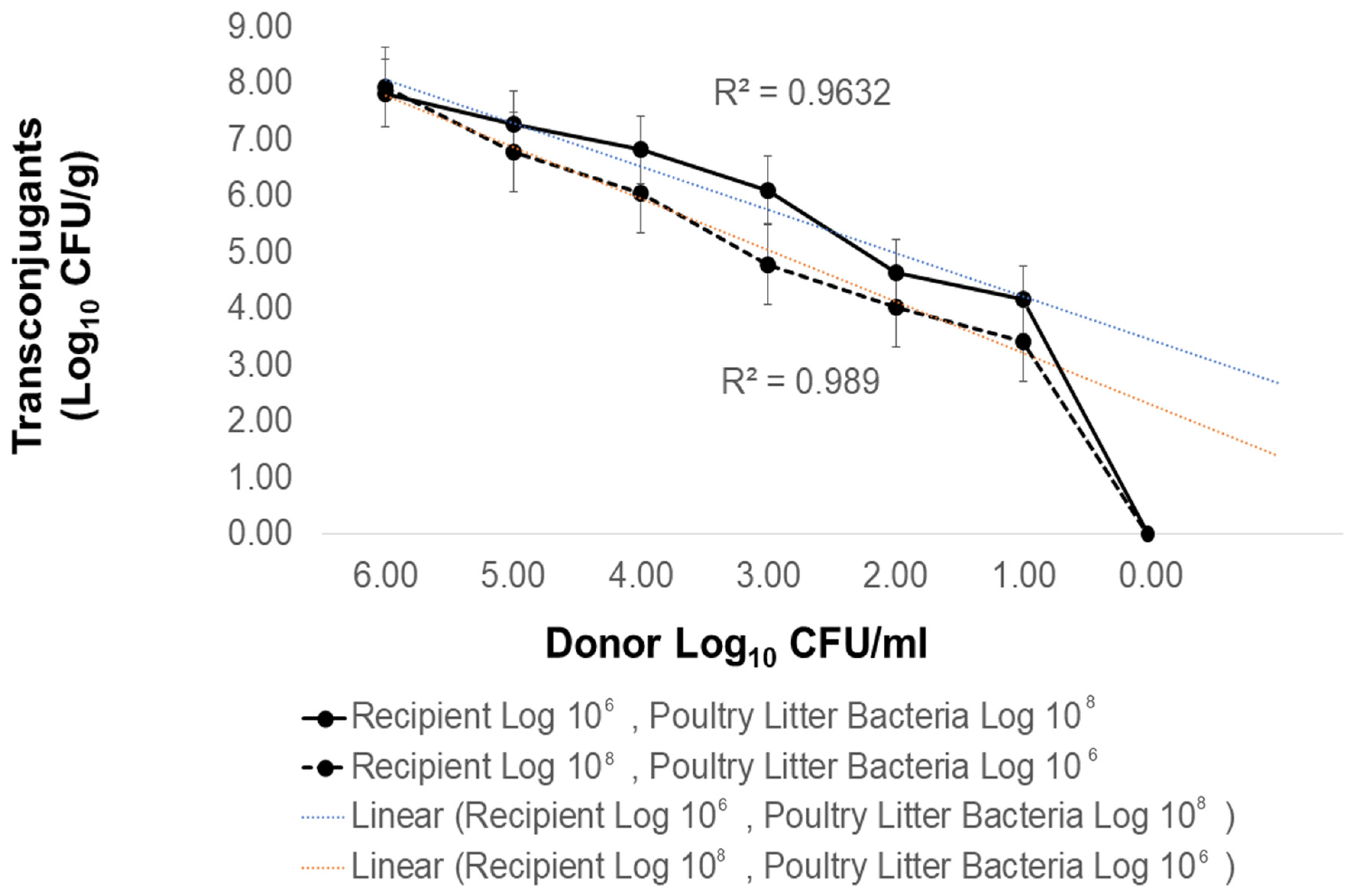
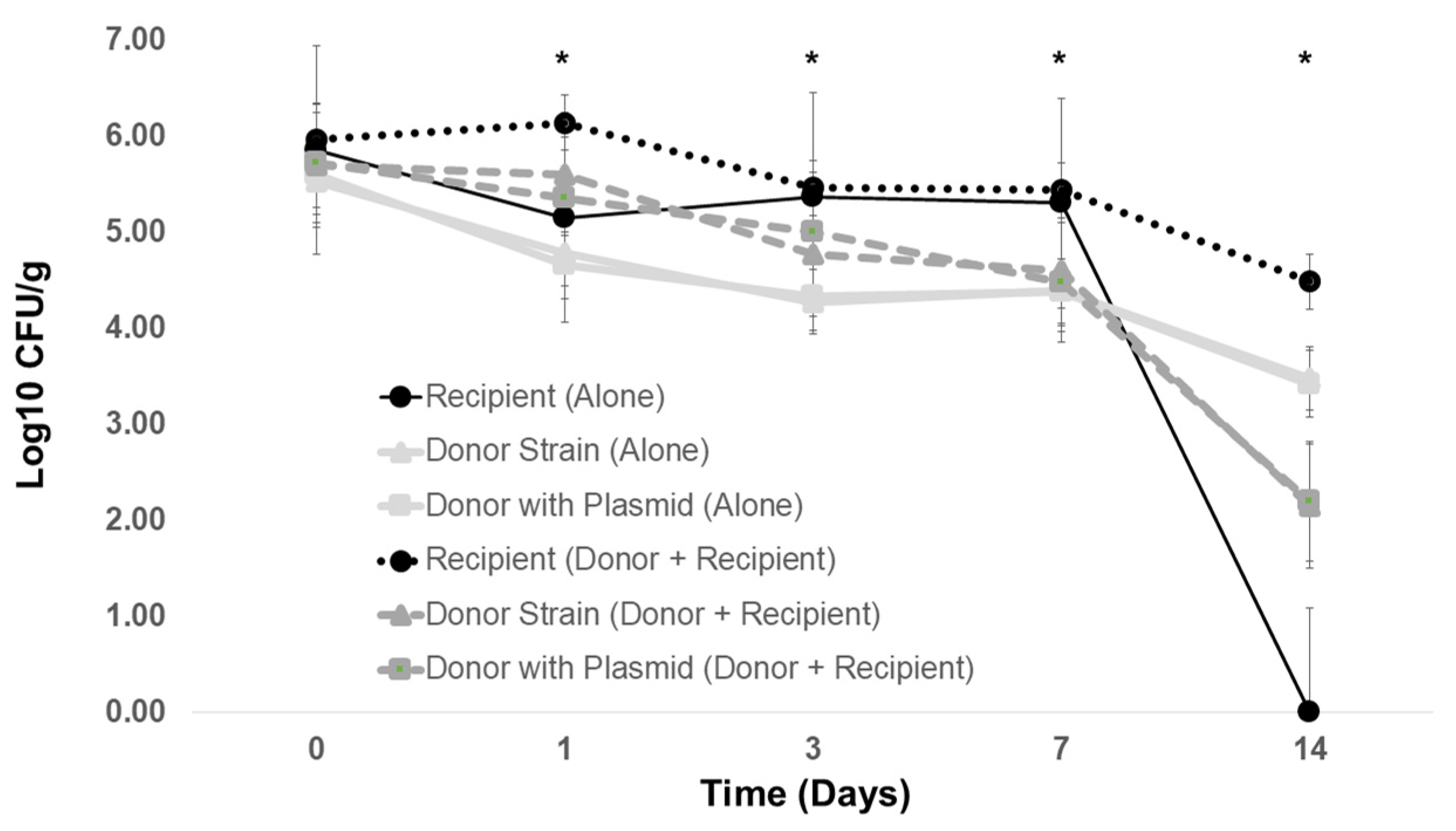
2.4. The Contribution of Donor Plasmid and Donor Strain Type to Antibiotic Resistance Transmission in Poultry Litter
3. Discussion
4. Materials and Methods
4.1. Extraction of Bacteria from Litter
4.2. Enumeration of Poultry Litter Bacteria: Total Aerobic Counts, Gram-Negative Enterics, and Antimicrobial-Resistant Bacterial Count
4.3. In Vitro Conjugation
4.4. DNA Extraction and qPCR
4.5. Poultry Litter Microcosm
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Henuk, Y.L.; Dingle, J.G. Poultry manure: Source of fertilizer, fuel and feed. World’s Poult. Sci. J. 2003, 59, 350–360. [Google Scholar] [CrossRef]
- Ashworth, A.J.; Chastain, J.P.; Moore, P.A. Nutrient Characteristics of Poultry Manure and Litter. In Animal Manure: Production, Characteristics, Environmental Concerns, and Management; Waldrip, H.M.P.H.P., He, Z., Eds.; Wiley Online Library: Hoboken, NJ, USA, 2020; pp. 63–87. [Google Scholar]
- Coleman, P. Guide for Organic Crop Producers. November 2012. Available online: https://www.ams.usda.gov/sites/default/files/media/GuideForOrganicCropProducers.pdf (accessed on 24 October 2022).
- King, G.M.J.P.; Brooks, S.; Brown, C.; Gerba, G.A.; O’Connor, I.L. Land Application of Organic Residuals: Public Health Threat or Environmental Benefit; American Society for Microbiology: Washington, DC, USA, 2011. [Google Scholar]
- Hutchison, M.L.; Walters, L.D.; Avery, S.M.; Munro, F.; Moore, A. Analyses of Livestock Production, Waste Storage, and Pathogen Levels and Prevalences in Farm Manures. Appl. Environ. Microbiol. 2005, 71, 1231–1236. [Google Scholar] [CrossRef] [PubMed]
- Pornsukarom, S.; Thakur, S. Horizontal Dissemination of Antimicrobial Resistance Determinants in Multiple Salmonella Serotypes following Isolation from the Commercial Swine Operation Environment after Manure Application. Appl. Environ. Microbiol. 2017, 83, e01503-17. [Google Scholar] [CrossRef]
- Bennett, S.D.; Sodha, S.V.; Ayers, T.L.; Lynch, M.F.; Gould, L.H.; Tauxe, R.V. Produce-associated foodborne disease outbreaks, USA, 1998–2013. Epidemiol. Infect. 2018, 146, 1397–1406. [Google Scholar] [CrossRef] [PubMed]
- Carstens, C.K.; Salazar, J.K.; Darkoh, C. Multistate Outbreaks of Foodborne Illness in the United States Associated With Fresh Produce From 2010 to 2017. Front. Microbiol. 2019, 10, 2667. [Google Scholar] [CrossRef] [PubMed]
- Sivapalasingam, S.; Friedman, C.R.; Cohen, L.; Tauxe, R.V. Fresh Produce: A Growing Cause of Outbreaks of Foodborne Illness in the United States, 1973 through 1997. J. Food Prot. 2004, 67, 2342–2353. [Google Scholar] [CrossRef]
- Luna, S.; Krishnasamy, V.; Saw, L.; Smith, L.; Wagner, J.; Weigand, J.; Tewell, M.; Kellis, M.; Penev, R.; McCullough, L.; et al. Outbreak of E. coli O157:H7 Infections Associated with Exposure to Animal Manure in a Rural Community—Arizona and Utah, June–July 2017. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 659–662. [Google Scholar] [CrossRef]
- Bottichio, L.; Keaton, A.; Thomas, D.; Fulton, T.; Tiffany, A.; Frick, A.; Mattioli, M.; Kahler, A.; Murphy, J.; Otto, M.; et al. Shiga Toxin-Producing Escherichia coli Infections Associated With Romaine Lettuce-United States, 2018. Clin. Infect. Dis. 2020, 71, e323–e330. [Google Scholar] [CrossRef]
- Söderström, A.; Österberg, P.; Lindqvist, A.; Jönsson, B.; Lindberg, A.; Ulander, S.B.; Welinder-Olsson, C.; Löfdahl, S.; Kaijser, B.; De Jong, B.; et al. A Large Escherichia coli O157 Outbreak in Sweden Associated with Locally Produced Lettuce. Foodborne Pathog. Dis. 2008, 5, 339–349. [Google Scholar] [CrossRef]
- Zalewska, M.; Błażejewska, A.; Czapko, A.; Popowska, M. Antibiotics and Antibiotic Resistance Genes in Animal Manure—Consequences of Its Application in Agriculture. Front. Microbiol. 2021, 12, 640. [Google Scholar] [CrossRef]
