Abstract
Providencia stuartii is a member of the Morganellaceae family, notorious for its intrinsic resistance to several antibiotics, including last-resort drugs such as colistin and tigecycline. Between February and March 2022, a four-patient outbreak sustained by P. stuartii occurred in a hospital in Rome. Phenotypic analyses defined these strains as eXtensively Drug-Resistant (XDR). Whole-genome sequencing was performed on the representative P. stuartii strains and resulted in fully closed genomes and plasmids. The genomes were highly related phylogenetically and encoded various virulence factors, including fimbrial clusters. The XDR phenotype was primarily driven by the presence of the blaNDM-1 metallo-β-lactamase alongside the rmtC 16S rRNA methyltransferase, conferring resistance to most β-lactams and every aminoglycoside, respectively. These genes were found on an IncC plasmid that was highly related to an NDM-IncC plasmid retrieved from a ST15 Klebsiella pneumoniae strain circulating in the same hospital two years earlier. Given its ability to acquire resistance plasmids and its intrinsic resistance mechanisms, P. stuartii is a formidable pathogen. The emergence of XDR P. stuartii strains poses a significant public health threat. It is essential to monitor the spread of these strains and develop new strategies for their control and treatment.
1. Introduction
The Providencia species has historically been described as a member of the Proteeae tribe [1]. Following a classification proposal for genera previously assigned to the Enterobacteriaceae family, the genus Providencia is now considered to belong to the Morganellaceae family in the Enterobacterales order [2]. From a biochemical viewpoint, bacteria belonging to this family are negative for oxidase, arginine decarboxylase, and Voges–Proskauer.
Providencia stuartii is an opportunistic pathogen capable of causing infection mainly of the respiratory and urinary tract [3], and it contributes significantly to biofilm formation in catheterized patients, also causing relevant outbreaks in hospital settings [3,4,5,6].
In the current antimicrobial resistance (AMR) crisis [7], bacteria belonging to the Morganellaceae family should be regarded as dangerous foes, displaying an intrinsic resistance to antibiotics such as colistin (due to the carriage of the arnBCADTEF and eptB genes, whose expression results in the addition of phosphoethanolamine and arabinose cationic groups on the Lipopolysaccharide, respectively [8]) and tigecycline [9], coupled with higher MICs for the carbapenem imipenem [10]. The use of the colistin and tigecycline as last-resort antimicrobial agents to treat Multi-Drug-Resistant (MDR) [11] strains is significantly promoting the spread of members of the Morganellaceae family [12].
Numerous cases of P. stuartii carrying relevant antibiotic resistance genes, located either on the chromosome or on plasmids, have been described. P. stuartii isolates carrying chromosomal-located blaNDM-1 and blaOXA-48 carbapenemase genes were identified in Switzerland in wound samples from a patient transferred from Macedonia, and several isolates carrying carbapenemases, such as KPC and VIM, have been described in Greece, Argentina, Brazil, and Saudi Arabia [6,13,14,15,16]. In recent years, a large number of carbapenem-resistant P. stuartii carrying blaNDM has been reported [3,15,17,18]. Several sequences of IncC-type plasmids carrying blaNDM isolated in P. stuartii strains are available [12,13]. These data raise attention to the relevant role of this microorganism in the spread of antimicrobial resistance genes.
This study reports the characterization of P. stuartii causing an outbreak in two Intensive Care Units (ICUs) at the Policlinico Umberto I Hospital in Rome (PUI). This outbreak was noteworthy because P. stuartii is a relatively uncommon pathogen and blaNDM, with the exception of an outbreak in Tuscany [19,20], is rarely reported in Italy [21]. Given the eXtensively Drug-Resistant (XDR) phenotype these isolates displayed, being susceptible only to the trimethoprim-sulfamethoxazole combination and aztreonam, a genomic investigation was conducted. The objectives of these analyses were: (1) to describe the outbreak strain; (2) to identify the genetic mechanisms at the basis of the blaNDM acquisition; (3) to detect if other relevant resistance mechanisms were co-acquired alongside the blaNDM; and (4) to compare Italian isolates at a global level. The study outcome was to gain a deeper knowledge about the diffusion and spread of an emerging MDR-resistant pathogen, P. stuartii.
2. Results and Discussion
2.1. An Outbreak of NDM-Producing Providencia Stuartii
Between February and March of 2022, four cases of colonization or infection sustained by NDM-producing P. stuartii were identified at PUI, and a total of seven isolates were collected from four patients (Table 1).

Table 1.
Antimicrobial susceptibility testing and sampling origin of the Providencia stuartii strains isolated in this study. Values in bold indicate resistance.
All patients were males, with a median age of 51 years and a median Intensive Care Unit (ICU) stay of 35 days. Two of them were hospitalized within the 90 days prior to the P. stuartii diagnosis. Within the 90 days before hospitalization, all patients received antibiotic therapy, but none of the antibiotics used were based on a carbapenem regimen (Table S1).
