Biocide Susceptibility and Antimicrobial Resistance of Escherichia coli Isolated from Swine Feces, Pork Meat and Humans in Germany
Abstract
1. Introduction
2. Results
2.1. Biocide Susceptibility
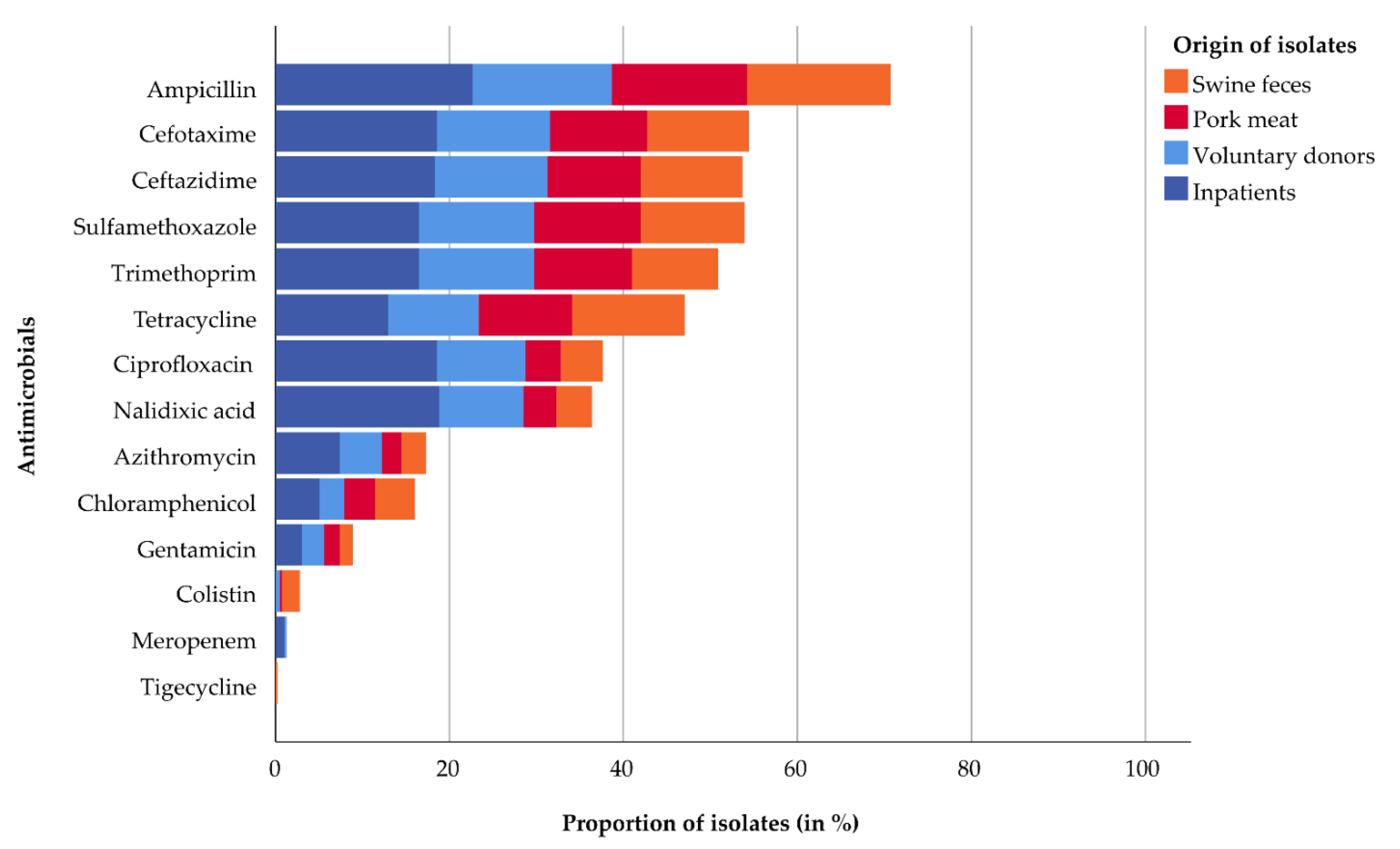
2.2. Antimicrobial Susceptibility
2.3. Association between Antimicrobial and Biocide Susceptibility
3. Discussion
4. Materials and Methods
4.1. E. coli Isolates
4.2. Biocides
4.3. Biocide Susceptibility Testing
4.4. Antimicrobial Susceptibility Testing
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McDonnell, G.; Russell, A.D. Antiseptics and disinfectants: Activity, action, and resistance. Clin. Microbiol. Rev. 1999, 12, 147–179. [Google Scholar] [CrossRef] [PubMed]
- Butucel, E.; Balta, I.; Ahmadi, M.; Dumitrescu, G.; Morariu, F.; Pet, I.; Stef, L.; Corcionivoschi, N. Biocides as biomedicines against foodborne pathogenic bacteria. Biomedicines 2022, 10, 379. [Google Scholar] [CrossRef] [PubMed]
- Kampf, G. Biocidal agents used for disinfection can enhance antibiotic resistance in gram-negative species. Antibiotics 2018, 7, 110. [Google Scholar] [CrossRef] [PubMed]
- Meade, E.; Slattery, M.A.; Garvey, M. Biocidal resistance in clinically relevant microbial species: A major public health risk. Pathogens 2021, 10, 598. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, A.R.; Safaee, M.M.; Wuest, W.M.; Furst, A.L. The silent pandemic: Emergent antibiotic resistances following the global response to SARS-CoV-2. iScience 2021, 24, 102304. [Google Scholar] [CrossRef]
- Hardy, K.; Sunnucks, K.; Gil, H.; Shabir, S.; Trampari, E.; Hawkey, P.; Webber, M. Increased usage of antiseptics is associated with reduced susceptibility in clinical isolates of Staphylococcus aureus. MBio 2018, 9, e00894-18. [Google Scholar] [CrossRef]
- Pidot, S.J.; Gao, W.; Buultjens, A.H.; Monk, I.R.; Guerillot, R.; Carter, G.P.; Lee, J.Y.H.; Lam, M.M.C.; Grayson, M.L.; Ballard, S.A.; et al. Increasing tolerance of hospital Enterococcus faecium to handwash alcohols. Sci. Transl. Med. 2018, 10, eaar6115. [Google Scholar] [CrossRef]
- Stein, C.; Vincze, S.; Kipp, F.; Makarewicz, O.; Al Dahouk, S.; Pletz, M.W. Carbapenem-resistant Klebsiella pneumoniae with low chlorhexidine susceptibility. Lancet Infect. Dis. 2019, 19, 932–933. [Google Scholar] [CrossRef]
- Dopcea, G.N.; Dopcea, I.; Nanu, A.E.; Diguţă, C.F.; Matei, F. Resistance and cross-resistance in Staphylococcus spp. strains following prolonged exposure to different antiseptics. J. Glob. Antimicrob. Resist. 2020, 21, 399–404. [Google Scholar] [CrossRef]
- Soumet, C.; Méheust, D.; Pissavin, C.; Le Grandois, P.; Frémaux, B.; Feurer, C.; Le Roux, A.; Denis, M.; Maris, P. Reduced susceptibilities to biocides and resistance to antibiotics in food-associated bacteria following exposure to quaternary ammonium compounds. J. Appl. Microbiol. 2016, 121, 1275–1281. [Google Scholar] [CrossRef]
- WOAH. World Organization for Animal Health. Antimicrobial Resistance. 2023. Available online: https://www.woah.org/en/what-we-do/global-initiatives/antimicrobial-resistance/ (accessed on 2 February 2023).
- WHO. World Health Organization. Antimicrobial Resistance. 2021. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 1 February 2023).
