Nisin Z Potential for the Control of Diabetic Foot Infections Promoted by Pseudomonas aeruginosa Persisters
Abstract
1. Introduction
2. Results
2.1. P. aeruginosa DFI Susceptibility to Ciprofloxacin
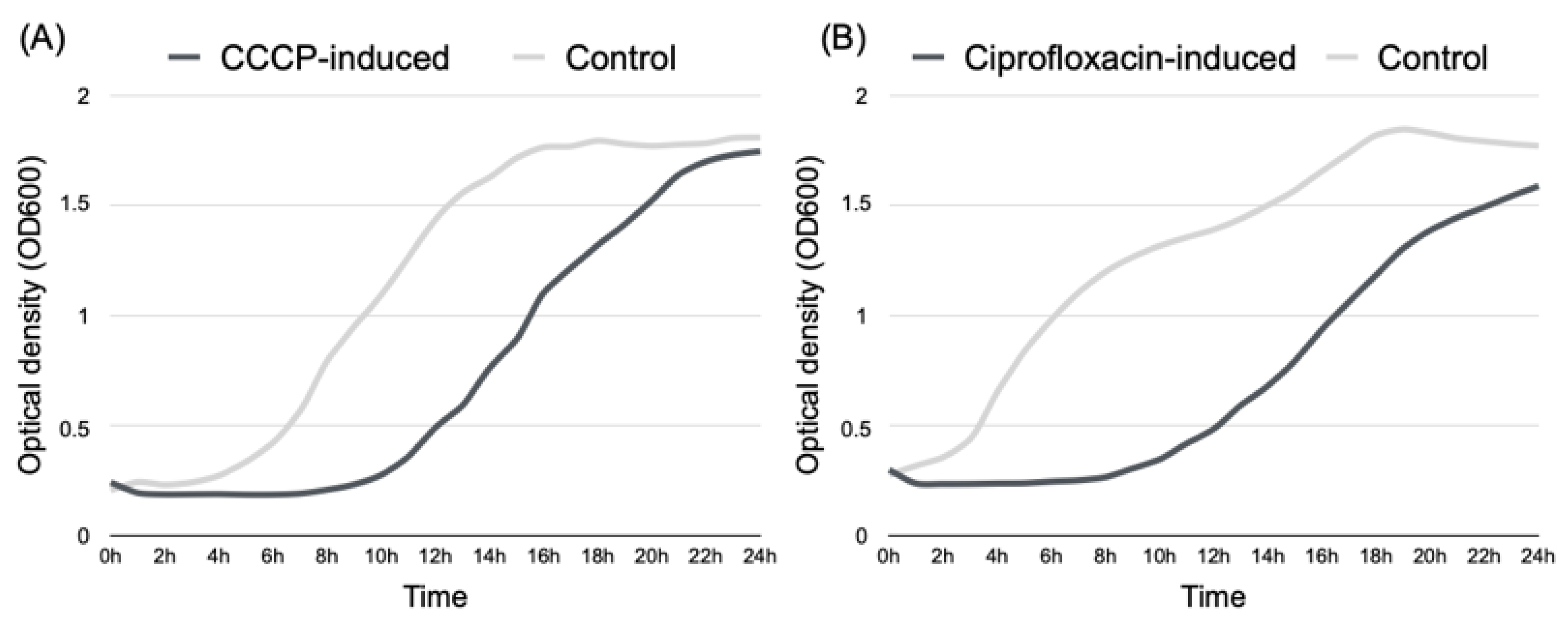
2.2. Induction of P. aeruginosa Persister Cells
2.3. Persisters’ Susceptibility to Ciprofloxacin
2.4. Efficacy of Persister Treatment with Nisin Z
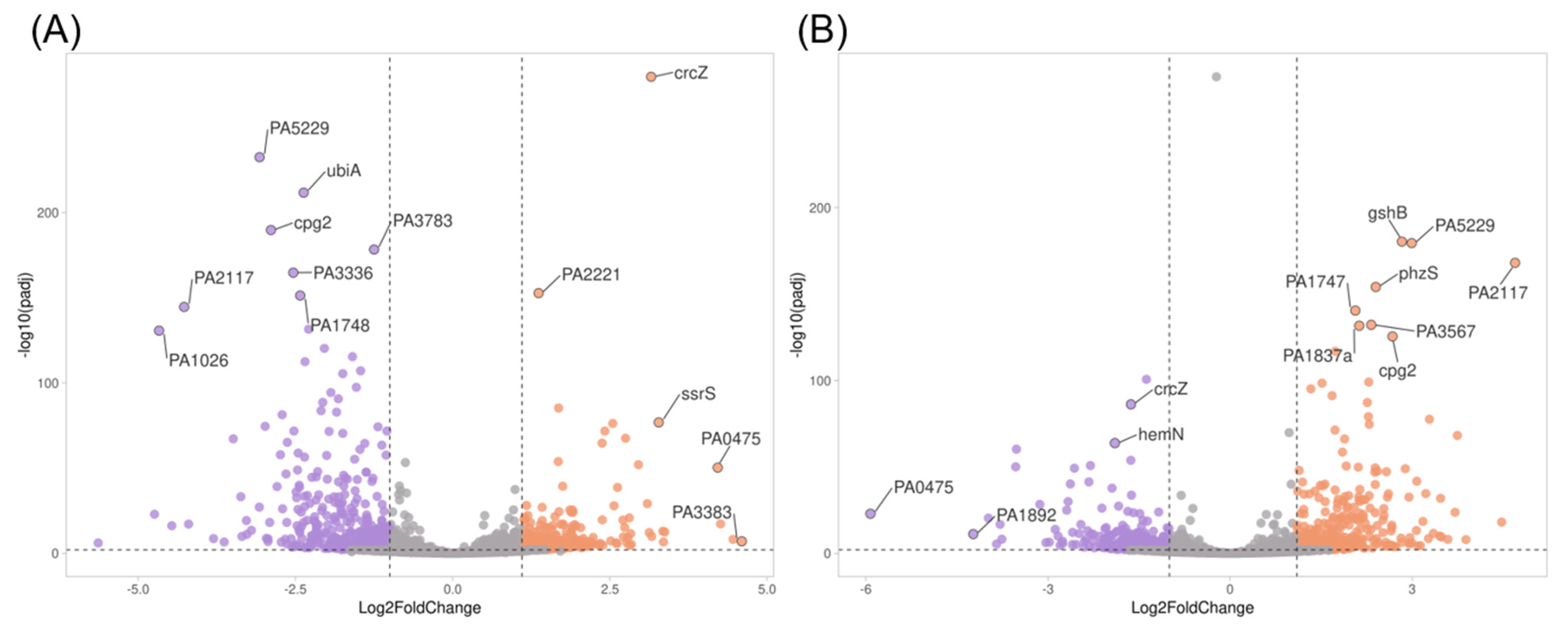
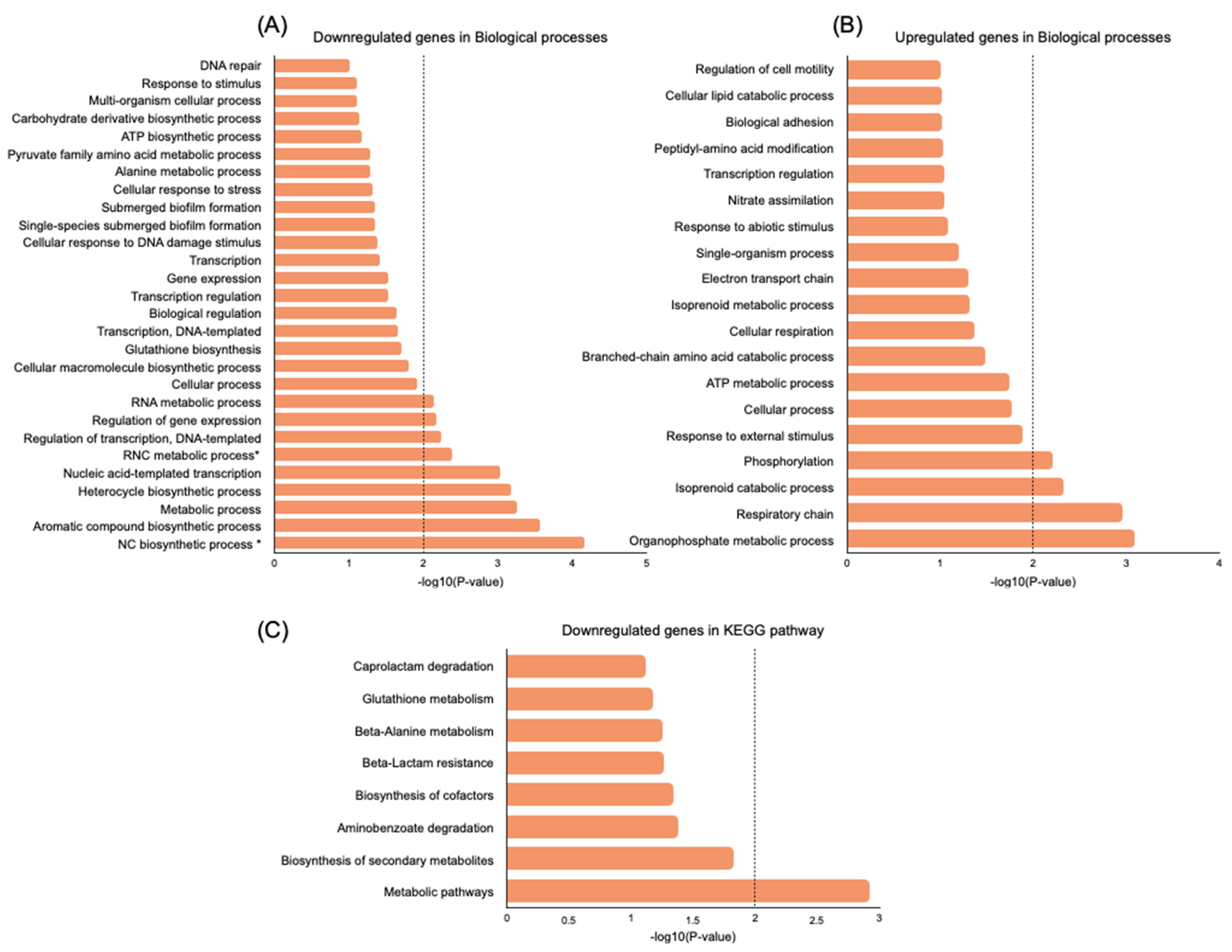
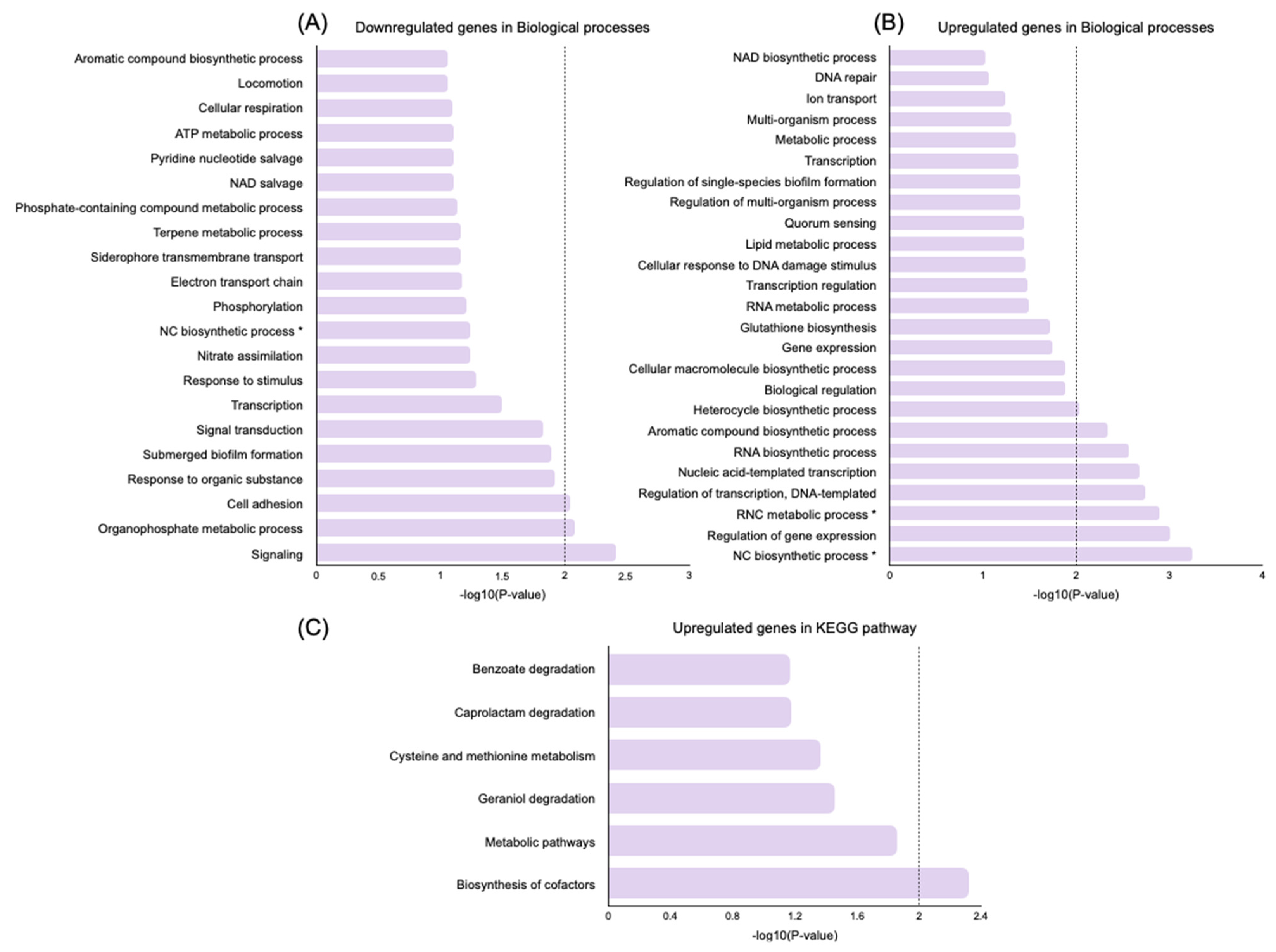
2.5. Transcriptome Evaluation of Persister Cell Gene Expression
Genes Related to Cell Wall Synthesis, Stress Response, Biofilm Formation
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Stock Solutions
4.3. Determination of the Minimum Inhibitory Concentration (MIC) and Minimum Biofilm Inhibitory Concentration (MBIC) of Ciprofloxacin towards P. aeruginosa
4.4. Induction of Persister Cells in P. aeruginosa Planktonic Cultures using CCCP
4.5. Induction of Persister Cells in P. aeruginosa Biofilms Using Ciprofloxacin
4.6. Evaluation of the Nisin Z Activity against Planktonic and Biofilm Persister Cells
4.7. RNA Extraction, Sequencing, and Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| diabetic foot ulcer | DFU |
| diabetic foot infection | DFI |
| carbonyl cyanide m-chlorophenylhydrazone | CCCP |
| antimicrobial peptides | AMPs |
| minimum inhibitory concentration | MIC |
| minimum biofilm inhibitory concentration | MBIC |
| minimum biofilm eradication concentration | MBEC |
| differentially expressed genes | DEGs |
| gene ontology | GO |
| Kyoto Encyclopedia of Genes and Genomes | KEGG |
| Database for Annotation, Visualization and Integrated discovery | DAVID |
| dimethyl sulfoxide | DMSO |
| Mueller-Hinton broth | MHb |
| tryptic soy broth | TSB |
| sodium chloride | NaCl |
| Luria-Bertani medium | LB |
| phosphate-buffered saline | PBS |
| tryptic soy agar | TSA |
| sequence alignment and map | SAM |
References