- Lu, J.; Sanchez, S.; Hofacre, C.; Maurer, J.J.; Harmon, B.G.; Lee, M.D. Evaluation of Broiler Litter with Reference to the Microbial Composition as Assessed by Using 16S rRNA and Functional Gene Markers. Appl. Environ. Microbiol. 2003, 69, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Mak, P.H.W.; Rehman, M.A.; Kiarie, E.G.; Topp, E.; Diarra, M.S. Production systems and important antimicrobial resistant-pathogenic bacteria in poultry: A review. J. Anim. Sci. Biotechnol. 2022, 13, 148. [Google Scholar] [CrossRef] [PubMed]
- Bass, L.; Liebert, C.A.; Lee, M.D.; Summers, A.O.; White, D.G.; Thayer, S.G.; Maurer, J.J. Incidence and Characterization of Integrons, Genetic Elements Mediating Multiple-Drug Resistance, in Avian Escherichia coli. Antimicrob. Agents Chemother. 1999, 43, 2925–2929. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, C.; Lee, M.D.; Sanchez, S.; Hudson, C.; Phillips, B.; Register, B.; Grady, M.; Liebert, C.; Summers, A.; White, D.G.; et al. Incidence of Class 1 and 2 Integrases in Clinical and Commensal Bacteria from Livestock, Companion Animals, and Exotics. Antimicrob. Agents Chemother. 2001, 45, 723–726. [Google Scholar] [CrossRef]
- Nandi, S.; Maurer, J.J.; Hofacre, C.; Summers, A.O. Gram-positive bacteria are a major reservoir of Class 1 antibiotic resistance integrons in poultry litter. Proc. Natl. Acad. Sci. USA 2004, 101, 7118–7122. [Google Scholar] [CrossRef]
- He, L.-Y.; Liu, Y.-S.; Su, H.-C.; Zhao, J.-L.; Liu, S.-S.; Chen, J.; Liu, W.-R.; Ying, G.-G. Dissemination of Antibiotic Resistance Genes in Representative Broiler Feedlots Environments: Identification of Indicator ARGs and Correlations with Environmental Variables. Environ. Sci. Technol. 2014, 48, 13120–13129. [Google Scholar] [CrossRef]
- Anonymous. Antibiotic Resistance Threats in the United States 2019. Center for Disease Control and Prevention. 2019. Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed on 23 November 2021).
- Scallan, E.; Mahon, B.E.; Hoekstra, R.M.; Griffin, P.M. Estimates of Illnesses, Hospitalizations and Deaths Caused by Major Bacterial Enteric Pathogens in Young Children in the United States. Pediatr. Infect. Dis. J. 2013, 32, 217–221. [Google Scholar] [CrossRef]
- Scallan, E.; Crim, S.M.; Runkle, A.; Henao, O.L.; Mahon, B.E.; Hoekstra, R.M.; Griffin, P.M. Bacterial Enteric Infections Among Older Adults in the United States: Foodborne Diseases Active Surveillance Network, 1996–2012. Foodborne Pathog. Dis. 2015, 12, 492–499. [Google Scholar] [CrossRef]
- Scallan, E.; Griffin, P.M.; McLean, H.Q.; Mahon, B.E. Hospitalisations due to bacterial gastroenteritis: A comparison of surveillance and hospital discharge data. Epidemiol. Infect. 2018, 146, 954–960. [Google Scholar] [CrossRef]
- Collard, J.-M.; Place, S.; Denis, O.; Rodriguez-Villalobos, H.; Vrints, M.; Weill, F.-X.; Baucheron, S.; Cloeckaert, A.; Struelens, M.; Bertrand, S. Travel-acquired salmonellosis due to Salmonella Kentucky resistant to ciprofloxacin, ceftriaxone and co-trimoxazole and associated with treatment failure. J. Antimicrob. Chemother. 2007, 60, 190–192. [Google Scholar] [CrossRef]
- Tribble, D.R. Resistant pathogens as causes of traveller’s diarrhea globally and impact(s) on treatment failure and recommendations. J. Travel Med. 2017, 24, S6–S12. [Google Scholar] [CrossRef]
- Duong, V.T.; Tuyen, H.T.; Van Minh, P.; Campbell, J.I.; Le Phuc, H.; Nhu, T.D.H.; Tu, L.T.P.; Chau, T.T.H.; Nhi, L.T.Q.; Hung, N.T.; et al. No Clinical Benefit of Empirical Antimicrobial Therapy for Pediatric Diarrhea in a High-Usage, High-Resistance Setting. Clin. Infect. Dis. 2017, 66, 504–511. [Google Scholar] [CrossRef]
- McCollister, B.; Kotter, C.V.; Frank, D.N.; Washburn, T.; Jobling, M.G. Whole-Genome Sequencing Identifies In Vivo Acquisition of a blaCTX-M-27-Carrying IncFII Transmissible Plasmid as the Cause of Ceftriaxone Treatment Failure for an Invasive Salmonella enterica Serovar Typhimurium Infection. Antimicrob. Agents Chemother. 2016, 60, 7224–7235. [Google Scholar] [CrossRef]