- Russell, A.D. Biocide use and antibiotic resistance: The relevance of laboratory findings to clinical and environmental situations. Lancet Infect. Dis. 2003, 3, 794–803. [Google Scholar] [CrossRef]
- Morrissey, I.; Oggioni, M.R.; Knight, D.; Curiao, T.; Coque, T.; Kalkanci, A.; Martinez, J.L. Evaluation of epidemiological cut-off values indicates that biocide resistant subpopulations are uncommon in natural isolates of clinically-relevant microorganisms. PLoS ONE 2014, 9, e86669. [Google Scholar] [CrossRef]
- Roedel, A.; Vincze, S.; Projahn, M.; Roesler, U.; Robé, C.; Hammerl, J.A.; Noll, M.; Al Dahouk, S.; Dieckmann, R. Genetic but no phenotypic associations between biocide tolerance and antibiotic resistance in Escherichia coli from German broiler fattening farms. Microorganisms 2021, 9, 651. [Google Scholar] [CrossRef]
- Feßler, A.T.; Scholtzek, A.D.; Schug, A.R.; Kohn, B.; Weingart, C.; Hanke, D.; Schink, A.-K.; Bethe, A.; Lübke-Becker, A.; Schwarz, S. Antimicrobial and biocide resistance among canine and feline Enterococcus faecalis, Enterococcus faecium, Escherichia coli, Pseudomonas aeruginosa, and Acinetobacter baumannii isolates from diagnostic submissions. Antibiotics 2022, 11, 152. [Google Scholar] [CrossRef]
- Feßler, A.T.; Scholtzek, A.D.; Schug, A.R.; Kohn, B.; Weingart, C.; Schink, A.-K.; Bethe, A.; Lübke-Becker, A.; Schwarz, S. Antimicrobial and biocide resistance among feline and canine Staphylococcus aureus and Staphylococcus pseudintermedius isolates from diagnostic submissions. Antibiotics 2022, 11, 127. [Google Scholar] [CrossRef]
- Kaesbohrer, A.; Schroeter, A.; Tenhagen, B.-A.; Alt, K.; Guerra, B.; Appel, B. Emerging antimicrobial resistance in commensal Escherichia coli with public health relevance. Zoonoses Public Health 2012, 59, 158–165. [Google Scholar] [CrossRef]
- Tadesse, D.A.; Zhao, S.; Tong, E.; Ayers, S.; Singh, A.; Bartholomew, M.J.; McDermott, P.F. Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950–2002. Emerg. Infect. Dis. 2012, 18, 741–749. [Google Scholar] [CrossRef]
- Nyirabahizi, E.; Tyson, G.H.; Dessai, U.; Zhao, S.; Kabera, C.; Crarey, E.; Womack, N.; Crews, M.K.; Strain, E.; Tate, H. Evaluation of Escherichia coli as an indicator for antimicrobial resistance in Salmonella recovered from the same food or animal ceca samples. Food Control 2020, 115, 107280. [Google Scholar] [CrossRef]
- van Elsas, J.D.; Semenov, A.V.; Costa, R.; Trevors, J.T. Survival of Escherichia coli in the environment: Fundamental and public health aspects. ISME J 2011, 5, 173–183. [Google Scholar] [CrossRef]
- Oniciuc, E.-A.; Likotrafiti, E.; Alvarez-Molina, A.; Prieto, M.; López, M.; Alvarez-Ordóñez, A. Food processing as a risk factor for antimicrobial resistance spread along the food chain. Curr. Opin. Food Sci. 2019, 30, 21–26. [Google Scholar] [CrossRef]
- Behnke, M.; Aghdassi, S.J.; Hansen, S.; Diaz, L.A.P.; Gastmeier, P.; Piening, B. The prevalence of nosocomial infection and antibiotic use in German hospitals. Dtsch. Arztebl. Int. 2017, 114, 851–857. [Google Scholar] [CrossRef] [PubMed]
- Ludden, C.; Coll, F.; Gouliouris, T.; Restif, O.; Blane, B.; Blackwell, G.A.; Kumar, N.; Naydenova, P.; Crawley, C.; Brown, N.M.; et al. Defining nosocomial transmission of Escherichia coli and antimicrobial resistance genes: A genomic surveillance study. Lancet Microbe 2021, 2, e472–e480. [Google Scholar] [CrossRef] [PubMed]