Antimicrobial susceptibility testing (AST) revealed that the strains were resistant to all tested β-lactams, except monobactams (ceftazidime, piperacillin/tazobactam, cefoxitin, cefuroxime, ceftazidime/avibactam, ceftolozane/tazobactam, imipenem, meropenem), aminoglycosides (amikacin), ciprofloxacin, and fosfomycin, but susceptible to co-trimoxazole (Table 1). In addition, P. stuartii is intrinsically resistant to colistin, tigecycline, gentamicin, and tobramycin.
Patients #3 and #4, who had a respiratory tract infection sustained by NDM-producing P. stuartii, were treated with the combination of aztreonam (AZT) + ceftazidime/avibactam (CZA) (Table S1). The rationale of this therapeutic regimen is that AZT is stable against Metallo-β-lactamases such as NDM, yet these strains may also encode for Extended Spectrum β-lactamases (ESBLs) and/or AmpC enzymes, which may hydrolyze AZT; thus, AZT alone has limited clinical utility against NDM-producing strains. The avibactam component of CZA, instead, has no ability to inhibit metallo-β-lactamases but can protect AZT from the hydrolysis of secondary ESBLs or AmpCs [22].
Of the seven isolates, three prototypical isolates (namely 65, 883, and 41) sampled from three different sources (urinary tract, respiratory tract, and rectal swab) in three different patients from two wards, were chosen to represent the complexity of this outbreak. The three strains underwent both Illumina and Nanopore sequencing.
All genomes were positive for the aac(6′)-Ib3, sul1, ΔqacE, rmtC, blaNDM-1, and blaCMY-6 genes located on IncC plasmids and the tet(B), catA3 and aac(2′)-Ia genes identified in the chromosome. Furthermore, isolate 41 carried a ColpVC plasmid encoding no resistance.
2.2. The NDM-Carrying IncC Plasmid
The first plasmid attributed to the IncC group (R16a) can be traced back to 1966 and was identified in a P. stuartii strain [23], suggesting a long-term relationship between IncC and the Providencia genus. However, in more recent years, some MDR plasmids have been reported in Providencia strains; among them, there are several IncC-type plasmids carrying blaNDM isolated from P. stuartii strains that have been described [17,18].
In the three sequenced Providencia isolates from our hospital, a 141,594-bp IncC plasmid was identified, highly conserved among them (100% identity, 100% coverage).
We compared one of these plasmids, belonging to isolate 41 (pVCpro_41, Acc. No. OQ750828), with another blaNDM-carrying IncC plasmid from Klebsiella pneumoniae isolate 0831 (p0831_NDM_IncC, Acc. No. MZ606383), sampled from our hospital in 2020 [24].
These two plasmids were almost identical, having a percentage of identity of 100% (Figure 1), but differed in length.

Figure 1.
Circos plot of two blaNDM-carrying IncC plasmids. Circos plot representing the synteny between two blaNDM-carrying IncC plasmids, pVCpro_41, and p0831_NDM_IncC, isolated from Providencia stuartii and a Klebsiella pneumoniae, respectively. The isolates were sampled from the Policlinico Umberto I University Hospital of Rome, Italy. Half circles represent the plasmids, color-coded according to the legend. The gray bands depict homology regions with 100% sequence identity between the plasmids, while white areas represent discontinuities (i.e., an ISEc23 in the case of the blue dot and the three genes represented by yellow arrows in the case of the yellow dot).
It was possible to observe the following:
- p0831_NDM_IncC carried an Insertion Sequence (IS, ISEc23), which was not present in the P. stuartii plasmid;
- pVCpro_41 carried three genes, which were not present in the p0831_NDM_IncC plasmid, encoding for an uncharacterized MFS-type transporter, and two uncharacterized proteins named YjiL and YjiM.
Performing a BLASTN search of the MFS-type transporter, yjiL and yjiM genes in the NCBI database, all results were from the chromosome of isolates belonging to the Klebsiella genus; on excluding this genus from the search, no perfect match (100% coverage, 100% identity) was obtained. No other IncC plasmid in the PLSDB, including those identified in Klebsiella, carried these extra genes in its scaffold. These three genes were also found in the chromosomes of NDM-producing ST15 K. pneumoniae strains involved in an outbreak that occurred in 2020 in the same hospital (BioProject PRJNA746265) [24]. Furthermore, NDM-IncC plasmids were not detected in any other bacterial strain isolated at the hospital in the period of 2020–2022.
Despite P. stuartii isolates expressing both NDM, CMY-2 and RmtC, have already been described in Egypt [10]; the presence of the three genes from the Klebsiella chromosome strongly supports the hypothesis that the IncC plasmid, before being transferred into P. stuartii, passed through ST15 outbreak K. pneumoniae strains. Nonetheless, before the P. stuartii outbreak in 2022, it is possible to speculate that this IncC plasmid was maintained in the hospital in hidden reservoirs for almost two years, probably from environmental sources.
2.3. Genomic Epidemiology of blaNDM-Carrying IncC Plasmids
IncC plasmids are commonly found in multi-drug-resistant Gram-negative bacteria of various species, indicating the wide host range of these plasmids. A total of 534 plasmids containing the IncC replicon, found in 18 different bacterial genera, were obtained from the PLSDB database [25] and screened using Kleborate [26]. Of these, 89 carried the blaNDM gene, mainly in its NDM-1 allele (84/89).
