Co-Existence of Oxazolidinone Resistance Genes cfr(D) and optrA on Two Streptococcus parasuis Isolates from Swine
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Bacterial Strains
2.2. Antimicrobial Susceptibility Testing
2.3. PCR Analysis
2.4. Whole-Genome Sequencing (WGS) and Analysis
2.5. Phylogenetic Analyses of S. parasuis Isolates
2.6. Transfer Experiments
2.7. Nucleotide Sequence Accession Numbers
3. Results
3.1. Identification of cfr(D) and optrA in the Streptococcus Isolates
3.2. Antibiotic Resistance and Resistance Determinants
3.3. WGS Analyses
3.4. Characterization of Plasmids Carrying cfr(D)
3.5. Genetic Environment of optrA in the Chromosomal DNA
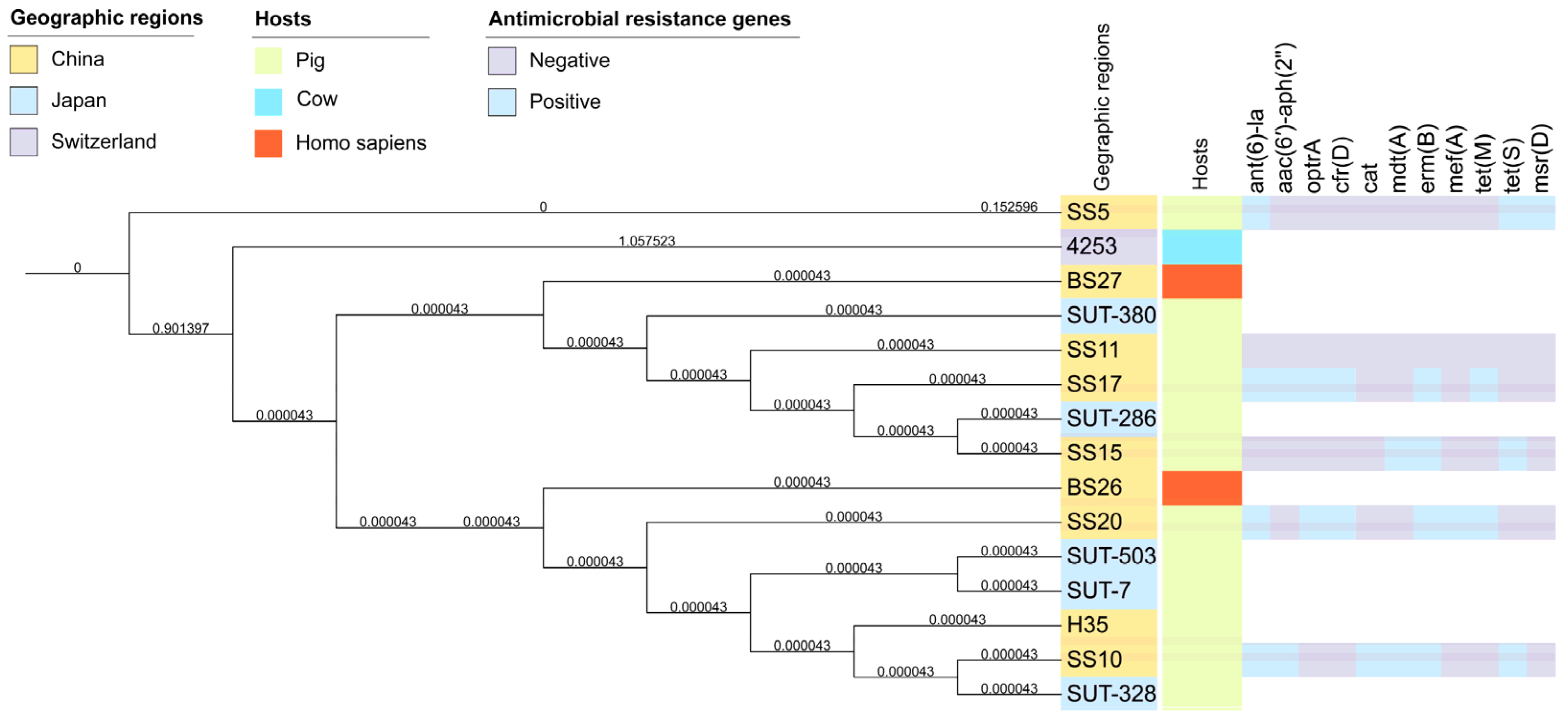
3.6. Phylogenetic Relatedness of Streptococcus parasuis Strains
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Almeida, L.M.; Lebreton, F.; Gaca, A.; Bispo, P.M.; Saavedra, J.T.; Calumby, R.N.; Grillo, L.M.; Nascimento, T.G.; Filsner, P.H.; Moreno, A.M.; et al. Transferable Resistance Gene in Enterococcus faecalis from Swine in Brazil. Antimicrob. Agents Chemother. 2020, 64, e00142-20. [Google Scholar] [CrossRef] [PubMed]
- Leach, K.L.; Brickner, S.J.; Noe, M.C.; Miller, P.F. Linezolid, the first oxazolidinone antibacterial agent. Ann. N. Y. Acad. Sci. 2011, 1222, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Klevens, R.M.; Morrison, M.A.; Nadle, J.; Petit, S.; Gershman, K.; Ray, S.; Harrison, L.H.; Lynfield, R.; Dumyati, G.; Townes, J.M.; et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 2007, 298, 1763–1771. [Google Scholar] [CrossRef]
- Hoyert, D.L.; Xu, J. Deaths: Preliminary data for 2011. Natl. Vital. Stat. Rep. 2012, 61, 1–65. [Google Scholar] [PubMed]
- Vehreschild, M.J.G.T.; Haverkamp, M.; Biehl, L.M.; Lemmen, S.; Fätkenheuer, G. Vancomycin-resistant enterococci (VRE): A reason to isolate? Infection 2019, 47, 7–11. [Google Scholar] [CrossRef]
- Locke, J.B.; Hilgers, M.; Shaw, K.J. Novel ribosomal mutations in Staphylococcus aureus strains identified through selection with the oxazolidinones linezolid and torezolid (TR-700). Antimicrob. Agents Chemother. 2009, 53, 5265–5274. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Zhang, W.; Du, X.-D.; Krüger, H.; Feßler, A.T.; Ma, S.; Zhu, Y.; Wu, C.; Shen, J.; Wang, Y. Mobile Oxazolidinone Resistance Genes in Gram-Positive and Gram-Negative Bacteria. Clin. Microbiol. Rev. 2021, 34, e0018820. [Google Scholar] [CrossRef]
- Jung, Y.-H.; Cha, M.-H.; Woo, G.-J.; Chi, Y.-M. Characterization of oxazolidinone and phenicol resistance genes in non-clinical enterococcal isolates from Korea. J. Glob. Antimicrob. Resist. 2021, 24, 363–369. [Google Scholar] [CrossRef]
- Meka, V.G.; Gold, H.S. Antimicrobial resistance to linezolid. Clin. Infect. Dis. 2004, 39, 1010–1015. [Google Scholar] [CrossRef]
- Aarestrup, F.M. The livestock reservoir for antimicrobial resistance: A personal view on changing patterns of risks, effects of interventions and the way forward. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140085. [Google Scholar] [CrossRef]