- Stewart, S.; Robertson, C.; Pan, J.; Kennedy, S.; Dancer, S.; Haahr, L.; Manoukian, S.; Mason, H.; Kavanagh, K.; Cook, B.; et al. Epidemiology of healthcare-associated infection reported from a hospital-wide incidence study: Considerations for infection prevention and control planning. J. Hosp. Infect. 2021, 114, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Meng, J.; McDermott, P.F.; Wang, F.; Yang, Q.; Cao, G.; Hoffmann, M.; Zhao, S. Presence of disinfectant resistance genes in Escherichia coli isolated from retail meats in the USA. J. Antimicrob. Chemother. 2014, 69, 2644–2649. [Google Scholar] [CrossRef] [PubMed]
- Deus, D.; Krischek, C.; Pfeifer, Y.; Sharifi, A.R.; Fiegen, U.; Reich, F.; Klein, G.; Kehrenberg, C. Comparative analysis of the susceptibility to biocides and heavy metals of extended-spectrum β-lactamase-producing Escherichia coli isolates of human and avian origin, Germany. Diagn. Microbiol. Infect. Dis. 2017, 88, 88–92. [Google Scholar] [CrossRef]
- Haley, B.J.; Kim, S.W.; Salaheen, S.; Hovingh, E.; Van Kessel, J.A.S. Virulome and genome analyses identify associations between antimicrobial resistance genes and virulence factors in highly drug-resistant Escherichia coli isolated from veal calves. PLoS ONE 2022, 17, e0265445. [Google Scholar] [CrossRef]
- Van den Poel, B.; Saegeman, V.; Schuermans, A. Increasing usage of chlorhexidine in health care settings: Blessing or curse? A narrative review of the risk of chlorhexidine resistance and the implications for infection prevention and control. Eur. J. Clin. Microbiol. Infect. 2022, 41, 349–362. [Google Scholar] [CrossRef]
- Oosterik, L.H.; Peeters, L.; Mutuku, I.; Goddeeris, B.M.; Butaye, P. Susceptibility of avian pathogenic Escherichia coli from laying hens in Belgium to antibiotics and disinfectants and integron prevalence. Avian Dis. 2014, 58, 271–278. [Google Scholar] [CrossRef]
- Maertens, H.; De Reu, K.; Meyer, E.; Van Coillie, E.; Dewulf, J. Limited association between disinfectant use and either antibiotic or disinfectant susceptibility of Escherichia coli in both poultry and pig husbandry. BMC Vet. Res. 2019, 15, 310. [Google Scholar] [CrossRef]
- Puangseree, J.; Jeamsripong, S.; Prathan, R.; Pungpian, C.; Chuanchuen, R. Resistance to widely-used disinfectants and heavy metals and cross resistance to antibiotics in Escherichia coli isolated from pigs, pork and pig carcass. Food Control 2021, 124, 107892. [Google Scholar] [CrossRef]
- Neuhaus, S.; Feßler, A.T.; Dieckmann, R.; Thieme, L.; Pletz, M.W.; Schwarz, S.; Al Dahouk, S. Towards a harmonized terminology: A glossary for biocide susceptibility testing. Pathogens 2022, 11, 1455. [Google Scholar] [CrossRef]
- Schug, A.R.; Scholtzek, A.D.; Turnidge, J.; Meurer, M.; Schwarz, S.; Feßler, A.T.; the Biocide Susceptibility Study Group. Development of quality control ranges for biocide susceptibility testing. Pathogens 2022, 11, 223. [Google Scholar] [CrossRef]
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing. MIC and Zone Diameter Distributions and ECOFFs. 2022. Available online: https://mic.eucast.org/search/?search%5Bmethod%5D=mic&search%5Bantibiotic%5D=-1&search%5Bspecies%5D=261&search%5Bdisk_content%5D=-1&search%5Blimit%5D=50 (accessed on 29 November 2022).