- Noor, S.; Zubair, M.; Ahmad, J. Diabetic foot ulcer-A review on pathophysiology, classification and microbial etiology. Diabetes Metab. Syndr. 2015, 9, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Noor, S.; Khan, R.U.; Ahmad, J. Understanding Diabetic Foot Infection and its Management. Diabetes Metab. Syndr. 2017, 11, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Oliver, T.I.; Mutluoglu, M. Diabetic Foot Ulcer. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Boulton, A.J.M.; Armstrong, D.G.; Hardman, M.J.; Malone, M.; Embil, J.M.; Attinger, C.E.; Lipsky, B.A.; Aragón-Sánchez, J.; Li, H.K.; Schultz, G.; et al. Diagnosis and Management of Diabetic Foot Infections. Compendia 2020, 2020. [Google Scholar] [CrossRef] [PubMed]
- Mendes, J.J.; Leandro, C.; Mottola, C.; Barbosa, R.; Silva, F.A.; Oliveira, M.; Vilela, C.L.; Melo-Cristino, J.; Górski, A.; Pimentel, M.; et al. In vitro design of a novel lytic bacteriophage cocktail with therapeutic potential against organisms causing diabetic foot infections. J. Med. Microbiol. 2014, 63, 1055–1065. [Google Scholar] [CrossRef]
- Cendra, M.D.M.; Torrents, E. Pseudomonas aeruginosa biofilms and their partners in crime. Biotechnol. Adv. 2021, 49, 107734. [Google Scholar] [CrossRef]
- Ciofu, O.; Tolker-Nielsen, T. Tolerance and Resistance of Pseudomonas aeruginosa Biofilms to Antimicrobial Agents-How P. aeruginosa Can Escape Antibiotics. Front. Microbiol. 2019, 10, 913. [Google Scholar] [CrossRef]
- Serrano, I.; Oliveira, M.; Santos, J.P.; Bilocq, F.; Leitão, A.; Tavares, L.; Pirnay, J.P.; De Vos, D. Antimicrobial resistance and genomic rep-PCR fingerprints of Pseudomonas aeruginosa strains from animals on the background of the global population structure. BMC Vet. Res. 2017, 13, 58. [Google Scholar] [CrossRef]
- Wu, W.; Jin, Y.; Bai, F.; Jin, S. Pseudomonas aeruginosa; Academic Press: Cambridge, MA, USA, 2015; pp. 753–767. [Google Scholar]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef]