- Park, H.-R.; Kim, D.-M.; Yun, N.-R.; Kim, C.-M. Identifying the mechanism underlying treatment failure for Salmonella Paratyphi A infection using next-generation sequencing—A case report. BMC Infect. Dis. 2019, 19, 191. [Google Scholar] [CrossRef]
- Piddock, L. Quinolone resistance and Campylobacter spp. J. Antimicrob. Chemother. 1995, 36, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Gibreel, A.; Taylor, D.E. Macrolide resistance in Campylobacter jejuni and Campylobacter coli. J. Antimicrob. Chemother. 2006, 58, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Chan, E.W.C.; Chen, S. Evolution and transmission of a conjugative plasmid encoding both ciprofloxacin and ceftriaxone resistance in Salmonella. Emerg. Microbes Infect. 2019, 8, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lynne, A.M.; David, D.E.; Tang, H.; Xu, J.; Nayak, R.; Kaldhone, P.; Logue, C.M.; Foley, S.L. DNA Sequence Analysis of Plasmids from Multidrug Resistant Salmonella enterica Serotype Heidelberg Isolates. PLoS ONE 2012, 7, e51160. [Google Scholar] [CrossRef]
- Na, S.H.; Moon, D.C.; Kang, H.Y.; Song, H.J.; Kim, S.J.; Choi, J.H.; Yoon, J.W.; Yoon, S.S.; Lim, S.K. Molecular characteristics of extended-spectrum beta-lactamase/AmpC-producing Salmonella enterica serovar Virchow isolated from food-producing animals during 2010–2017 in South Korea. Int. J. Food Microbiol. 2020, 322, 108572. [Google Scholar] [CrossRef]
- Bogomazova, A.N.; Gordeeva, V.D.; Krylova, E.V.; Soltynskaya, I.V.; Davydova, E.E.; Ivanova, O.E.; Komarov, A.A. Mega-plasmid found worldwide confers multiple antimicrobial resistance in Salmonella Infantis of broiler origin in Russia. Int. J. Food Microbiol. 2020, 319, 108497. [Google Scholar] [CrossRef]
- Jeon, H.Y.; Bin Kim, Y.; Lim, S.-K.; Lee, Y.J.; Seo, K.W. Characteristics of cephalosporin-resistant Salmonella isolates from poultry in Korea, 2010–2017. Poult. Sci. 2019, 98, 957–965. [Google Scholar] [CrossRef] [PubMed]
- Camarda, A.; Pugliese, N.; Pupillo, A.; Oliva, M.; Circella, E.; Dionisi, A.M.; Ricci, A.; Legretto, M.; Caroli, A.; Pazzani, C. Resistance genes, phage types and pulsed field gel electrophoresis pulsotypes in Salmonella enterica strains from laying hen farms in southern Italy. Int. J. Environ. Res. Public Health 2013, 10, 3347–3362. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and Barriers to, Horizontal Gene Transfer between Bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef]
- Fricke, W.F.; McDermott, P.F.; Mammel, M.K.; Zhao, S.; Johnson, T.J.; Rasko, D.A.; Fedorka-Cray, P.J.; Pedroso, A.; Whichard, J.M.; LeClerc, J.E.; et al. Antimicrobial Resistance-Conferring Plasmids with Similarity to Virulence Plasmids from Avian Pathogenic Escherichia coli Strains in Salmonella enterica Serovar Kentucky Isolates from Poultry. Appl. Environ. Microbiol. 2009, 75, 5963–5971. [Google Scholar] [CrossRef]
- Vingopoulou, E.I.; Siarkou, V.I.; Batzias, G.; Kaltsogianni, F.; Sianou, E.; Tzavaras, I.; Koutinas, A.; Saridomichelakis, M.N.; Sofianou, D.; Tzelepi, E.; et al. Emergence and maintenance of multidrug-resistant Escherichia coli of canine origin harbouring a blaCMY-2-IncI1/ST65 plasmid and topoisomerase mutations. J. Antimicrob. Chemother. 2014, 69, 2076–2080. [Google Scholar] [CrossRef] [PubMed]
- Cox, K.E.; Schildbach, J.F. Sequence of the R1 plasmid and comparison to F and R100. Plasmid 2017, 91, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Liljebjelke, K.A.; Hofacre, C.L.; White, D.G.; Ayers, S.; Lee, M.D.; Maurer, J.J. Diversity of Antimicrobial Resistance Phenotypes in Salmonella Isolated from Commercial Poultry Farms. Front. Vet. Sci. 2017, 4, 96. [Google Scholar] [CrossRef]
- Bythwood, T.N.; Soni, V.; Lyons, K.; Hurley-Bacon, A.; Lee, M.D.; Hofacre, C.; Sanchez, S.; Maurer, J.J. Antimicrobial Resistant Salmonella enterica Typhimurium Colonizing Chickens: The Impact of Plasmids, Genotype, Bacterial Communities, and Antibiotic Administration on Resistance. Front. Sustain. Food Syst. 2019, 3, 20. [Google Scholar] [CrossRef]