Due to the high levels of correlation between blaNDM gene presence and 16S rRNA methyltransferase (16RMTases) gene co-presence on the same plasmid, IncC plasmids carrying blaNDM have a higher likelihood of carrying a 16RMTases gene (39.8% vs. 10.1%, p < 0.0001). Instead, there are no significant differences between IncC plasmids carrying blaNDM and the ones not carrying it, both in terms of length (mean 162.1 kb vs. 162.9 kb) and coding sequences (CDSs, mean 206.4 of which 62 are “core” vs. 199.4 of which 30 are “core”), defining a specific IncC plasmid Core Genome (pCG).
The PLSDB database [12] shows that complete IncC plasmids encoding for NDM are exclusively found within the Enterobacteriaceae family, with Escherichia coli and K. pneumoniae being the most identified host species (31 and 27 isolates, respectively). A geospatial analysis performed on a subset of 44 NDM-encoding IncC plasmids, 39 from PLSDB with available coordinates, 3 from P. stuartii, and 2 from K. pneumoniae identified at the PUI, revealed that their distribution is not uniform. These plasmids are more widely distributed in Southeast Asia, Central Europe, and North America, with a greater prevalence in these regions (Figure 2).
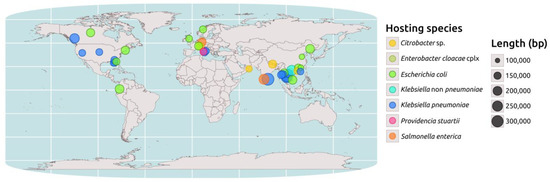
Figure 2.
Geospatial analysis of the blaNDM-carrying IncC plasmids. Map representing the distribution of a selected set of 39 IncC plasmids carrying the blaNDM gene retrieved from the PLSDB database, for which geographical coordinates were available. Additionally, the three Providencia stuartii (pink dot) plasmids described in this study and the two found in ST15 Klebsiella pneumoniae (blue dot) in the same hospital were added to the map. Plasmid sequences were screened through the Kleborate tool. The map was created using the “maps” and “ggplot2” packages in the R programming language. The circles in the map represent the IncC plasmids, which are color-coded based on the hosting species. The size of the circles is proportional to the plasmid size.
To investigate the relationship between the pCG in the NDM-encoding IncC plasmids and the species carrying them, a phylogenetic analysis was conducted on the above-mentioned 89 plasmids, as well as on the 3 additional IncC plasmids obtained from P. stuartii in the current study and the 2 IncC plasmids from the ST15 K. pneumoniae previous study, performed in the same hospital [24].
This analysis divided the NDM-encoding IncC plasmids into two main branches. The first branch included the P. stuartii plasmids from this study, which clustered together with 59 closely related plasmids (Figure 3).
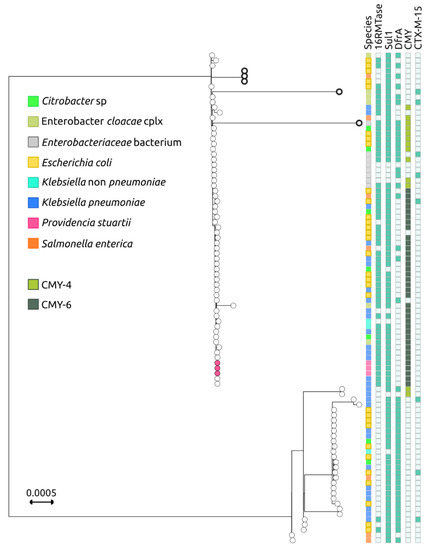
Figure 3.
Phylogenetic tree of NDM-encoding IncC plasmids based on plasmid Core genome (pCG). Phylogenetic tree of 93 IncC plasmids (89 retrieved from the PLSDB database, 3 from this study, and plasmids p0831_NDM_IncC (MZ606383) and p1027_NDM_IncC (MZ606384) from ST15 K. pneumoniae isolated in our hospital carrying the blaNDM carbapenemase, based on the concatenation of the 60 core genes, which constitute the plasmid Core genome (pCG). Metadata colors depicting the hosting species (first column) and the CMY variant (fifth column) are explained in the legend. The three IncC plasmids identified in the isolates sequenced in this study are indicated by magenta dots. The five divergent IncC plasmids in the most populated branch, discussed in the text are represented by bold circles.
The first branch, except for a small cluster of three plasmids from Southeast Asia (CP031297 found in E. coli from Vietnam, MN604267 found in Salmonella enterica serovar London, and MN604268 found in E. coli from Singapore [27]) and two highly divergent plasmids (CP041052 found in Enterobacter hormaechei and MN657252 found in an Enterobacteriaceae [28]), showed an extremely well-conserved pCG. The second branch is less populated and composed of 30 plasmids, which were found to be more heterogeneous in terms of pCG. Additionally, 28 out of the 30 plasmids in this branch did not have genes encoding for 16RMTases, and 28 out of the 30 did not encode for CMY. IncC plasmids are one of the main routes for the diffusion of blaCMY cephalosporinase genes [29], which, in association with blaNDM, confer resistance to most β-lactams. Overall, the findings suggest that the NDM-encoding IncC plasmids are diverse in terms of their pCG and host species. The first branch of plasmids appears to be highly conserved, while the second branch is less conserved and lacks the 16RMTases gene in most cases; both plasmid lineages can be hosted by multiple species (Figure 3).