- Grassi, L.; Di Luca, M.; Maisetta, G.; Rinaldi, A.C.; Esin, S.; Trampuz, A.; Batoni, G. Generation of Persister Cells of Pseudomonas aeruginosa and Staphylococcus aureus by Chemical Treatment and Evaluation of Their Susceptibility to Membrane-Targeting Agents. Front. Microbiol. 2017, 8, 1917. [Google Scholar] [CrossRef]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Rybtke, M.; Hultqvist, L.D.; Givskov, M.; Tolker-Nielsen, T. Pseudomonas aeruginosa Biofilm Infections: Community Structure, Antimicrobial Tolerance and Immune Response. J. Mol. Biol. 2015, 427, 3628–3645. [Google Scholar] [CrossRef]
- Soares, A.; Alexandre, K.; Etienne, M. Tolerance and Persistence of Pseudomonas aeruginosa in Biofilms Exposed to Antibiotics: Molecular Mechanisms, Antibiotic Strategies and Therapeutic Perspectives. Front. Microbiol. 2020, 11, 2057. [Google Scholar] [CrossRef]
- Fisher, R.A.; Gollan, B.; Helaine, S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017, 15, 453–464. [Google Scholar] [CrossRef]
- Mulcahy, L.R.; Burns, J.L.; Lory, S.; Lewis, K. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 2010, 192, 6191–6199. [Google Scholar] [CrossRef]
- Bahar, A.A.; Ren, D. Antimicrobial Peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef]
- Gomes, D.; Santos, R.; S.Soares, R.; Reis, S.; Carvalho, S.; Rego, P.; C.Peleteiro, M.; Tavares, L.; Oliveira, M. Pexiganan in Combination with Nisin to Control Polymicrobial Diabetic Foot Infections. Antibiotics 2020, 9, 128. [Google Scholar] [CrossRef]
- Scheper, H.; Wubbolts, J.M.; Verhagen, J.A.M.; de Visser, A.W.; van der Wal, R.J.P.; Visser, L.G.; de Boer, M.G.J.; Nibbering, P.H. SAAP-148 Eradicates MRSA Persisters within Mature Biofilm Models Simulating Prosthetic Joint Infection. Front. Microbiol. 2021, 12, 625952. [Google Scholar] [CrossRef]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef]
- Le Blay, G.; Lacroix, C.; Zihler, A.; Fliss, I. In vitro inhibition activity of nisin A, nisin Z, pediocin PA-1 and antibiotics against common intestinal bacteria. Lett. Appl. Microbiol. 2007, 45, 252–257. [Google Scholar] [CrossRef]