- Větrovský, T.; Baldrian, P. The Variability of the 16S rRNA Gene in Bacterial Genomes and Its Consequences for Bacterial Community Analyses. PLoS ONE 2013, 8, e57923. [Google Scholar] [CrossRef]
- Sysoeva, T.A.; Kim, Y.; Rodriguez, J.; Lopatkin, A.J.; You, L. Growth-stage-dependent regulation of conjugation. AIChE J. 2020, 66, e16848. [Google Scholar] [CrossRef]
- Shafieifini, M.; Sun, Y.; Staley, Z.R.; Riethoven, J.-J.; Li, X. Effects of Nutrient Level and Growth Rate on the Conjugation Process That Transfers Mobile Antibiotic Resistance Genes in Continuous Cultures. Appl. Environ. Microbiol. 2022, 88, e01121-22. [Google Scholar] [CrossRef] [PubMed]
- Molin, S.; Tolker-Nielsen, T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 2003, 14, 255–261. [Google Scholar] [CrossRef]
- Berghaus, R.D.; Thayer, S.G.; Law, B.F.; Mild, R.M.; Hofacre, C.L.; Singer, R.S. Enumeration of Salmonella and Campylobacter spp. in Environmental Farm Samples and Processing Plant Carcass Rinses from Commercial Broiler Chicken Flocks. Appl. Environ. Microbiol. 2013, 79, 4106–4114. [Google Scholar] [CrossRef] [PubMed]
- Deaton, J.W.; Branton, S.L.; Simmons, J.D.; Lott, B.D. The Effect of Brooding Temperature on Broiler Performance. Poult. Sci. 1996, 75, 1217–1220. [Google Scholar] [CrossRef]
- Ghigo, J.-M. Natural conjugative plasmids induce bacterial biofilm development. Nature 2001, 412, 442–445. [Google Scholar] [CrossRef]
- Llosa, M.; Bolland, S.; De La Cruz, F. Structural and functional analysis of the origin of conjugal transfer of the broad-host-range IneW plasmid R388 and comparison with the related IncN plasmid R46. Mol. Genet. Genom. 1991, 226, 473–483. [Google Scholar] [CrossRef]
- Maurer, J.J.; Lee, M.D.; Lobsinger, C.; Brown, T.; Maier, M.; Thayer, S.G. Molecular typing of avian Escherichia coli isolates by random amplification of polymorphic DNA. Avian Dis. 1998, 42, 431. [Google Scholar] [CrossRef] [PubMed]
- Wooley, R.; Dickerson, H.; Simmons, K.; Shotts, E.; Brown, J. Effect of EDTA-TRIS on an Escherichia coli isolate containing R plasmids. Vet. Microbiol. 1986, 12, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Błażejewska, A.; Zalewska, M.; Grudniak, A.; Popowska, M. A Comprehensive Study of the Microbiome, Resistome, and Physical and Chemical Characteristics of Chicken Waste from Intensive Farms. Biomolecules 2022, 12, 1132. [Google Scholar] [CrossRef]
- Gupta, C.L.; Blum, S.E.; Kattusamy, K.; Daniel, T.; Druyan, S.; Shapira, R.; Krifucks, O.; Zhu, Y.-G.; Zhou, X.-Y.; Su, J.-Q. Longitudinal study on the effects of growth-promoting and therapeutic antibiotics on the dynamics of chicken cloacal and litter microbiomes and resistomes. Microbiome 2021, 9, 178. [Google Scholar] [CrossRef]
- Mazhar, S.H.; Li, X.; Rashid, A.; Su, J.; Xu, J.; Brejnrod, A.D.; Su, J.-Q.; Wu, Y.; Zhu, Y.-G.; Zhou, S.G.; et al. Co-selection of antibiotic resistance genes, and mobile genetic elements in the presence of heavy metals in poultry farm environments. Sci. Total Environ. 2021, 755, 142702. [Google Scholar] [CrossRef]
- Forgetta, V.; Rempel, H.; Malouin, F.; Vaillancourt, R.; Topp, E.; Dewar, K.; Diarra, M. Pathogenic and multidrug-resistant Escherichia fergusonii from broiler chicken. Poult. Sci. 2012, 91, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Mataseje, L.F.; Baudry, P.J.; Zhanel, G.G.; Morck, D.W.; Read, R.R.; Louie, M.; Mulvey, M.R. Comparison of CMY-2 plasmids isolated from human, animal, and environmental Escherichia coli and Salmonella spp. from Canada. Diagn. Microbiol. Infect. Dis. 2010, 67, 387–391. [Google Scholar] [CrossRef]
- Ahmer, B.M.M.; Tran, M.; Heffron, F. The Virulence Plasmid of Salmonella typhimurium Is Self-Transmissible. J. Bacteriol. 1999, 181, 1364–1368. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Astorga, A.; Muela, A.; Cisterna, R.; Iriberri, J.; Barcina, I. Biotic and abiotic factors affecting plasmid transfer in Escherichia coli strains. Appl. Environ. Microbiol. 1992, 58, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Cook, L.C.C.; Shu, C.-C.; Chen, Y.; Manias, D.A.; Ramkrishna, D.; Dunny, G.M.; Hu, W.-S. Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer. Proc. Natl. Acad. Sci. USA 2013, 110, 7086–7090. [Google Scholar] [CrossRef]