2.4. Anatomy of Three Providencia Stuartii Isolates Carrying the IncC Plasmid
Given the high conservation of the IncC plasmids among them and with those identified in ST15 K. pneumoniae, an in-depth characterization of the three P. stuartii strains was needed to discern between horizontal transfer of IncC in three different P. stuartii recipients and vertical expansion of one single P. stuartii NDM-positive clone.
The three sequenced isolates from PUI have highly similar genomes, differing by 2–5 SNPs on 4120 core genes (Figure S1, dataset File S1). One of these SNPs differentiated the isolate 41 from isolates 65 and 883, causing a frameshift variant of the efflux transporter periplasmic adaptor sub-unit MexH, often implied in AMR in members of the Pseudomonas genus [30]. The contribution of this mutation to the AMR profile could not be evaluated due to the substantial number of resistance genes already carried by these isolates.
A total of 17 different virulence-related genes were identified, encoding for the biosynthesis of the O-antigen and LPS (pgi, lpxC, msbA, lpxB, galE, rmlB, wecA), the flagellum (fliG, flgG, flgE, cheA), and the numerous secretion systems present in the P. stuartii genome (ysaC and escR genes, belonging to a type 3 secretion system, and four copies of the hcp-2 gene for the type 6 secretion system). Several gene clusters belonging to the chaperone-usher fimbriae and crucial virulence factors [31] were identified in the sequenced strains.
Overall, 21 ushers were identified in the 3 sequenced strains of P. stuartii. These protein sequences were compared to those from other species available in the GenBank database. The resulting phylogenetic tree showed that the ushers from P. stuartii clustered into six distinct branches (Figure 4A). Except for one usher in fimbrial cluster 4, which was also found in Proteus mirabilis, all other ushers were specific to P. stuartii. Conducting a detailed analysis of these clusters, significant variations even within the same phylogroup were detected (Figure 4B).
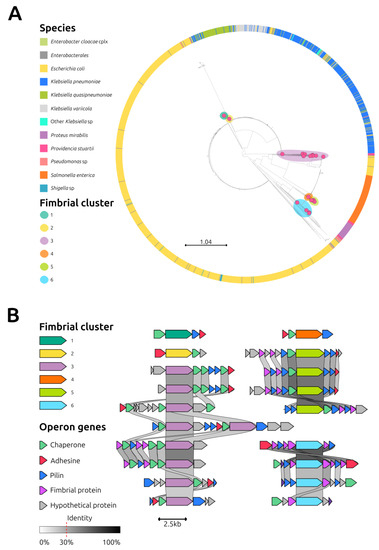
Figure 4.
Analysis of fimbrial clusters. (A): Phylogenetic analysis of 1881 usher proteins (1860 downloaded from the RefSeq NCBI Protein DataBase, https://www.ncbi.nlm.nih.gov/protein, accessed on 31 January 2023, using as query “Fimbria” AND “Usher” and 21 identified in isolate 41). Metadata colors depicting the hosting species are explained in the legend. The 21 usher proteins identified in isolate 41 are indicated by magenta dots; the colored halos around magenta dots indicate the corresponding fimbrial cluster. (B): Synteny analysis of the operons belonging to the 6 fimbrial clusters identified in Providencia stuartii isolate 41. Arrows represent genes and are color-coded according to the legend. Grey links represent the nucleotide homology between genes. The nucleotide identity threshold was set to 30%, and higher identity is reflected by a darker shade of gray.
A CAS-TypeIF bacterial defense system, located within the cluster encoding for the flagellum, was identified. Nonetheless, not all the CRISPR regions were located within the flagellar cluster, with some dispersed in other sites of the genome. Furthermore, only two of the identified spacers could be associated with bacteriophages.
Three prophages could be localized in the analyzed P. stuartii chromosomal sequences; specifically, one belonged to the Myoviridae family, one to the Siphoviridae family, and one could not be clearly classified.
3. Materials and Methods
3.1. Bacterial Isolation and Antimicrobial Susceptibility Testing
Between February and March 2022, seven Providencia stuartii isolates were retrieved from four patients at the PUI (Table 1).
Bacteria were isolated from samples collected during routine microbiologic processes. Isolated colonies were identified as P. stuartii by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) system (Bruker Daltonics GmbH, Bremen, Germany). The antimicrobial susceptibility test was carried out using the MicroScan WalkAway system (Beckman Coulter, Inc., Brea, CA, USA), testing the following antibiotics: amikacin, gentamicin, tobramycin, piperacillin/tazobactam cefuroxime, ceftazidime, cefoxitin, ceftazidime/avibactam, ceftolozane/tazobactam, imipenem, meropenem, ciprofloxacin, co-trimoxazole, fosfomycin, colistin, and tigecycline.