- Wu, J.; Hu, S.; Cao, L. Therapeutic effect of nisin Z on subclinical mastitis in lactating cows. Antimicrob. Agents Chemother. 2007, 51, 3131–3135. [Google Scholar] [CrossRef]
- Bolocan, A.S.; Pennone, V.; O’Connor, P.M.; Coffey, A.; Nicolau, A.I.; McAuliffe, O.; Jordan, K. Inhibition of Listeria monocytogenes biofilms by bacteriocin-producing bacteria isolated from mushroom substrate. J. Appl. Microbiol. 2017, 122, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Mu, D.; Qiao, W.; Zhu, D.; Wang, X.; Liu, F.; Xu, H.; Saris, P.; Kuipers, O.P.; Qiao, M. Co-expression of Nisin Z and Leucocin C as a Basis for Effective Protection Against Listeria monocytogenes in Pasteurized Milk. Front. Microbiol. 2018, 9, 547. [Google Scholar] [CrossRef] [PubMed]
- Niaz, T.; Shabbir, S.; Noor, T.; Abbasi, R.; Raza, Z.A.; Imran, M. Polyelectrolyte Multicomponent Colloidosomes Loaded with Nisin Z for Enhanced Antimicrobial Activity against Foodborne Resistant Pathogens. Front. Microbiol. 2017, 8, 2700. [Google Scholar] [CrossRef] [PubMed]
- Akerey, B.; Le-Lay, C.; Fliss, I.; Subirade, M.; Rouabhia, M. In vitro efficacy of nisin Z against Candida albicans adhesion and transition following contact with normal human gingival cells. J. Appl. Microbiol. 2009, 107, 1298–1307. [Google Scholar] [CrossRef]
- Kart, D.; Reçber, T.; Nemutlu, E.; Sagiroglu, M. Sub-Inhibitory Concentrations of Ciprofloxacin Alone and Combinations with Plant-Derived Compounds against P. aeruginosa Biofilms and Their Effects on the Metabolomic Profile of P. aeruginosa Biofilms. Antibiotics 2021, 10, 414. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Kwan, B.W.; Valenta, J.A.; Benedik, M.J.; Wood, T.K. Arrested protein synthesis increases persister-like cell formation. Antimicrob. Agents Chemother. 2013, 57, 1468–1473. [Google Scholar] [CrossRef]