- Zheng, J.; Chen, T.; Chen, H. Antibiotic resistome promotion in drinking water during biological activated carbon treatment: Is it influenced by quorum sensing? Sci. Total Environ. 2018, 612, 1–8. [Google Scholar] [CrossRef]
- Hegde, M.; Englert, D.L.; Schrock, S.; Cohn, W.B.; Vogt, C.; Wood, T.K.; Manson, M.D.; Jayaraman, A. Chemotaxis to the Quorum-Sensing Signal AI-2 Requires the Tsr Chemoreceptor and the Periplasmic LsrB AI-2-Binding Protein. J. Bacteriol. 2011, 193, 768–773. [Google Scholar] [CrossRef]
- Glessner, A.; Smith, R.S.; Iglewski, B.H.; Robinson, J.B. Roles of Pseudomonas aeruginosa las and rhl Quorum-Sensing Systems in Control of Twitching Motility. J. Bacteriol. 1999, 181, 1623–1629. [Google Scholar] [CrossRef]
- Daniels, R.; Vanderleyden, J.; Michiels, J. Quorum sensing and swarming migration in bacteria. FEMS Microbiol. Rev. 2004, 28, 261–289. [Google Scholar] [CrossRef]
- van Gestel, J.; Bareia, T.; Tenennbaum, B.; Dal Co, A.; Guler, P.; Aframian, N.; Puyesky, S.; Grinberg, I.; D’Souza, G.G.; Erez, Z.; et al. Short-Range quorum sensing controls horizontal gene transfer at micron scale in bacterial communities. Nat. Commun. 2021, 12, 2324. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.G. In vivo transfer of R factors between Escherichia coli strains inoculated into the rumen of sheep. J. Hyg. 1975, 75, 363–370. [Google Scholar] [CrossRef]
- Oladeinde, A.; Abdo, Z.; Press, M.O.; Cook, K.; Cox, N.A.; Zwirzitz, B.; Woyda, R.; Lakin, S.M.; Thomas, I.V.J.C.; Looft, T. Horizontal gene transfer is the main driver of antimicrobial resistance in broiler chicks infected with Salmonella enterica serovar Heidelberg. Msystems 2021, 6, e00729-21. [Google Scholar] [CrossRef]
- Neil, K.; Allard, N.; Grenier, F.; Burrus, V.; Rodrigue, S. Highly efficient gene transfer in the mouse gut microbiota is enabled by the Incl2 conjugative plasmid TP114. Commun. Biol. 2020, 3, 523. [Google Scholar] [CrossRef] [PubMed]
- Ronda, C.; Chen, S.P.; Cabral, V.; Yaung, S.J.; Wang, H.H. Metagenomic engineering of the mammalian gut microbiome in situ. Nat. Methods 2019, 16, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Ott, L.C.; Stromberg, Z.R.; Redweik, G.A.J.; Wannemuehler, M.J.; Mellata, M. Mouse Genetic Background Affects Transfer of an Antibiotic Resistance Plasmid in the Gastrointestinal Tract. Msphere 2020, 5, e00847-19. [Google Scholar] [CrossRef]
- Stecher, B.; Denzler, R.; Maier, L.; Bernet, F.; Sanders, M.J.; Pickard, D.J.; Barthel, M.; Westendorf, A.M.; Krogfelt, K.A.; Walker, A.W.; et al. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc. Natl. Acad. Sci. USA 2012, 109, 1269–1274. [Google Scholar] [CrossRef]
- Mizan, S.; Lee, M.D.; Harmon, B.G.; Tkalcic, S.; Maurer, J.J. Acquisition of Antibiotic Resistance Plasmids by Enterohemorrhagic Escherichia coli O157:H7 within Rumen Fluid. J. Food Prot. 2002, 65, 1038–1040. [Google Scholar] [CrossRef]
- Oladeinde, A.; Abdo, Z.; Zwirzitz, B.; Woyda, R.; Lakin, S.M.; Press, M.O.; Cox, N.A.; Thomas, I.V.J.C.; Looft, T.; Rothrock, M.J., Jr. Litter commensal bacteria can limit the horizontal gene transfer of antimicrobial resistance to Salmonella in chickens. Appl. Environ. Microbiol. 2022, 88, e02517-21. [Google Scholar] [CrossRef]
- Dimitriu, T.; Marchant, L.; Buckling, A.; Raymond, B. Bacteria from natural populations transfer plasmids mostly towards their kin. Proc. R. Soc. B Boil. Sci. 2019, 286, 20191110. [Google Scholar] [CrossRef]
- Mølbak, L.; Licht, T.R.; Kvist, T.; Kroer, N.; Andersen, S.R. Plasmid transfer from Pseudomonas putida to the indigenous bacteria on alfalfa sprouts: Characterization, direct quantification, and in situ location of transconjugant cells. Appl. Environ. Microbiol. 2003, 69, 5536–5542. [Google Scholar] [CrossRef]