- Liu, B.G.; Yuan, X.L.; He, D.D.; Hu, G.Z.; Miao, M.S.; Xu, E.P. Research progress on the oxazolidinone drug linezolid resistance. Eur. Rev. Med. Pharm. Sci. 2020, 24, 9274–9281. [Google Scholar] [CrossRef]
- Deshpande, L.M.; Castanheira, M.; Flamm, R.K.; Mendes, R.E. Evolving oxazolidinone resistance mechanisms in a worldwide collection of enterococcal clinical isolates: Results from the SENTRY Antimicrobial Surveillance Program. J. Antimicrob. Chemother. 2018, 73, 2314–2322. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gong, S.; Dong, X.; Li, J.; Grenier, D.; Yi, L. Mixed Biofilm of and Impacts Antibiotic Susceptibility and Modulates Virulence Factor Gene Expression. Front. Microbiol. 2020, 11, 507. [Google Scholar] [CrossRef] [PubMed]
- Sharkey, L.K.R.; O’Neill, A.J. Antibiotic Resistance ABC-F Proteins: Bringing Target Protection into the Limelight. ACS Infect. Dis. 2018, 4, 239–246. [Google Scholar] [CrossRef]
- Locke, J.B.; Finn, J.; Hilgers, M.; Morales, G.; Rahawi, S.; Kedar, G.C.; Picazo, J.J.; Im, W.; Shaw, K.J.; Stein, J.L. Structure-activity relationships of diverse oxazolidinones for linezolid-resistant Staphylococcus aureus strains possessing the cfr methyltransferase gene or ribosomal mutations. Antimicrob. Agents Chemother. 2010, 54, 5337–5343. [Google Scholar] [CrossRef]
- Stojkovic, V.; Ulate, M.F.; Hidalgo-Villeda, F.; Aguilar, E.; Monge-Cascante, C.; Pizarro-Guajardo, M.; Tsai, K.; Tzoc, E.; Camorlinga, M.; Paredes-Sabja, D.; et al. cfr(B), cfr(C), and a New cfr-Like Gene, cfr(E), in Clostridium difficile Strains Recovered across Latin America. Antimicrob. Agents Chemother. 2019, 64, e01074-19. [Google Scholar] [CrossRef]
- Huang, J.; Sun, J.; Wu, Y.; Chen, L.; Duan, D.; Lv, X.; Wang, L. Identification and pathogenicity of an XDR Streptococcus suis isolate that harbours the phenicol-oxazolidinone resistance genes optrA and cfr, and the bacitracin resistance locus bcrABDR. Int. J. Antimicrob. Agents 2019, 54, 43–48. [Google Scholar] [CrossRef]
- Nomoto, R.; Maruyama, F.; Ishida, S.; Tohya, M.; Sekizaki, T.; Osawa, R. Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22 and 26: Streptococcus parasuis sp. nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 438–443. [Google Scholar] [CrossRef]
- Wang, J.; Yi, X.; Liang, P.; Tao, Y.; Wang, Y.; Jin, D.; Luo, B.; Yang, J.; Zheng, H. Investigation of the Genomic and Pathogenic Features of the Potentially Zoonotic. Pathogens 2021, 10, 834. [Google Scholar] [CrossRef]
- Nomoto, R.; Ishida-Kuroki, K.; Nakagawa, I.; Sekizaki, T. Complete Genome Sequences of Four Streptococcus parasuis Strains Obtained from Saliva of Domestic Pigs in Japan. Microbiol. Resour. Announc. 2022, 11, e0124521. [Google Scholar] [CrossRef]
- Zhu, Y.; Yang, Q.; Schwarz, S.; Yang, W.; Xu, Q.; Wang, L.; Liu, S.; Zhang, W. Identification of a Streptococcus parasuis isolate co-harbouring the oxazolidinone resistance genes cfr(D) and optrA. J. Antimicrob. Chemother. 2021, 76, 3059–3061. [Google Scholar] [CrossRef]