- Tkhilaishvili, T.; Lombardi, L.; Klatt, A.B.; Trampuz, A.; Di Luca, M. Bacteriophage Sb-1 enhances antibiotic activity against biofilm, degrades exopolysaccharide matrix and targets persisters of Staphylococcus aureus. Int. J. Antimicrob. Agents 2018, 52, 842–853. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, Z.; Xu, Y.; Lu, J.; Tang, W.; Zhang, J. Effect of new carbonyl cyanide aromatic hydrazones on biofilm inhibition against methicillin resistant Staphylococcus aureus. RSC Adv. 2020, 10, 17854–17861. [Google Scholar] [CrossRef]
- Ghoul, M.; Pommepuy, M.; Moillo-Batt, A.; Cormier, M. Effect of carbonyl cyanide m-chlorophenylhydrazone on Escherichia coli halotolerance. Appl. Environ. Microbiol. 1989, 55, 1040–1043. [Google Scholar] [CrossRef]
- Dörr, T.; Vulić, M.; Lewis, K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010, 8, e1000317. [Google Scholar] [CrossRef]
- Conlon, B.P.; Rowe, S.E.; Lewis, K. Persister cells in biofilm associated infections. Adv. Exp. Med. Biol. 2015, 831, 1–9. [Google Scholar] [CrossRef]
- Wood, T.K.; Knabel, S.J.; Kwan, B.W. Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 2013, 79, 7116–7121. [Google Scholar] [CrossRef]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar]
- Shai, Y. Mode of action of membrane active antimicrobial peptides. Biopolymers 2002, 66, 236–248. [Google Scholar] [CrossRef]
- Serrano, I.; Alhinho, B.; Cunha, E.; Tavares, L.; Trindade, A.; Oliveira, M. Bacteriostatic and Antibiofilm Efficacy of a Nisin Z Solution against Co-Cultures of Staphylococcus aureus and Pseudomonas aeruginosa from Diabetic Foot Infections. Life 2023, 13, 504. [Google Scholar] [CrossRef]
- Thöming, J.G.; Tomasch, J.; Preusse, M.; Koska, M.; Grahl, N.; Pohl, S.; Willger, S.D.; Kaever, V.; Müsken, M.; Häussler, S. Parallel evolutionary paths to produce more than one Pseudomonas aeruginosa biofilm phenotype. NPJ Biofilms Microbiomes 2020, 6, 2. [Google Scholar] [CrossRef]