- Klümper, U.; Riber, L.; Dechesne, A.; Sannazzarro, A.; Hansen, L.H.; Sørensen, S.J.; Smets, B.F. Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME J. 2014, 9, 934–945. [Google Scholar] [CrossRef]
- Getino, M.; de la Cruz, F. Natural and Artificial Strategies to Control the Conjugative Transmission of Plasmids. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Oluwadare, M.; Lee, M.D.; Grim, C.J.; Lipp, E.K.; Cheng, Y.; Maurer, J.J. The Role of the Salmonella spvB IncF Plasmid and Its Resident Entry Exclusion Gene traS on Plasmid Exclusion. Front. Microbiol. 2020, 11, 949. [Google Scholar] [CrossRef]
- Loftie-Eaton, W.; Bashford, K.; Quinn, H.; Dong, K.; Millstein, J.; Hunter, S.; Thomason, M.K.; Merrikh, H.; Ponciano, J.M.; Top, E.M. Compensatory mutations improve general permissiveness to antibiotic resistance plasmids. Nat. Ecol. Evol. 2017, 1, 1354–1363. [Google Scholar] [CrossRef]
- Heß, S.; Kneis, D.; Virta, M.; Hiltunen, T. The spread of the plasmid RP4 in a synthetic bacterial community is dependent on the particular donor strain. FEMS Microbiol. Ecol. 2021, 97, fiab147. [Google Scholar] [CrossRef] [PubMed]
- Kottara, A.; Hall, J.P.J.; Brockhurst, M. The proficiency of the original host species determines community-level plasmid dynamics. FEMS Microbiol. Ecol. 2021, 97, fiab026. [Google Scholar] [CrossRef] [PubMed]
- Virolle, C.; Goldlust, K.; Djermoun, S.; Bigot, S.; Lesterlin, C. Plasmid Transfer by Conjugation in Gram-Negative Bacteria: From the Cellular to the Community Level. Genes 2020, 11, 1239. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, M.M.S.; Silva, N.M.V.; Ferreira, V.A.; Filho, A.L.B.M.; Givisiez, P.E.N.; Neto, O.C.F.; Júnior, A.B.; Gebreyes, W.A.; de Oliveira, C.J.B. Residual concentrations of antimicrobial growth promoters in poultry litter favour plasmid conjugation among Escherichia coli. Lett. Appl. Microbiol. 2022, 74, 831–838. [Google Scholar] [CrossRef]
- Saraiva, M.M.; Filho, A.L.M.; Vasconcelos, P.C.; Nascimento, P.V.; Azevedo, P.S.; Neto, O.C.F.; Givisiez, P.E.; Gebreyes, W.A.; Oliveira, C.J. Chemical treatment of poultry litter affects the conjugation of plasmid-mediated extended-spectrum beta-lactamase resistance genes in E. coli. J. Appl. Poult. Res. 2019, 29, 197–203. [Google Scholar] [CrossRef]
- Guan, J.; Wasty, A.; Grenier, C.; Chan, M. Influence of Temperature on Survival and Conjugative Transfer of Multiple Antibiotic-Resistant Plasmids in Chicken Manure and Compost Microcosms. Poult. Sci. 2007, 86, 610–613. [Google Scholar] [CrossRef]
- Tecon, R.; Ebrahimi, A.; Kleyer, H.; Levi, S.E.; Or, D. Cell-to-cell bacterial interactions promoted by drier conditions on soil surfaces. Proc. Natl. Acad. Sci. USA 2018, 115, 9791–9796. [Google Scholar] [CrossRef]
- Lu, J.; Idris, U.; Harmon, B.; Hofacre, C.; Maurer, J.J.; Lee, M.D. Diversity and Succession of the Intestinal Bacterial Community of the Maturing Broiler Chicken. Appl. Environ. Microbiol. 2003, 69, 6816–6824. [Google Scholar] [CrossRef] [PubMed]
- Provence, D.L.; Curtiss, R., III. Gene Transfer in Gram-Negative Bacteria; ASM Press: Washington, DC, USA, 1994. [Google Scholar]
- Anthony, K.G.; Klimke, W.A.; Manchak, J.; Frost, L.S. Comparison of Proteins Involved in Pilus Synthesis and Mating Pair Stabilization from the Related Plasmids F and R100-1: Insights into the Mechanism of Conjugation. J. Bacteriol. 1999, 181, 5149–5159. [Google Scholar] [CrossRef] [PubMed]
- Kehoe, M.A.; Foster, T.J. Genetic analysis of mutations in the transfer genes of pDU202 tra::Tn10 plasmids, caused by the excision of Tn10. Mol. Genet. Genom. 1977, 157, 109–118. [Google Scholar] [CrossRef]
- Suzuki, M.T.; Taylor, L.T.; DeLong, E.F. Quantitative Analysis of Small-Subunit rRNA Genes in Mixed Microbial Populations via 5′-Nuclease Assays. Appl. Environ. Microbiol. 2000, 66, 4679–4687. [Google Scholar] [CrossRef]