- Sjölund, M.; Bengtsson, S.; Bonnedahl, J.; Hernandez, J.; Olsen, B.; Kahlmeter, G. Antimicrobial susceptibility in Escherichia coli of human and avian origin—A comparison of wild-type distributions. Clin. Microbiol. Infect. 2009, 15, 461–465. [Google Scholar] [CrossRef]
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 13.0. 2022. Available online: https://www.eucast.org/clinical_breakpoints/ (accessed on 12 December 2022).
- Galler, H.; Luxner, J.; Petternel, C.; Reinthaler, F.F.; Habib, J.; Haas, D.; Kittinger, C.; Pless, P.; Feierl, G.; Zarfel, G. Multiresistant bacteria isolated from intestinal faeces of farm animals in Austria. Antibiotics 2021, 10, 466. [Google Scholar] [CrossRef]
- Musa, L.; Proietti, P.C.; Marenzoni, M.L.; Stefanetti, V.; Kika, T.S.; Blasi, F.; Magistrali, C.F.; Toppi, V.; Ranucci, D.; Branciari, R.; et al. Susceptibility of commensal E. coli isolated from conventional, antibiotic-free, and organic meat chickens on farms and at slaughter toward antimicrobials with public health relevance. Antibiotics 2021, 10, 1321. [Google Scholar] [CrossRef]
- Costa, M.M.; Cardo, M.; Soares, P.; Cara d’Anjo, M.; Leite, A. Multi-drug and β-lactam resistance in Escherichia coli and food-borne pathogens from animals and food in Portugal, 2014–2019. Antibiotics 2022, 11, 90. [Google Scholar] [CrossRef]
- Ayandele, A.; Oladipo, E.; Oyebisi, O.; Kaka, M. Prevalence of multi-antibiotic resistant Escherichia coli and Klebsiella species obtained from a tertiary medical institution in Oyo State, Nigeria. Qatar Med. J. 2020, 2020, 9. [Google Scholar] [CrossRef]
- El Mekes, A.; Zahlane, K.; Ait Said, L.; Tadlaoui Ouafi, A.; Barakate, M. The clinical and epidemiological risk factors of infections due to multi-drug resistant bacteria in an adult intensive care unit of University Hospital Center in Marrakesh-Morocco. J. Infect. Public Health 2020, 13, 637–643. [Google Scholar] [CrossRef]
- Benaissa, E.; Elmrimar, N.; Belouad, E.; Mechal, Y.; Ghazouani, M.; Bsaibiss, F.; Benlahlou, Y.; Chadli, M.; Touil, N.; Lemnaouer, A.; et al. Update on the resistance of Escherichia coli isolated from urine specimens in a Moroccan hospital: A review of a 7-year period. Germs 2021, 11, 189–198. [Google Scholar] [CrossRef]
- Di Carlo, P.; Serra, N.; Lo Sauro, S.; Carelli, V.M.; Giarratana, M.; Signorello, J.C.; Lucchesi, A.; Manta, G.; Napolitano, M.S.; Rea, T.; et al. Epidemiology and pattern of resistance of gram-negative bacteria isolated from blood samples in hospitalized patients: A single center retrospective analysis from southern Italy. Antibiotics 2021, 10, 1402. [Google Scholar] [CrossRef]
- Piazza, A.; Principe, L.; Comandatore, F.; Perini, M.; Meroni, E.; Mattioni Marchetti, V.; Migliavacca, R.; Luzzaro, F. Whole-genome sequencing investigation of a large nosocomial outbreak caused by ST131 H30Rx KPC-producing Escherichia coli in Italy. Antibiotics 2021, 10, 718. [Google Scholar] [CrossRef] [PubMed]
- RKI. Robert Koch Institute. Antibiotika-Resistenz-Surveillance. 2021. Available online: https://ars.rki.de/Content/Database/ResistanceOverview.aspx (accessed on 17 January 2023).