- D’Arpa, P.; Karna, S.L.R.; Chen, T.; Leung, K.P. Pseudomonas aeruginosa transcriptome adaptations from colonization to biofilm infection of skin wounds. Sci. Rep. 2021, 11, 20632. [Google Scholar] [CrossRef]
- Brazas, M.D.; Hancock, R.E. Ciprofloxacin induction of a susceptibility determinant in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2005, 49, 3222–3227. [Google Scholar] [CrossRef]
- Murray, J.L.; Kwon, T.; Marcotte, E.M.; Whiteley, M. Intrinsic Antimicrobial Resistance Determinants in the Superbug Pseudomonas aeruginosa. mBio 2015, 6, e01603–e01615. [Google Scholar] [CrossRef]
- Zhou, J.W.; Muhammad, J.; Sun, B.; Yang, R.; Wadood, A.; Wang, J.S.; Jia, A.Q. Metabolomic analysis of quorum sensing inhibitor hordenine on Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2019, 103, 6271–6285. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ji, P.C.; Qi, Y.H.; Chen, S.J.; Wang, C.Y.; Yang, Y.J.; Zhao, X.Y.; Zhou, J.W. Inactivation of Pseudomonas aeruginosa biofilms by thymoquinone in combination with nisin. Front. Microbiol. 2022, 13, 1029412. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, M.T.; Schiller, N.L. Alginate lyase (AlgL) activity is required for alginate biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 2005, 187, 3869–3872. [Google Scholar] [CrossRef] [PubMed]
- Ryder, C.; Byrd, M.; Wozniak, D.J. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 2007, 10, 644–648. [Google Scholar] [CrossRef]
- Soares, A.; Roussel, V.; Pestel-Caron, M.; Barreau, M.; Caron, F.; Bouffartigues, E.; Chevalier, S.; Etienne, M. Understanding Ciprofloxacin Failure in Pseudomonas aeruginosa Biofilm: Persister Cells Survive Matrix Disruption. Front. Microbiol. 2019, 10, 2603. [Google Scholar] [CrossRef]
- Pagès, J.M.; Masi, M.; Barbe, J. Inhibitors of efflux pumps in Gram-negative bacteria. Trends Mol. Med. 2005, 11, 382–389. [Google Scholar] [CrossRef]
- Chen, H.; Hu, J.; Chen, P.R.; Lan, L.; Li, Z.; Hicks, L.M.; Dinner, A.R.; He, C. The Pseudomonas aeruginosa multidrug efflux regulator MexR uses an oxidation-sensing mechanism. Proc. Natl. Acad. Sci. USA 2008, 105, 13586–13591. [Google Scholar] [CrossRef]