3.2. Whole-Genome Sequencing and Assembly
Among the seven above-mentioned isolates, three prototypical strains (namely 41, 65, and 883) isolated from three patients (Table 1) were further characterized by Whole-Genome Sequencing (WGS).
Genomic DNA extracted using the Bioline kit was used as input for the Illumina MiSeq instrument (Illumina, Inc., San Diego, CA, USA), following the Nextera XT DNA sample preparation kit, which generated paired-end libraries with the 2 × 300 PE protocol (Illumina). High-molecular-weight genomic DNA was purified using a proteinase K-phenol-chloroform extraction method and used for Oxford Nanopore Technologies (ONT) sequencing, as previously described [32]. Illumina reads and ONT assemblies were integrated by the Unicycler tool version 0.4.8.0 using a bold bridging mode [33].
3.3. Genomic and Phylogenetic Analyses
The mobilome and resistome of the three P. stuartii genomes sequenced in this study were, respectively, analyzed using PlasmidFinder [34] and ResFinder [35] at the Center for Genomic Epidemiology website (http://www.genomicepidemiology.org/services/ (accessed on 31 May 2022)).
Their genomes were annotated using RAST [36] and Prokka [37]. The presence of CRISPR loci was assessed by CRISPR-Cas finder [38] and the ones of phages by Phigaro [39].
Manual curation was performed on the annotated genomes to identify the genes encoding for fimbrial clusters using the EDGAR 3.0 server [40] and Bakta [41]. To classify and evaluate the identified fimbriae, a comparative analysis of the amino acid sequence of the fimbrial usher proteins of isolate 41 was performed in comparison with those obtained from the NCBI Protein Database. Specifically, a phylogenetic relatedness analysis of the fimbrial usher protein [42] was performed with 1860 usher proteins and the relative metadata downloaded from the RefSeq NCBI Protein Database (https://www.ncbi.nlm.nih.gov/protein (accessed on 31 January 2023)) using as query “Fimbria” AND “Usher”. The synteny among the identified fimbrial operons was visualized using the Clinker tool [43] and adjusted using the open-source InkScape software.
A total of 60 genomes belonging to P. stuartii (3 sequenced in this study and 57 downloaded from the NCBI database) were annotated using Prokka [37]. A core-gene alignment was built using Roary [44] from the respective GFF, with a minimum percentage identity for BLASTP of 95% and a percentage of isolates a gene must be in to be core of 95%. A consensus phylogenetic tree based on 1000 ultrafast bootstraps [45] was generated with IQ-TREE [46] using the GTR + F + I + G4 substitution model. All phylogenetic analyses were carried out using the Galaxy Europe instance (https://usegalaxy.eu/ (accessed on 28 February 2023)). The phylogenetic tree and its metadata were visualized using Microreact [47] and adjusted using the open-source InkScape software. To enhance the clarity of the tree, 10 genomes downloaded from the GenBank database were excluded from its final version because their significant phylogenetic distance made it harder to interpret the relationships between our isolates and the rest of the tree.
The analysis of virulence genes was carried out using the Virulence Finder Database [48], taking into account genes with a percent identity and coverage greater than 70%.
The R programming language (Version 4.2.2) and two related packages (maps and ggplot2) were utilized to obtain world map data using the ‘map_data’ function from the maps package.
3.4. IncC Plasmids Analysis
Plasmids carrying the IncC replicon were downloaded from the PLSDB [25] database version 2021_06_23_v2, filtered for the presence of the PlasmidFinder [34] IncC replicon and screened for the presence of blaNDM using Kleborate [26]. A phylogenetic analysis based on a core-gene alignment of the blaNDM-positive IncC plasmid was performed using Roary [44] from the respective GFF, with a minimum percentage identity for BLASTP of 90% and a percentage of isolates a gene must be in to be core of 95%. The resulting core genome has been defined as the IncC plasmid Core Genome (pCG). A consensus phylogenetic tree based on 1000 ultrafast bootstraps [45] was generated with IQ-TREE [46] using the GTR + F + G4 substitution model. The tree and metadata were visualized using Microreact [47] and adjusted using the open-source InkScape software verison 1.2.2.
3.5. Statistical Analyses
Statistical analyses were performed using JASP version 0.17.1. To assess the relationships between categorical variables and to determine differences between continuous variables, χ2 and Mann–Whitney U tests were deployed, respectively. The χ2 test compares observed and expected frequencies to determine if there is a significant association between the two categorical variables. The Mann–Whitney U test compares the medians of two independent samples to determine if they come from the same population or not. Both tests were used to evaluate the significance of the results at p < 0.01.
4. Conclusions
A small outbreak sustained by NDM-1-producing Klebsiella pneumoniae occurred in 2020 at the PUI in Rome. In those isolates, we identified an IncC plasmid carrying the blaNDM and rmtC genes. This plasmid was not reported in other strains from this hospital in the following two years. In 2022, an NDM-1-encoding IncC plasmid carrying three genes from the K. pneumoniae chromosome was identified in Providencia stuartii, causing a small outbreak in the hospital.
The presence of an IncC plasmid harboring blaNDM in a member of the Morganellaceae family, intrinsically resistant to several last-resort antibiotics such as colistin or tigecycline, is of the uttermost relevance.