- Yamada, R.; Tien, L.H.T.; Arai, S.; Tohya, M.; Ishida-Kuroki, K.; Nomoto, R.; Kim, H.; Suzuki, E.; Osawa, R.; Watanabe, T.; et al. Development of PCR for identifying Streptococcus parasuis, a close relative of Streptococcus suis. J. Vet. Med. Sci. 2018, 80, 1101–1107. [Google Scholar] [CrossRef] [PubMed]
- Stevens, M.J.A.; Cernela, N.; Corti, S.; Stephan, R. Draft Genome Sequence of Streptococcus parasuis 4253, the First Available for the Species. Microbiol. Resour. Announc. 2019, 8, e00203-19. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Performance Standards for an Timicrobial Susceptibility Testing; Twenty-Sixth Informational Supplement M100-S26; CLSI: Wayne, PA, USA, 2016. [Google Scholar]
- Clinical and laboratory Standards Institute. Performance Standards for Antimi Crobial Disk and Diffusion Susceptibility Tests for Bacteria Isolated from Animals; Second Informational Supplement VET01-S2; CLSI: Wayne, PA, USA, 2013. [Google Scholar]
- Wang, Y.; Lv, Y.; Cai, J.; Schwarz, S.; Cui, L.; Hu, Z.; Zhang, R.; Li, J.; Zhao, Q.; He, T.; et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 2015, 70, 2182–2190. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, W.; Wang, J.; Wu, C.; Shen, Z.; Fu, X.; Yan, Y.; Zhang, Q.; Schwarz, S.; Shen, J. Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob. Agents Chemother. 2012, 56, 1485–1490. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.-S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef]
- Bitar, I.; Moussa, J.; Abboud, E.; Hrabak, J.; Tokajian, S. Integration of two pKPX-2-derived antibiotic resistance islands in the genome of an ESBL-producing Klebsiella pneumoniae ST3483 from Lebanon. J. Glob. Antimicrob. Resist. 2019, 18, 257–259. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Stamatakis, A. RaxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Yu, R.; Zhang, Y.; Xu, Y.; Schwarz, S.; Li, X.-S.; Shang, Y.-H.; Du, X.-D. Emergence of a (M) Variant Conferring Resistance to Tigecycline in. Front. Vet. Sci. 2021, 8, 709327. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, X.Y.; Schwarz, S.; Yang, M.; Zhang, S.M.; Hao, W.; Du, X.D. Tn6674 Is a Novel Enterococcal optrA-Carrying Multiresistance Transposon of the Tn554 Family. Antimicrob. Agents Chemother. 2019, 63, e00142-20. [Google Scholar] [CrossRef]
- Yao, T.G.; Li, B.Y.; Luan, R.D.; Wang, H.N.; Lei, C.W. Whole genome sequence of Enterococcus gallinarum EG81, a porcine strain harbouring the oxazolidinone-phenicol resistance gene optrA with chromosomal and plasmid location. J. Glob. Antimicrob. Resist. 2020, 22, 598–600. [Google Scholar] [CrossRef]
- He, Y.Z.; Li, X.P.; Miao, Y.Y.; Lin, J.; Sun, R.Y.; Wang, X.P.; Guo, Y.Y.; Liao, X.P.; Liu, Y.H.; Feng, Y.; et al. The ISApl1 2 Dimer Circular Intermediate Participates in mcr-1 Transposition. Front. Microbiol. 2019, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Lyras, D.; Rood, J.I. Transposition of Tn4451 and Tn4453 involves a circular intermediate that forms a promoter for the large resolvase, TnpX. Mol. Microbiol. 2000, 38, 588–601. [Google Scholar] [CrossRef]