- Yeh, Y.-C.; Chang, K.-C.; Yang, J.-C.; Fang, C.-T.; Wang, J.-T. Association of Metronidazole Resistance and Natural Competence in Helicobacter pylori. Antimicrob. Agents Chemother. 2002, 46, 1564–1567. [Google Scholar] [CrossRef] [PubMed]
- Stanczak-Mrozek, K.I.; Manne, A.; Knight, G.M.; Gould, K.; Witney, A.A.; Lindsay, J.A. Within-host diversity of MRSA antimicrobial resistances. J. Antimicrob. Chemother. 2015, 70, 2191–2198. [Google Scholar] [CrossRef]
- Simjee, S.; McDermott, P.F.; White, D.G.; Hofacre, C.; Berghaus, R.D.; Carter, P.J.; Stewart, L.; Liu, T.; Maier, M.; Maurer, J.J. Antimicrobial Susceptibility and Distribution of Antimicrobial-Resistance Genes Among Enterococcus and Coagulase-Negative Staphylococcus Isolates Recovered from Poultry Litter. Avian Dis. 2007, 51, 884–892. [Google Scholar] [CrossRef]
- Chee-Sanford, J.C.; Aminov, R.I.; Krapac, I.J.; Garrigues-Jeanjean, N.; Mackie, R.I. Occurrence and Diversity of Tetracycline Resistance Genes in Lagoons and Groundwater Underlying Two Swine Production Facilities. Appl. Environ. Microbiol. 2001, 67, 1494–1502. [Google Scholar] [CrossRef]
- Fairchild, A.S.; Smith, J.L.; Idris, U.; Lu, J.; Sanchez, S.; Purvis, L.B.; Hofacre, C.; Lee, M.D. Effects of Orally Administered Tetracycline on the Intestinal Community Structure of Chickens and on tet Determinant Carriage by Commensal Bacteria and Campylobacter jejuni. Appl. Environ. Microbiol. 2005, 71, 5865–5872. [Google Scholar] [CrossRef]
- McMillan, E.A.; Jackson, C.R.; Frye, J.G. Transferable Plasmids of Salmonella enterica Associated With Antibiotic Resistance Genes. Front. Microbiol. 2020, 11, 562181. [Google Scholar] [CrossRef]
- Bucher, M.G.; Zwirzitz, B.; Oladeinde, A.; Cook, K.; Plymel, C.; Zock, G.; Lakin, S.; Aggrey, S.E.; Ritz, C.; Looft, T.; et al. Reused poultry litter microbiome with competitive exclusion potential against Salmonella Heidelberg. J. Environ. Qual. 2020, 49, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Valeris-Chacin, R.; Pieters, M.; Hwang, H.; Johnson, T.J.; Singer, R.S. Association of Broiler Litter Microbiome Composition and Campylobacter Isolation. Front. Vet. Sci. 2021, 8, 654927. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Idler, C.; Ammon, C.; Amon, T. Effects of the C/N ratio and moisture content on the survival of ESBL-producing Escherichia coli during chicken manure composting. Waste Manag. 2020, 105, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Nazmi, A.; Pitesky, M.; Gallardo, R.; Pandey, P. Thermal Inactivation of Escherichia coli and Salmonella Typhimurium in Poultry Carcass and Litter at Thermophilic Temperatures. J. Appl. Poult. Res. 2019, 28, 307–317. [Google Scholar] [CrossRef]
| Sample a | Total Aerobic Counts (Log10 CFU/g) b | Gram-Negative Bacteria (Log10 CFU/g) c | Antimicrobial Resistance (Log10 CFU/g) d | AMR Gene Abundance e | ||||
|---|---|---|---|---|---|---|---|---|
| Cm | Ap | Sm | Tc | aadA1/BG g | sul1/BG g | |||
| 1 | 8.56 + 7.58 | <2.99 f | 4.46 ± 3.47 | <2.99 | 6.20 ± 5.36 | 7.30 ± 6.48 | −2.37 ± −3.29 | −0.46 ± −1.37 |
| (Range) | 8.00–8.90 h | - | 4.00–4.78 i | - | 5.90–6.60 h | 6.73–7.70 j | −2.22–2.62 i | −0.92–0.61 h |
| 2 | 8.81 ± 8.10 | 4.63 ± 4.12 | - | 7.57 ± 7.29 | 7.19 ± 6.93 | 7.25 ± 6.60 | −3.42 ± −3.95 | −1.15 ± −2.20 |
| (Range) | 8.15–9.11 j | 3.00–4.90 j | <2.99–3.78 | 4.00–8.30 j | 4.60–8.30 j | 5.50–7.78 j | −4.00–2.91 j | −1.45–0.91 i |
| t-test, p = k | 0.0200 | ND | ND | ND | 0.0604 | 0.3196 | 1.22 × 10−5 | 3.43 × 10−5 |
| Mating Mix Combination | Recipients (Log10) | Transconjugants (Log10) a | Conjugation Frequency b | ||
|---|---|---|---|---|---|
| Cm c | Sm c | Tc c | |||
| Salmonella LT2 (recipient control) d | 9.14 + 8.14 | <2.99 e | <2.99 e | <2.99 e | 0.0 |
| Escherichia coli R100 (donor control) | <2.99 e | <2.99 e | <2.99 e | <2.99 e | 0.0 |
| Salmonella LT2 + E. coli R100 f | 9.03 ± 8.25 | 7.56 ± 6.70 | 7.61 ± 6.77 | 7.61 ± 6.72 | 3.32 × 10−2 |
| Salmonella LT2 + Litter 1 g | 9.16 ± 6.24 | <2.99 e | <2.99 e | <2.99 e | 0.0 |