- Rhomberg, P.R.; Jones, R.N. Summary trends for the meropenem yearly susceptibility test information collection program: A 10-year experience in the United States (1999–2008). Diagn. Microbiol. Infect. Dis. 2009, 65, 414–426. [Google Scholar] [CrossRef] [PubMed]
- WHO. World Health Organization, Electronic Essential Medicines List (eEML). Available online: https://list.essentialmeds.org/ (accessed on 6 December 2022).
- Kelesidis, T.; Karageorgopoulos, D.E.; Kelesidis, I.; Falagas, M.E. Tigecycline for the treatment of multidrug-resistant Enterobacteriaceae: A systematic review of the evidence from microbiological and clinical studies. J. Antimicrob. Chemother. 2008, 62, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Bundesamt für Verbraucherschutz und Lebensmittelsicherheit (BVL). BVL-Report 15.2 Berichte zur Lebensmittelsicherheit Zoonosen-Monitoring 2019. 2020. Available online: https://www.bvl.bund.de/SharedDocs/Downloads/01_Lebensmittel/04_Zoonosen_Monitoring/Zoonosen_Monitoring_Bericht_2019.pdf?__blob=publicationFile&v=5 (accessed on 15 December 2022).
- Guenther, S.; Falgenhauer, L.; Semmler, T.; Imirzalioglu, C.; Chakraborty, T.; Roesler, U.; Roschanski, N. Environmental emission of multiresistant Escherichia coli carrying the colistin resistance gene mcr-1 from German swine farms. J. Antimicrob. Chemother. 2017, 72, 1289–1292. [Google Scholar] [CrossRef]
- Abdalla, S.E.; Abia, A.L.K.; Amoako, D.G.; Perrett, K.; Bester, L.A.; Essack, S.Y. From farm-to-fork: E. coli from an intensive pig production system in South Africa shows high resistance to critically important antibiotics for human and animal use. Antibiotics 2021, 10, 178. [Google Scholar] [CrossRef]
- Ketkhao, P.; Thongratsakul, S.; Poolperm, P.; Poolkhet, C.; Amavisit, P. Antimicrobial resistance profiles of Escherichia coli from swine farms using different antimicrobials and management systems. Vet. World 2021, 14, 689–695. [Google Scholar] [CrossRef]
- Martelli, F.; AbuOun, M.; Cawthraw, S.; Storey, N.; Turner, O.; Ellington, M.; Nair, S.; Painset, A.; Teale, C.; Anjum, M.F. Detection of the transferable tigecycline resistance gene tet(X4) in Escherichia coli from pigs in the United Kingdom. J. Antimicrob. Chemother. 2021, 77, 846–848. [Google Scholar] [CrossRef]
- SCENIHR. Scientific Committee on Emerging and Newly Identified Health Risks. Assessment of the Antibiotic Resistance Effects of Biocides. 2009. Available online: https://health.ec.europa.eu/scientific-committees/former-scientific-committees/scientific-committee-emerging-and-newly-identified-health-risks-scenihr_en (accessed on 15 December 2022).
- Rezasoltani, S.; Yadegar, A.; Hatami, B.; Asadzadeh Aghdaei, H.; Zali, M.R. Antimicrobial resistance as a hidden menace lurking behind the COVID-19 outbreak: The global impacts of too much hygiene on AMR. Front. Microbiol. 2020, 11, 590683. [Google Scholar] [CrossRef]
- Ansari, S.; Hays, J.P.; Kemp, A.; Okechukwu, R.; Murugaiyan, J.; Ekwanzala, M.D.; Ruiz Alvarez, M.J.; Paul-Satyaseela, M.; Iwu, C.D.; Balleste-Delpierre, C.; et al. The potential impact of the COVID-19 pandemic on global antimicrobial and biocide resistance: An AMR Insights global perspective. JAC Antimicrob. Resist. 2021, 3, dlab038. [Google Scholar] [CrossRef]
- Rizvi, S.G.; Ahammad, S.Z. COVID-19 and antimicrobial resistance: A cross-study. Sci. Total Environ. 2022, 807, 150873. [Google Scholar] [CrossRef]