- Manganelli, R.; Romano, L.; Ricci, S.; Zazzi, M.; Pozzi, G. Dosage of Tn916 circular intermediates in Enterococcus faecalis. Plasmid 1995, 34, 48–57. [Google Scholar] [CrossRef]
- García-Sánchez, L.; Melero, B.; Diez, A.M.; Jaime, I.; Rovira, J. Characterization of Campylobacter species in Spanish retail from different fresh chicken products and their antimicrobial resistance. Food Microbiol. 2018, 76, 457–465. [Google Scholar] [CrossRef]
- Susilawathi, N.M.; Tarini, N.M.A.; Fatmawati, N.N.D.; Mayura, P.I.B.; Suryapraba, A.A.A.; Subrata, M.; Sudewi, A.A.R.; Mahardika, G.N. Streptococcus suis-Associated Meningitis, Bali, Indonesia, 2014–2017. Emerg. Infect. Dis. 2019, 25, 2235–2242. [Google Scholar] [CrossRef] [PubMed]
- Segura, M.; Fittipaldi, N.; Calzas, C.; Gottschalk, M. Critical Streptococcus suis Virulence Factors: Are They All Really Critical? Trends Microbiol. 2017, 25, 585–599. [Google Scholar] [CrossRef]
- Haenni, M.; Lupo, A.; Madec, J.-Y. Antimicrobial Resistance in spp. Microbiol. Spectr. 2018, 6, 585–599. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.; Wang, Z.; Li, Q.; Yu, Y.; Li, Y.; Tan, Z.; Zhang, W. Genomic characterization of Streptococcus parasuis, a close relative of Streptococcus suis and also a potential opportunistic zoonotic pathogen. BMC Genom. 2022, 23, 469. [Google Scholar] [CrossRef]
- Mishra, S.; Klümper, U.; Voolaid, V.; Berendonk, T.U.; Kneis, D. Simultaneous estimation of parameters governing the vertical and horizontal transfer of antibiotic resistance genes. Sci. Total Environ. 2021, 798, 149174. [Google Scholar] [CrossRef]
- Biggel, M.; Nüesch-Inderbinen, M.; Jans, C.; Stevens, M.J.A.; Stephan, R. Genetic Context of and in Florfenicol-Resistant Enterococci Isolated from Flowing Surface Water in Switzerland. Antimicrob. Agents Chemother. 2021, 65, e0108321. [Google Scholar] [CrossRef] [PubMed]
- Freitas, A.R.; Tedim, A.P.; Novais, C.; Lanza, V.F.; Peixe, L. Comparative genomics of global optrA-carrying uncovers a common chromosomal hotspot for acquisition within a diversity of core and accessory genomes. Microb. Genom. 2020, 6, AAC0108321. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.; Li, D.; Feßler, A.T.; Wu, C.; Schwarz, S.; Wang, Y. Distribution of optrA and cfr in florfenicol-resistant Staphylococcus sciuri of pig origin. Vet. Microbiol. 2017, 210, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Werckenthin, C.; Kehrenberg, C. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 2000, 44, 2530–2533. [Google Scholar] [CrossRef]
- Kehrenberg, C.; Ojo, K.K.; Schwarz, S. Nucleotide sequence and organization of the multiresistance plasmid pSCFS1 from Staphylococcus sciuri. J. Antimicrob. Chemother. 2004, 54, 936–939. [Google Scholar] [CrossRef]
- Kehrenberg, C.; Aarestrup, F.M.; Schwarz, S. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob. Agents Chemother. 2007, 51, 483–487. [Google Scholar] [CrossRef]