The limitation of this study is the small number of outbreak isolates and the lack of intermediate strains demonstrating the hypothesis of a passage of blaNDM-1 IncC plasmids between K. pneumoniae and P. stuartii. However, the IncC plasmid of P. stuartii acquired chromosomal genes from K. pneumoniae, and these genes were not present in other IncC plasmids, suggesting that horizontal transfer between the two species occurred.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics12050943/s1, Figure S1: Phylogenetic tree of Providencia stuartii; Table S1: Clinical information. File S1: Supplementary Dataset.
Author Contributions
Conceptualization, G.A. (Gabriele Arcari), A.O., G.R., G.A. (Guido Antonelli) and A.C.; methodology, V.C., G.A. (Gabriele Arcari), F.S. and G.M.; software, R.P. and L.F.; validation, V.C., G.A. (Gabriele Arcari) and A.C.; formal analysis, V.C., R.P. and G.M.; investigation, A.O.; resources, F.S., G.R. and G.A. (Gabriele Arcari); data curation, G.A. (Gabriele Arcari) and A.C.; writing—original draft preparation, V.C., G.A. (Gabriele Arcari) and A.C.; writing—review and editing, A.O., F.S., G.M., L.F., R.P., G.R. and G.A. (Guido Antonelli); supervision, A.C., funding acquisition, A.O. and A.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by EU funding to Alessandra Carattoli within the NextGeneration EU-MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases (Project no. PE00000007, PE13 INF-ACT, Spoke 3). Riccardo Polani was supported by the PNRR PHD SCHOLARSHIP (EX M.D 351/22) financed by the Rome Technopole project.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by Ethics Committee of Sapienza University of Rome (protocol code 449/19 and date of approval 20 June 2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
1. A BioProject has been released at DDBJ/ENA/GenBank, no.: PRJNA948429 (https://www.ncbi.nlm.nih.gov/sra/PRJNA948429, accessed on 1 May 2023); 2. Circular complete pVCpro_41 plasmid has been released under accession no. OQ750828.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ewing, W.H. The Tribe Proteeae: Its Nomenclature and Taxonomy. Int. Bull. Bacteriol. Nomencl. Taxon. 1962, 12, 93–102. [Google Scholar] [CrossRef]
- Adeolu, M.; Alnajar, S.; Naushad, S.; Gupta, R.S. Genome-Based Phylogeny and Taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales Ord. Nov. Divided into the Families Enterobacteriaceae, Erwiniaceae Fam. Nov., Pectobacteriaceae Fam. Nov., Yersiniaceae Fam. Nov., Hafniaceae Fam. Nov., Morganellaceae Fam. Nov., and Budviciaceae Fam. Nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5575–5599. [Google Scholar] [CrossRef] [PubMed]
- Molnár, S.; Flonta, M.M.M.; Almaş, A.; Buzea, M.; Licker, M.; Rus, M.; Földes, A.; Székely, E. Dissemination of NDM-1 Carbapenemase-Producer Providencia stuartii Strains in Romanian Hospitals: A Multicentre Study. J. Hosp. Infect. 2019, 103, 165–169. [Google Scholar] [CrossRef]
- Mnif, B.; Ktari, S.; Chaari, A.; Medhioub, F.; Rhimi, F.; Bouaziz, M.; Hammami, A. Nosocomial Dissemination of Providencia stuartii Isolates Carrying BlaOXA-48, BlaPER-1, BlaCMY-4 and QnrA6 in a Tunisian Hospital. J. Antimicrob. Chemother. 2013, 68, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Rakov, C.; Ben Porat, S.; Alkalay-Oren, S.; Yerushalmy, O.; Abdalrhman, M.; Gronovich, N.; Huang, L.; Pride, D.; Coppenhagen-Glazer, S.; Nir-Paz, R.; et al. Targeting Biofilm of MDR Providencia stuartii by Phages Using a Catheter Model. Antibiotics 2021, 10, 375. [Google Scholar] [CrossRef]
- Douka, E.; Perivolioti, E.; Kraniotaki, E.; Fountoulis, K.; Economidou, F.; Tsakris, A.; Skoutelis, A.; Routsi, C. Emergence of a Pandrug-Resistant VIM-1-Producing Providencia stuartii Clonal Strain Causing an Outbreak in a Greek Intensive Care Unit. Int. J. Antimicrob. Agents 2015, 45, 533–536. [Google Scholar] [CrossRef]
- Ventola, C.L. The Antibiotic Resistance Crisis: Part 1: Causes and Threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Gogry, F.A.; Siddiqui, M.T.; Sultan, I.; Haq, Q. Mohd.R. Current Update on Intrinsic and Acquired Colistin Resistance Mechanisms in Bacteria. Front. Med. 2021, 8, 677720. [Google Scholar] [CrossRef]