- Wang, D.; Seeve, C.; Pierson, L.S., 3rd; Pierson, E.A. Transcriptome profiling reveals links between ParS/ParR, MexEF-OprN, and quorum sensing in the regulation of adaptation and virulence in Pseudomonas aeruginosa. BMC Genomics 2013, 14, 618. [Google Scholar] [CrossRef]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef]
- Moscoso, J.A.; Mikkelsen, H.; Heeb, S.; Williams, P.; Filloux, A. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 2011, 13, 3128–3138. [Google Scholar] [CrossRef]
- Sugawara, E.; Nagano, K.; Nikaido, H. Alternative folding pathways of the major porin OprF of Pseudomonas aeruginosa. Febs j 2012, 279, 910–918. [Google Scholar] [CrossRef]
- Cornforth, D.M.; Dees, J.L.; Ibberson, C.B.; Huse, H.K.; Mathiesen, I.H.; Kirketerp-Møller, K.; Wolcott, R.D.; Rumbaugh, K.P.; Bjarnsholt, T.; Whiteley, M. Pseudomonas aeruginosa transcriptome during human infection. Proc. Natl. Acad. Sci. USA 2018, 115, E5125–E5134. [Google Scholar] [CrossRef]
- Mendes, J.J.; Marques-Costa, A.; Vilela, C.; Neves, J.; Candeias, N.; Cavaco-Silva, P.; Melo-Cristino, J. Clinical and bacteriological survey of diabetic foot infections in Lisbon. Diabetes Res. Clin. Pract. 2012, 95, 153–161. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI Supplement M100: Wayne, PA, USA, 2020. [Google Scholar]

| P. aeruginosa Z25.1 | P. aeruginosa ATCC 27583 | |
|---|---|---|
| MIC | 4.889 ± 0.588 µg/mL | 0.250 ± 0 µg/mL |
| MBIC | 2.444 ± 0.294 µg/mL | 0.361 ± 0.044 µg/mL |
| Condition | Biological Process | Gene | Functional Protein | Log2 Fold Change (*) |
|---|---|---|---|---|
| PCs vs. C | Cell wall synthesis | algK | Alginate biosynthesis protein | −1.96 |
| algL | Alginate lyase | −2.23 | ||
| Stress response | mexR | Multidrug resistance operon repressor | −1.12 | |
| oprN | Multidrug efflux outer membrane protein | −1.27 | ||
| parS | Two-component sensor | 1.59 | ||