- Li, S.-M.; Zhou, Y.-F.; Li, L.; Fang, L.-X.; Duan, J.-H.; Liu, F.-R.; Liang, H.-Q.; Wu, Y.-T.; Gu, W.-Q.; Liao, X.-P.; et al. Characterization of the Multi-Drug Resistance Gene in Methicillin-Resistant (MRSA) Strains Isolated from Animals and Humans in China. Front. Microbiol. 2018, 9, 2925. [Google Scholar] [CrossRef]
- Guerin, F.; Sassi, M.; Dejoies, L.; Zouari, A.; Schutz, S.; Potrel, S.; Auzou, M.; Collet, A.; Lecointe, D.; Auger, G.; et al. Molecular and functional analysis of the novel cfr(D) linezolid resistance gene identified in Enterococcus faecium. J. Antimicrob. Chemother. 2020, 75, 1699–1703. [Google Scholar] [CrossRef]
- Brenciani, A.; Morroni, G.; Vincenzi, C.; Manso, E.; Mingoia, M.; Giovanetti, E.; Varaldo, P.E. Detection in Italy of two clinical Enterococcus faecium isolates carrying both the oxazolidinone and phenicol resistance gene optrA and a silent multiresistance gene cfr. J. Antimicrob. Chemother. 2016, 71, 1118–1119. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.-X.; Duan, J.-H.; Chen, M.-Y.; Deng, H.; Liang, H.-Q.; Xiong, Y.Q.; Sun, J.; Liu, Y.-H.; Liao, X.-P. Prevalence of cfr in Enterococcus faecalis strains isolated from swine farms in China: Predominated cfr-carrying pCPPF5-like plasmids conferring “non-linezolid resistance” phenotype. Infect. Genet. Evol. 2018, 62, 188–192. [Google Scholar] [CrossRef] [PubMed]

| Isolate | Species | Source | MLST | MIC (mg/L) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ERY | CLI | MEM | CEF | LZD | VAN | TET | PEN | AMP | ENR | SXT | FFC | AMO | Resistance Genes | ||||
| SS17 | S. parasuis | nasal swab | NA | 32 | >32 | ≤0.03 | 0.5 | 1 | 0.12 | 64 | 2 | 1 | 4 | 32 | 64 | 0.5 | ant(6)-la, aac(6′)-aph(2″), optrA, erm(B), tet(M), cfr(D) |
| SS20 | S. parasuis | nasal swab | NA | 32 | >32 | ≤0.03 | 0.5 | 1 | 0.12 | 64 | 2 | 1 | 4 | 64 | >64 | 1 | erm(B), ant(6)-la, optrA, mef(A), tet(M), cfr(D) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, N.; Li, J.; Wan, P.; Pan, Y.; Xu, T.; Xiong, W.; Zeng, Z. Co-Existence of Oxazolidinone Resistance Genes cfr(D) and optrA on Two Streptococcus parasuis Isolates from Swine. Antibiotics 2023, 12, 825. https://doi.org/10.3390/antibiotics12050825
Han N, Li J, Wan P, Pan Y, Xu T, Xiong W, Zeng Z. Co-Existence of Oxazolidinone Resistance Genes cfr(D) and optrA on Two Streptococcus parasuis Isolates from Swine. Antibiotics. 2023; 12(5):825. https://doi.org/10.3390/antibiotics12050825
Chicago/Turabian StyleHan, Ning, Jie Li, Peng Wan, Yu Pan, Tiantian Xu, Wenguang Xiong, and Zhenling Zeng. 2023. "Co-Existence of Oxazolidinone Resistance Genes cfr(D) and optrA on Two Streptococcus parasuis Isolates from Swine" Antibiotics 12, no. 5: 825. https://doi.org/10.3390/antibiotics12050825
APA StyleHan, N., Li, J., Wan, P., Pan, Y., Xu, T., Xiong, W., & Zeng, Z. (2023). Co-Existence of Oxazolidinone Resistance Genes cfr(D) and optrA on Two Streptococcus parasuis Isolates from Swine. Antibiotics, 12(5), 825. https://doi.org/10.3390/antibiotics12050825