- Wand, M.E.; Bock, L.J.; Bonney, L.C.; Sutton, J.M. Mechanisms of increased resistance to chlorhexidine and cross-resistance to colistin following exposure of Klebsiella pneumoniae clinical isolates to chlorhexidine. Antimicrob. Agents Chemother. 2017, 61, e01162-16. [Google Scholar] [CrossRef]
- Fernando, D.M.; Xu, W.; Loewen, P.C.; Zhanel, G.G.; Kumar, A. Triclosan can select for an AdeIJK-overexpressing mutant of Acinetobacter baumannii ATCC 17978 that displays reduced susceptibility to multiple antibiotics. Antimicrob. Agents Chemother. 2014, 58, 6424–6431. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef]
- Lavilla Lerma, L.; Benomar, N.; Casado Muñoz, M.d.C.; Gálvez, A.; Abriouel, H. Correlation between antibiotic and biocide resistance in mesophilic and psychrotrophic Pseudomonas spp. isolated from slaughterhouse surfaces throughout meat chain production. Food Microbiol. 2015, 51, 33–44. [Google Scholar] [CrossRef]
- Oggioni, M.R.; Coelho, J.R.; Furi, L.; Knight, D.R.; Viti, C.; Orefici, G.; Martinez, J.L.; Freitas, A.T.; Coque, T.M.; Morrissey, I. Significant differences characterise the correlation coefficients between biocide and antibiotic susceptibility profiles in Staphylococcus aureus. Curr. Pharm. Des. 2015, 21, 2054–2057. [Google Scholar] [CrossRef]
- Adkin, P.; Hitchcock, A.; Smith, L.J.; Walsh, S.E. Priming with biocides: A pathway to antibiotic resistance? J. Appl. Microbiol. 2022, 133, 830–841. [Google Scholar] [CrossRef]
- Christensen, E.G.; Gram, L.; Kastbjerg, V.G. Sublethal Triclosan Exposure Decreases Susceptibility to Gentamicin and Other Aminoglycosides in Listeria monocytogenes. Antimicrob. Agents Chemother. 2011, 55, 4064–4071. [Google Scholar] [CrossRef]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 10th ed.; CLSI Document Ms07-A10; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015. [Google Scholar]
| Biocide | Origin of Isolates | Number of Isolates with MIC Values (mg/L) of | MIC95 | p-Value | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | 1024 | 2048 | 4096 | 8192 | 16,384 | 32,768 | 65,536 | 131,072 | 262,144 | ||||
| GDA | Swine feces | - | - | 18 | 81 | 1 | - | - | - | 512 | 0.002 | |||||||||||||
| Pork meat | - | - | 15 | 82 | 2 | - | - | - | 512 | |||||||||||||||
| Voluntary donors | - | - | 16 | 67 | 13 | - | - | - | 1024 | |||||||||||||||
| Inpatients | - | - | 3 | 88 | 6 | 1 | - | - | 1024 | |||||||||||||||
| CHG | Swine feces | - | 12 | 79 | 9 | - | - | - | - | 4 | <0.0001 | |||||||||||||
| Pork meat | 7 | 50 | 39 | 2 | 1 | - | - | - | 2 | |||||||||||||||
| Voluntary donors | - | 7 | 77 | 9 | 1 | 2 | - | - | 4 | |||||||||||||||
| Inpatients | - | 11 | 81 | 3 | 3 | - | - | - | 4 | |||||||||||||||
| BAC | Swine feces | - | - | - | 30 | 67 | 3 | - | - | 32 | 0.239 | |||||||||||||
| Pork meat | - | - | - | 18 | 81 | - | - | - | 32 | |||||||||||||||
| Voluntary donors | - | - | 3 | 28 | 62 | 3 | - | - | 32 | |||||||||||||||
| Inpatients | - | - | 1 | 29 | 54 | 13 | 1 | - | 64 | |||||||||||||||
| OCT | Swine feces | - | - | 4 | 96 | - | - | - | - | 2 | 0.088 | |||||||||||||
| Pork meat | - | - | 1 | 96 | 2 | - | - | - | 2 | |||||||||||||||
| Voluntary donors | - | - | 1 | 92 | 3 | - | - | - | 2 | |||||||||||||||
| Inpatients | - | - | 1 | 94 | 3 | - | - | - | 2 | |||||||||||||||
| IPA | Swine feces | - | - | - | 4 | 54 | 40 | 2 | - | 65,536 | 0.022 | |||||||||||||
| Pork meat | - | - | - | 1 | 58 | 35 | 5 | - | 131,072 | |||||||||||||||
| Voluntary donors | - | - | - | 5 | 51 | 39 | 1 | - | 65,536 | |||||||||||||||
| Inpatients | - | - | - | 1 | 38 | 58 | 1 | - | 65,536 | |||||||||||||||