| Biofilm formation | lasR | Transcriptional regulator | −1.7 | |
| pilJ | Twitching motility protein | 1.18 | ||
| retS | Sensor histidine kinase MifS | 1.41 | ||
| NPCs vs. PCs | Cell wall synthesis | algB | Alginate biosynthesis transcriptional regulatory protein AlgB | −1.15 |
| algL | Alginate lyase | 1.97 | ||
| algP | Transcriptional regulatory protein ALgP | 1.08 | ||
| algX | Alginate biosynthesis protein AlgX | 1.01 | ||
| Stress response | mexR | Multidrug resistance operon repressor | 1.49 | |
| oprF | Outer membrane porin F | 1.7 | ||
| oprG | Outer membrane protein OprG | 1.14 | ||
| oprI | Outer membrane lipoprotein I | 3.57 | ||
| oprN | Multidrug efflux outer membrane protein | 1.75 | ||
| parS | Two-component sensor | −1.68 | ||
| Biofilm formation | lasR | Transcriptional regulator | 1.45 | |
| retS | Sensor histidine kinase MifS | −2.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zina, R.; Cunha, E.; Serrano, I.; Silva, E.; Tavares, L.; Oliveira, M. Nisin Z Potential for the Control of Diabetic Foot Infections Promoted by Pseudomonas aeruginosa Persisters. Antibiotics 2023, 12, 794. https://doi.org/10.3390/antibiotics12050794
Zina R, Cunha E, Serrano I, Silva E, Tavares L, Oliveira M. Nisin Z Potential for the Control of Diabetic Foot Infections Promoted by Pseudomonas aeruginosa Persisters. Antibiotics. 2023; 12(5):794. https://doi.org/10.3390/antibiotics12050794
Chicago/Turabian StyleZina, Rafaela, Eva Cunha, Isa Serrano, Elisabete Silva, Luís Tavares, and Manuela Oliveira. 2023. "Nisin Z Potential for the Control of Diabetic Foot Infections Promoted by Pseudomonas aeruginosa Persisters" Antibiotics 12, no. 5: 794. https://doi.org/10.3390/antibiotics12050794
APA StyleZina, R., Cunha, E., Serrano, I., Silva, E., Tavares, L., & Oliveira, M. (2023). Nisin Z Potential for the Control of Diabetic Foot Infections Promoted by Pseudomonas aeruginosa Persisters. Antibiotics, 12(5), 794. https://doi.org/10.3390/antibiotics12050794