- Yaghoubi, S.; Zekiy, A.O.; Krutova, M.; Gholami, M.; Kouhsari, E.; Sholeh, M.; Ghafouri, Z.; Maleki, F. Tigecycline Antibacterial Activity, Clinical Effectiveness, and Mechanisms and Epidemiology of Resistance: Narrative Review. Eur. J. Clin. Microbiol. Infect. Dis. 2022, 41, 1003–1022. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). CLSI Supplement M100: Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Hayakawa, K.; Marchaim, D.; Divine, G.W.; Pogue, J.M.; Kumar, S.; Lephart, P.; Risko, K.; Sobel, J.D.; Kaye, K.S. Growing Prevalence of Providencia stuartii Associated with the Increased Usage of Colistin at a Tertiary Health Care Center. Int. J. Infect. Dis. 2012, 16, e646–e648. [Google Scholar] [CrossRef] [PubMed]
- Moser, A.I.; Keller, P.M.; Campos-Madueno, E.I.; Poirel, L.; Nordmann, P.; Endimiani, A. A Patient With Multiple Carbapenemase Producers Including an Unusual Citrobacter sedlakii Hosting an IncC blaNDM-1- and ArmA-Carrying Plasmid. Pathog. Immun. 2021, 6, 119–134. [Google Scholar] [CrossRef] [PubMed]
- Hoard, A.; Montaña, S.; Moriano, A.; Fernandez, J.S.; Traglia, G.M.; Quiroga, C.; Franchi, A.; Cohen, E.; Corigliano, C.; Almuzara, M.; et al. Genomic Analysis of Two NDM-1 Providencia stuartii Strains Recovered from a Single Patient. Curr. Microbiol. 2020, 77, 4029–4036. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, M.; Alhababi, R.; Alqudah, N.; Aldyyat, B.; Alharthy, A. First Report of Carbapenem-Resistant Providencia stuartii in Saudi Arabia. New Microbes New Infect. 2018, 26, 107–109. [Google Scholar] [CrossRef]
- Tavares, C.P.; Pereira, P.S.; Marques, E.d.A.; Faria, C.; de Souza, M.d.P.A.H.; de Almeida, R.; Alves, C.d.F.M.; Asensi, M.D.; Carvalho-Assef, A.P.D. Molecular Epidemiology of KPC-2-Producing Enterobacteriaceae (Non-Klebsiella pneumoniae) Isolated from Brazil. Diagn. Microbiol. Infect. Dis. 2015, 82, 326–330. [Google Scholar] [CrossRef]
- Mc Gann, P.; Hang, J.; Clifford, R.J.; Yang, Y.; Kwak, Y.I.; Kuschner, R.A.; Lesho, E.P.; Waterman, P.E. Complete Sequence of a Novel 178-Kilobase Plasmid Carrying BlaNDM-1 in a Providencia stuartii Strain Isolated in Afghanistan. Antimicrob. Agents Chemother. 2012, 56, 1673–1679. [Google Scholar] [CrossRef]
- Manageiro, V.; Sampaio, D.A.; Pereira, P.; Rodrigues, P.; Vieira, L.; Palos, C.; Caniça, M. Draft Genome Sequence of the First NDM-1-Producing Providencia stuartii Strain Isolated in Portugal. Genome Announc. 2015, 3, e01077-15. [Google Scholar] [CrossRef]
- Di Pilato, V.; Henrici De Angelis, L.; Aiezza, N.; Baccani, I.; Niccolai, C.; Parisio, E.M.; Giordano, C.; Camarlinghi, G.; Barnini, S.; Forni, S.; et al. Resistome and Virulome Accretion in an NDM-1-Producing ST147 Sublineage of Klebsiella pneumoniae Associated with an Outbreak in Tuscany, Italy: A Genotypic and Phenotypic Characterisation. Lancet Microbe 2022, 3, e224–e234. [Google Scholar] [CrossRef]
- Martin, M.J.; Corey, B.W.; Sannio, F.; Hall, L.R.; MacDonald, U.; Jones, B.T.; Mills, E.G.; Harless, C.; Stam, J.; Maybank, R.; et al. Anatomy of an Extensively Drug-Resistant Klebsiella pneumoniae Outbreak in Tuscany, Italy. Proc. Natl. Acad. Sci. USA 2021, 118, e2110227118. [Google Scholar] [CrossRef]
- Di Pilato, V.; Errico, G.; Monaco, M.; Giani, T.; Del Grosso, M.; Antonelli, A.; David, S.; Lindh, E.; Camilli, R.; Aanensen, D.M.; et al. The Changing Epidemiology of Carbapenemase-Producing Klebsiella pneumoniae in Italy: Toward Polyclonal Evolution with Emergence of High-Risk Lineages. J. Antimicrob. Chemother. 2021, 76, 355–361. [Google Scholar] [CrossRef]
- Wu, W.; Feng, Y.; Tang, G.; Qiao, F.; McNally, A.; Zong, Z. NDM Metallo-β-Lactamases and Their Bacterial Producers in Health Care Settings. Clin. Microbiol. Rev. 2019, 32, e00115-18. [Google Scholar] [CrossRef] [PubMed]
- Ambrose, S.J.; Harmer, C.J.; Hall, R.M. Evolution and Typing of IncC Plasmids Contributing to Antibiotic Resistance in Gram-Negative Bacteria. Plasmid 2018, 99, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Sacco, F.; Raponi, G.; Oliva, A.; Bibbolino, G.; Mauro, V.; Di Lella, F.M.; Volpicelli, L.; Antonelli, G.; Venditti, M.; Carattoli, A.; et al. An Outbreak Sustained by ST15 Klebsiella pneumoniae Carrying 16S RRNA Methyltransferases and BlaNDM: Evaluation of the Global Dissemination of These Resistance Determinants. Int. J. Antimicrob. Agents 2022, 60, 106615. [Google Scholar] [CrossRef]