| NaOCl | Swine feces | - | - | 15 | 79 | 6 | - | - | 1024 | 0.016 | ||||||||||||||
| Pork meat | - | - | 22 | 76 | 1 | - | - | 512 | ||||||||||||||||
| Voluntary donors | - | - | 2 | 66 | 6 | - | - | 1024 | ||||||||||||||||
| Inpatients | - | - | 5 | 93 | - | - | - | 512 | ||||||||||||||||
| PCMC | Swine feces | - | - | 57 | 42 | 1 | - | - | - | 512 | 0.001 | |||||||||||||
| Pork meat | - | - | 81 | 18 | - | - | - | - | 512 | |||||||||||||||
| Voluntary donors | - | 1 | 68 | 27 | - | - | - | - | 512 | |||||||||||||||
| Inpatients | - | 2 | 73 | 23 | - | - | - | - | 512 | |||||||||||||||
| Biocide | Origin of Isolates | Number of Isolates with MBC Values (mg/L) of | MBC95 | p-Value | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | 1024 | 2048 | 4096 | 8192 | 16,384 | 32,768 | 65,536 | 131,072 | 262,144 | ||||
| GDA | Swine feces | - | - | 9 | 87 | 4 | - | - | - | 512 | 0.004 | |||||||||||||
| Pork meat | - | - | 6 | 90 | 3 | - | - | - | 512 | |||||||||||||||
| Voluntary donors | - | - | 11 | 68 | 16 | 1 | - | - | 1024 | |||||||||||||||
| Inpatients | - | - | 3 | 78 | 16 | 1 | - | - | 1024 | |||||||||||||||
| CHG | Swine feces | - | 10 | 73 | 14 | 2 | 1 | - | - | 4 | <0.0001 | |||||||||||||
| Pork meat | 6 | 49 | 37 | 6 | 1 | - | - | - | 4 | |||||||||||||||
| Voluntary donors | - | 4 | 75 | 12 | 3 | 2 | - | - | 8 | |||||||||||||||
| Inpatients | - | 9 | 57 | 24 | 7 | - | 1 | - | 8 | |||||||||||||||
| BAC | Swine feces | - | - | - | 13 | 71 | 16 | - | - | 64 | 0.069 | |||||||||||||
| Pork meat | - | - | - | 7 | 74 | 16 | 2 | - | 64 | |||||||||||||||
| Voluntary donors | - | - | 1 | 22 | 60 | 13 | - | - | 64 | |||||||||||||||
| Inpatients | - | - | 1 | 23 | 52 | 20 | 2 | - | 64 | |||||||||||||||
| OCT | Swine feces | - | - | 3 | 82 | 12 | 3 | - | - | 4 | 0.704 | |||||||||||||
| Pork meat | - | - | - | 83 | 11 | 5 | - | - | 8 | |||||||||||||||
| Voluntary donors | - | - | 1 | 82 | 9 | 4 | - | - | 4 | |||||||||||||||
| Inpatients | - | - | - | 81 | 11 | 6 | - | - | 8 | |||||||||||||||
| IPA | Swine feces | - | - | - | - | 4 | 19 | 73 | 3 | 131,072 | 0.009 | |||||||||||||
| Pork meat | - | - | - | - | 4 | 12 | 65 | 18 | 262,144 | |||||||||||||||
| Voluntary donors | - | - | - | 1 | 3 | 19 | 68 | 5 | 262,144 | |||||||||||||||
| Inpatients | - | - | - | - | 5 | 18 | 73 | 2 | 131,072 | |||||||||||||||
| NaOCl | Swine feces | - | - | - | 86 | 13 | 1 | - | 1024 | 0.178 | ||||||||||||||
| Pork meat | - | - | - | 91 | 8 | - | - | 1024 | ||||||||||||||||
| Voluntary donors | - | - | 5 | 72 | 18 | 1 | - | 1024 | ||||||||||||||||
| Inpatients | - | - | - | 92 | 6 | - | - | 1024 | ||||||||||||||||
| PCMC | Swine feces | - | - | 1 | 90 | 9 | - | - | - | 1024 | 0.022 | |||||||||||||
| Pork meat | - | - | - | 99 | - | - | - | - | 512 | |||||||||||||||
| Voluntary donors | - | - | 2 | 84 | 10 | - | - | - | 1024 | |||||||||||||||
| Inpatients | - | 1 | 2 | 92 | 3 | - | - | - | 512 | |||||||||||||||
| Antimicrobials | Swine Feces, n (%) | Pork Meat, n (%) | Voluntary Donors, n (%) | Inpatients, n (%) | Total, n (%) | ||||
|---|---|---|---|---|---|---|---|---|---|
| ESBL (n = 48) | Non-ESBL (n = 52) | ESBL (n = 44) | Non-ESBL (n = 55) | ESBL (n = 51) | Non-ESBL (n = 45) | ESBL (n = 73) | Non-ESBL (n = 25) | E. coli (n = 393) | |
| Ampicillin | 48 (100) | 17 (33) | 44 (100) | 17 (31) | 51 (100) | 12 (27) | 73 (100) | 16 (64) | 278 (71) |
| Cefotaxime | 48 (100) | 0 | 44 (100) | 0 | 51 (100) | 0 | 73 (100) | 0 | 214 (55) |