| Salmonella LT2 + Litter 1 g + E. coli R100 f | 9.30 ± 8.39 | 7.61 ± 7.14 | 7.46 ± 7.06 | 7.44 ± 6.61 | 1.2 × 10−2 |
| Salmonella LT2 + Litter 2 g | 7.42 ± 5.59 h | <2.99 e | <2.99 e | <2.99 e | 0.0 |
| Salmonella LT2 + Litter 2 g + E. coli R100 f | 7.41 + 6.67 h | 3.43 + 2.71 h | 3.43 + 2.71 h | 3.43 + 2.95 h | 9.30 × 10−5 |
| Mating Mix Combination | Conjugation Frequency a | |||
|---|---|---|---|---|
| 25 °C | 37 °C | |||
| 2 H | 24 H | 2 H | 24 H | |
| Salmonella LT2 (recipient alone) | 0.00 | 0.00 | 0.00 | 0.00 |
| Escherichia coli pR100 (donor alone) | 0.00 | 0.00 | 0.00 | 0.00 |
| Salmonella LT2 + E. coli pR100 | 0.00 | 0.00 | 0.00 | 1.43 × 10−1 |
| Salmonella LT2 + Litter 1 b | 0.00 | 0.00 | 0.00 | 0.00 |
| Escherichia coli Strain | Plasmid 3 | Temperature | |
|---|---|---|---|
| 25 °C 4 | 37 °C 4 | ||
| MC4100 | pR100 | 0.00 | 1.43 × 10−1 |
| 1932 1 | pRSA | 1.36 × 10−4 | 4.90 × 10−3 |
| 5651 2 | pRSA | 3.59 × 10−5 | ND |
| 8762 2 | pRSA | 0.00 | ND |
| 9270 2 | pRSA | 0.00 | ND |
| 1932 1 | pSDb1 | 8.05 × 10−4 | 1.73 × 10−7 |
| 1932 1 | pSDb2 | 0.00 | 0.00 |
| 1932 1 | pSHb1 | 2.10 × 10−5 | 2.32 × 10−6 |
| 1932 1 | pSKy1 | 5.97 × 10−9 | 2.23 × 10−8 |
| 1932 1 | pSKy2 | 2.90 × 10−9 | 1.04 × 10−4 |
| 1932 1 | pSNp1 | 0.00 | 1.69 × 10−4 |
| 1932 1 | pSTm1 | 4.73 × 10−4 | 2.24 × 10−6 |
| Bacterial Strain or Plasmid | Description a | References |
|---|---|---|
| Salmonella enterica | ||
| pSLT− | S. Typhimurium LT2 strain; Rifr, lacking the spvB− virulence plasmid | [78] |
| Escherichia coli | ||
| MC4100 | Smr; F− supE44 ∆lacU169 (ϕ80 lacZ∆M15) hsdR17 recA1 endA1 gyrA96 thi01 relA1; Nalr | |
| 1932 | Human isolate; Nalr | [52] |
| 5651 | Chicken isolate; Nalr | [51] |
| Plasmids | ||
| pR00 | IncFII plasmid; Cmr Far Smr Spr Sur Tcr, Tra+ conjugative | [89] |
| pDU202 | tetracycline-sensitive derivative of R100-1; intI1, aadA1, sul1 qPCR control | [90] |
| pRSA | Broad-host-range, conjugative IncW plasmid encoding Cmr, Kmr | [49] |
| pSDb1 | S. Dublin strain CVM 22429; IncA/C, IncI1 plasmid encoding extended spectrum cephasporinase, Cmr, Tcr, Smr | |
| pSDb2 | S. Dublin strain N16S275; IncA/C, IncFII, IncX plasmid encoding extended spectrum β-lactamase, Cmr, Smr, Tcr | |
| pSHb1 | S. Heidelberg strain N16S201; IncA/C, IncI1 plasmid encoding extended spectrum β-lactamase, Smr, Tcr | |
| pSKy1 | S. Kentucky strain N162104; IncFII, IncF1b, IncI1, IncXI plasmid encoding extended spectrum β-lactamase, Smr, Tcr | |
| pSKy2 | S. Kentucky strain N173914; incFIB, IncFII, IncI1, IncX1 plasmid encoding extended spectrum cephasporinase, Kmr, Tcr | |
| pSNp1 | S. Newport strain N17S1196; IncA/C plasmid encoding extended spectrum β-lactamase, Cmr, Smr, Tcr | |
| pSTm1 | S. Typhimurium strain N17S520 IncI1 plasmid encoding extended spectrum cephasporinase, Smr, Sur, Tcr |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oxendine, A.; Walsh, A.A.; Young, T.; Dixon, B.; Hoke, A.; Rogers, E.E.; Lee, M.D.; Maurer, J.J. Conditions Necessary for the Transfer of Antimicrobial Resistance in Poultry Litter. Antibiotics 2023, 12, 1006. https://doi.org/10.3390/antibiotics12061006
Oxendine A, Walsh AA, Young T, Dixon B, Hoke A, Rogers EE, Lee MD, Maurer JJ. Conditions Necessary for the Transfer of Antimicrobial Resistance in Poultry Litter. Antibiotics. 2023; 12(6):1006. https://doi.org/10.3390/antibiotics12061006
Chicago/Turabian StyleOxendine, Aaron, Allison A. Walsh, Tamesha Young, Brandan Dixon, Alexa Hoke, Eda Erdogan Rogers, Margie D. Lee, and John J. Maurer. 2023. "Conditions Necessary for the Transfer of Antimicrobial Resistance in Poultry Litter" Antibiotics 12, no. 6: 1006. https://doi.org/10.3390/antibiotics12061006
APA StyleOxendine, A., Walsh, A. A., Young, T., Dixon, B., Hoke, A., Rogers, E. E., Lee, M. D., & Maurer, J. J. (2023). Conditions Necessary for the Transfer of Antimicrobial Resistance in Poultry Litter. Antibiotics, 12(6), 1006. https://doi.org/10.3390/antibiotics12061006