- Galata, V.; Fehlmann, T.; Backes, C.; Keller, A. PLSDB: A Resource of Complete Bacterial Plasmids. Nucleic Acids Res. 2019, 47, D195–D202. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.M.C.; Wick, R.R.; Watts, S.C.; Cerdeira, L.T.; Wyres, K.L.; Holt, K.E. A Genomic Surveillance Framework and Genotyping Tool for Klebsiella pneumoniae and Its Related Species Complex. Nat. Commun. 2021, 12, 4188. [Google Scholar] [CrossRef]
- Octavia, S.; Chew, K.L.; Chew, K.L.; Lin, R.T.P.; Teo, J.W.P. Multidrug-Resistant Salmonella enterica Serovar London Carrying blaNDM-1 Encoding Plasmid from Singapore. Clin. Microbiol. Infect. 2020, 26, 963–966. [Google Scholar] [CrossRef]
- Weber, R.E.; Pietsch, M.; Frühauf, A.; Pfeifer, Y.; Martin, M.; Luft, D.; Gatermann, S.; Pfennigwerth, N.; Kaase, M.; Werner, G.; et al. IS26-Mediated Transfer of blaNDM–1 as the Main Route of Resistance Transmission During a Polyclonal, Multispecies Outbreak in a German Hospital. Front. Microbiol. 2019, 10, 2817. [Google Scholar] [CrossRef]
- Carattoli, A.; Villa, L.; Poirel, L.; Bonnin, R.A.; Nordmann, P. Evolution of IncA/C blaCMY-2-Carrying Plasmids by Acquisition of the blaNDM-1 Carbapenemase Gene. Antimicrob. Agents Chemother. 2012, 56, 783–786. [Google Scholar] [CrossRef]
- Schweizer, H.P. Efflux as a Mechanism of Resistance to Antimicrobials in Pseudomonas aeruginosa and Related Bacteria: Unanswered Questions. Genet. Mol. Res. 2003, 2, 48–62. [Google Scholar]
- Kolenda, R.; Ugorski, M.; Grzymajlo, K. Everything You Always Wanted to Know About Salmonella Type 1 Fimbriae, but Were Afraid to Ask. Front. Microbiol. 2019, 10, 1017. [Google Scholar] [CrossRef]
- Carattoli, A.; Arcari, G.; Bibbolino, G.; Sacco, F.; Tomolillo, D.; Di Lella, F.M.; Trancassini, M.; Faino, L.; Venditti, M.; Antonelli, G.; et al. Evolutionary Trajectories toward Ceftazidime-Avibactam Resistance in Klebsiella pneumoniae Clinical Isolates. Antimicrob. Agents Chemother. 2021, 65, e0057421. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for Predictions of Phenotypes from Genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an Update of CRISRFinder, Includes a Portable Version, Enhanced Performance and Integrates Search for Cas Proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef]
- Starikova, E.V.; Tikhonova, P.O.; Prianichnikov, N.A.; Rands, C.M.; Zdobnov, E.M.; Ilina, E.N.; Govorun, V.M. Phigaro: High-Throughput Prophage Sequence Annotation. Bioinformatics 2020, 36, 3882–3884. [Google Scholar] [CrossRef]
- Dieckmann, M.A.; Beyvers, S.; Nkouamedjo-Fankep, R.C.; Hanel, P.H.G.; Jelonek, L.; Blom, J.; Goesmann, A. EDGAR3.0: Comparative Genomics and Phylogenomics on a Scalable Infrastructure. Nucleic Acids Res. 2021, 49, W185–W192. [Google Scholar] [CrossRef]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and Standardized Annotation of Bacterial Genomes via Alignment-Free Sequence Identification. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef]
- Nuccio, S.-P.; Bäumler, A.J. Evolution of the Chaperone/Usher Assembly Pathway: Fimbrial Classification Goes Greek. Microbiol. Mol. Biol. Rev. 2007, 71, 551–575. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, C.L.M.; Chooi, Y.-H. Clinker & Clustermap. Js: Automatic Generation of Gene Cluster Comparison Figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar] [CrossRef] [PubMed]
- Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis|Bioinformatics|Oxford Academic. Available online: https://academic.oup.com/bioinformatics/article/31/22/3691/240757?login=true (accessed on 9 September 2022).
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Argimón, S.; Abudahab, K.; Goater, R.J.E.; Fedosejev, A.; Bhai, J.; Glasner, C.; Feil, E.J.; Holden, M.T.G.; Yeats, C.A.; Grundmann, H.; et al. Microreact: Visualizing and Sharing Data for Genomic Epidemiology and Phylogeography. Microb. Genom. 2016, 2, e000093. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A General Classification Scheme for Bacterial Virulence Factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).