| Ceftazidime | 48 (100) | 0 | 42 (95) | 0 | 51 (100) | 0 | 72 (99) | 0 | 211 (54) |
| Meropenem | 0 | 0 | 0 | 0 | 1 (2) | 0 | 4 (5) | 0 | 5 (1) |
| Nalidixic acid | 11 (23) | 5 (10) | 12 (27) | 3 (5) | 32 (63) | 6 (13) | 60 (82) | 14 (56) | 143 (36) |
| Ciprofloxacin | 12 (25) | 7 (13) | 13 (30) | 3 (5) | 33 (65) | 7 (16) | 59 (81) | 14 (56) | 148 (38) |
| Gentamicin | 4 (8) | 2 (4) | 6 (14) | 1 (2) | 7 (14) | 3 (7) | 11 (15) | 1 (4) | 35 (9) |
| Tetracycline | 30 (63) | 21 (40) | 22 (50) | 20 (36) | 30 (59) | 11 (24) | 41 (56) | 10 (40) | 185 (47) |
| Tigecycline | 1 (2) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (0.3) |
| Colistin | 7 (15) | 1 (2) | 0 | 1 (2) | 0 | 2 (4) | 0 | 0 | 11 (3) |
| Sulfamethoxazole | 31 (65) | 16 (31) | 30 (68) | 18 (33) | 42 (82) | 10 (22) | 53 (73) | 12 (48) | 212 (54) |
| Trimethoprim | 28 (58) | 11 (21) | 26 (59) | 18 (33) | 43 (84) | 9 (20) | 52 (71) | 13 (52) | 200 (51) |
| Chloramphenicol | 13 (27) | 5 (10) | 5 (11) | 9 (16) | 8 (16) | 3 (7) | 16 (22) | 4 (16) | 63 (16) |
| Azithromycin | 10 (21) | 1 (2) | 9 (21) | 0 | 16 (31) | 3 (7) | 24 (33) | 5 (20) | 68 (17) |
| Antimicrobials | Biocides | |||||
|---|---|---|---|---|---|---|
| Glutaraldehyde | Chlorhexidine Digluconate | Benzalkonium Chloride | ||||
| MIC | MBC | MIC | MBC | MIC | MBC | |
| Ampicillin | - | - | - | - | 0.119 ** | 0.121 * |
| Cefotaxime | - | 0.100 * | - | 0.117 * | - | - |
| Ceftazidime | - | - | - | - | 0.111 * | - |
| Meropenem | 0.127 * | 0.110 * | - | - | - | - |
| Nalidixic acid | 0.113 * | - | 0.198 ** | 0.280 ** | 0.136 ** | - |
| Ciprofloxacin | 0.109 * | - | 0.208 ** | 0.276 ** | 0.144 ** | - |
| Gentamicin | 0.164 ** | - | 0.228 ** | 0.195 ** | - | |
| Tetracycline | - | - | 0.229 ** | 0.246 ** | 0.100 * | - |
| Tigecycline | - | - | - | - | - | 0.121 * |
| Sulfamethoxazole | - | - | - | - | 0.155 ** | 0.128 * |
| Trimethoprim | - | - | - | - | 0.144 ** | 0.155 ** |
| Chloramphenicol | - | - | 0.109 * | 0.103 * | - | 0.099 * |
| Azithromycin | - | - | 0.152 ** | 0.138 ** | 0.173 ** | 0.152** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Silva, D.A.V.; Dieckmann, R.; Makarewicz, O.; Hartung, A.; Bethe, A.; Grobbel, M.; Belik, V.; Pletz, M.W.; Al Dahouk, S.; Neuhaus, S. Biocide Susceptibility and Antimicrobial Resistance of Escherichia coli Isolated from Swine Feces, Pork Meat and Humans in Germany. Antibiotics 2023, 12, 823. https://doi.org/10.3390/antibiotics12050823
da Silva DAV, Dieckmann R, Makarewicz O, Hartung A, Bethe A, Grobbel M, Belik V, Pletz MW, Al Dahouk S, Neuhaus S. Biocide Susceptibility and Antimicrobial Resistance of Escherichia coli Isolated from Swine Feces, Pork Meat and Humans in Germany. Antibiotics. 2023; 12(5):823. https://doi.org/10.3390/antibiotics12050823
Chicago/Turabian Styleda Silva, David Attuy Vey, Ralf Dieckmann, Oliwia Makarewicz, Anita Hartung, Astrid Bethe, Mirjam Grobbel, Vitaly Belik, Mathias W. Pletz, Sascha Al Dahouk, and Szilvia Neuhaus. 2023. "Biocide Susceptibility and Antimicrobial Resistance of Escherichia coli Isolated from Swine Feces, Pork Meat and Humans in Germany" Antibiotics 12, no. 5: 823. https://doi.org/10.3390/antibiotics12050823
APA Styleda Silva, D. A. V., Dieckmann, R., Makarewicz, O., Hartung, A., Bethe, A., Grobbel, M., Belik, V., Pletz, M. W., Al Dahouk, S., & Neuhaus, S. (2023). Biocide Susceptibility and Antimicrobial Resistance of Escherichia coli Isolated from Swine Feces, Pork Meat and Humans in Germany. Antibiotics, 12(5), 823. https://doi.org/10.3390/antibiotics12050823







