Targeting BAM for Novel Therapeutics against Pathogenic Gram-Negative Bacteria
Abstract
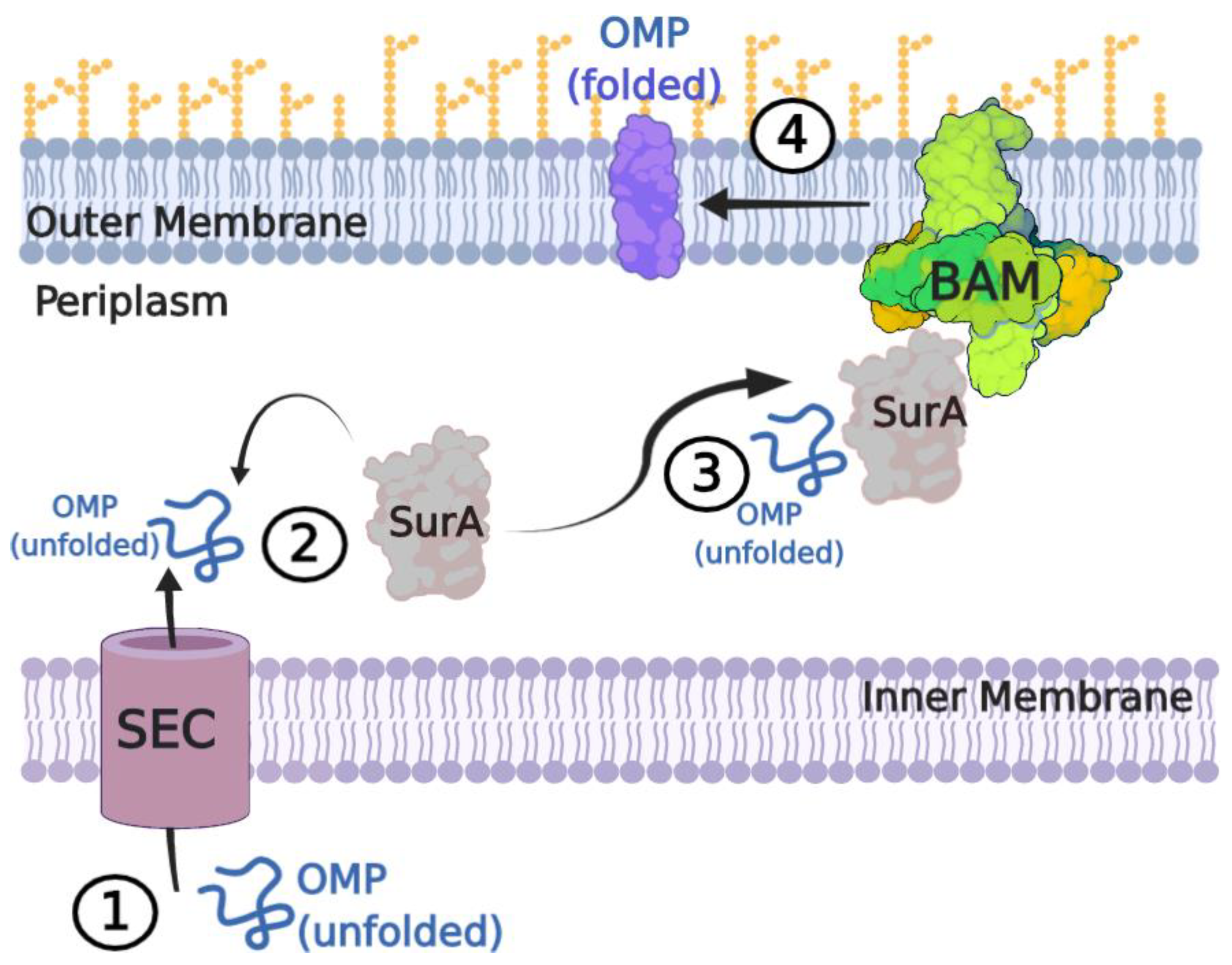
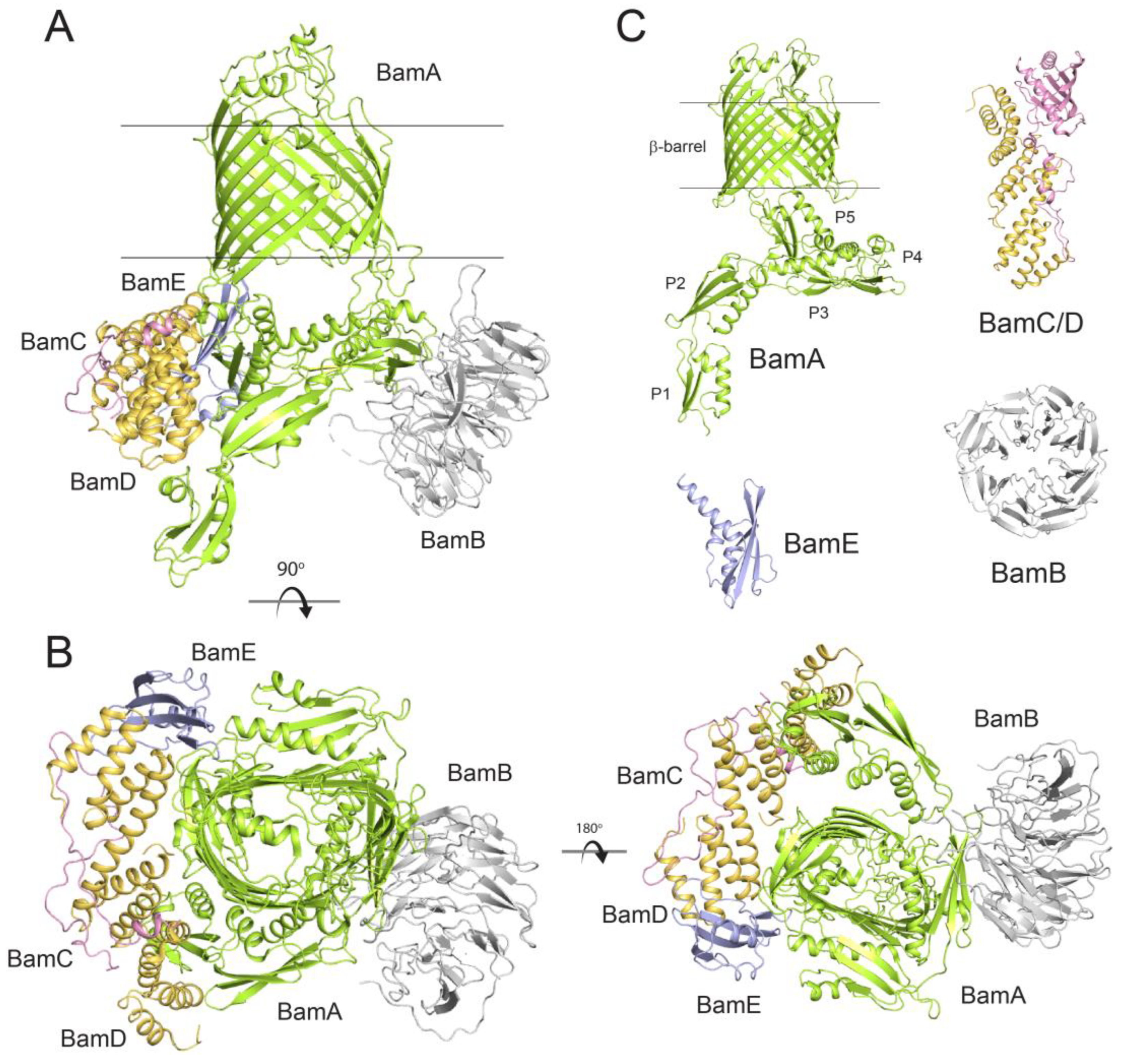
1. Introduction
2. Mutations in BAM Demonstrate Druggability and Enhancement of Antibiotic Sensitivity
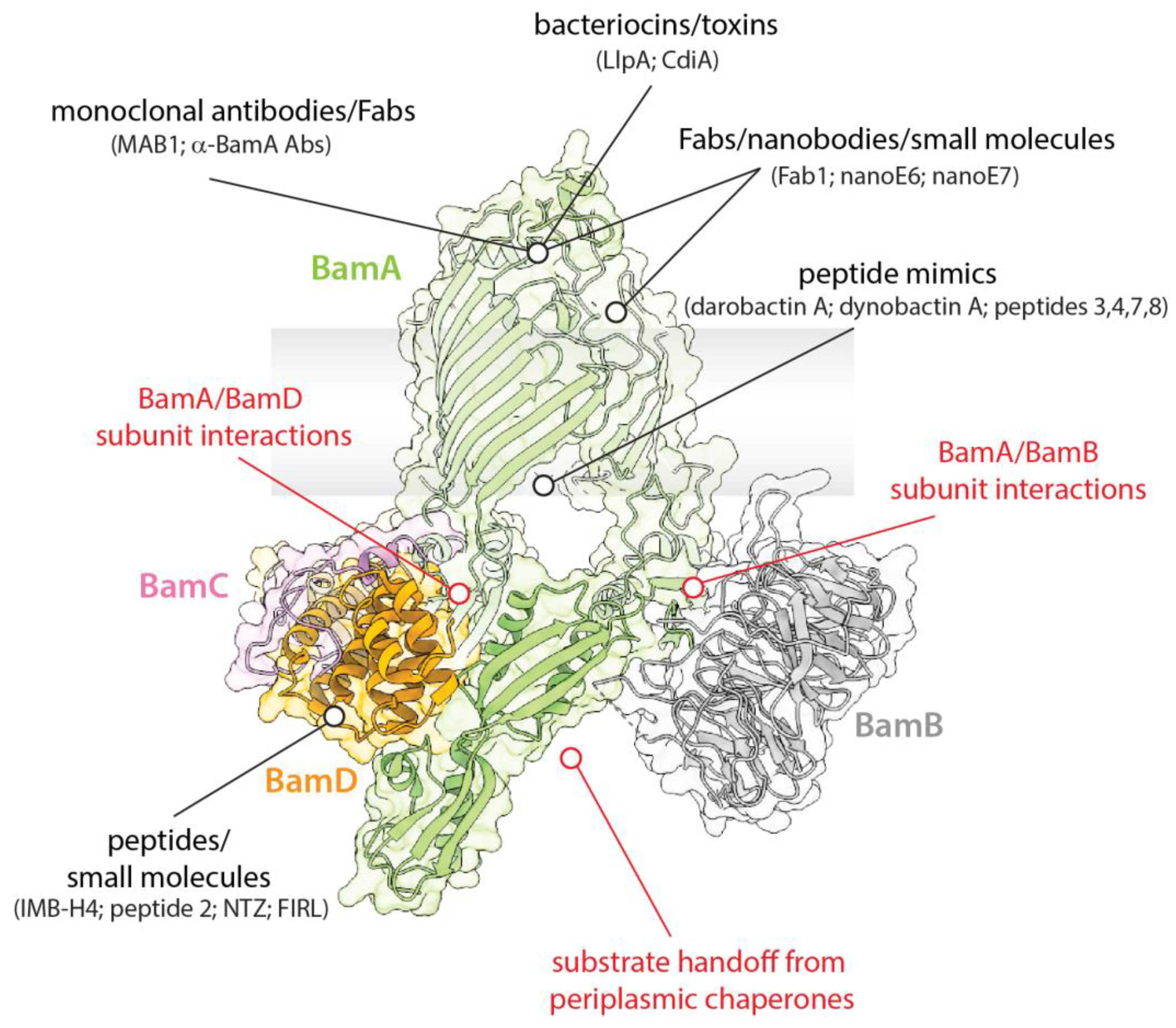
3. Targeting BAM with Small Molecules
4. Targeting BAM with Peptides and Proteins
5. Bacterial Warfare Using BAM: Lectin-like Bacteriocins and Contact-Dependent Growth Inhibition
6. Targeting BAM for Vaccines
7. Summary and Future Outlook
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- CDC. Antibiotic Resistance Threats in the United States; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019.
- Lin, J.; Huang, S.; Zhang, Q. Outer membrane proteins: Key players for bacterial adaptation in host niches. Microbes Infect. 2002, 4, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Ricci, D.P.; Silhavy, T.J. The Bam machine: A molecular cooper. Biochim. Biophys. Acta 2012, 1818, 1067–1084. [Google Scholar] [CrossRef] [PubMed]
- Bos, M.P.; Robert, V.; Tommassen, J. Biogenesis of the gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 2007, 61, 191–214. [Google Scholar] [CrossRef]
- Knowles, T.J.; Scott-Tucker, A.; Overduin, M.; Henderson, I.R. Membrane protein architects: The role of the BAM complex in outer membrane protein assembly. Nat. Rev. Microbiol. 2009, 7, 206–214. [Google Scholar] [CrossRef]
- Noinaj, N.; Rollauer, S.E.; Buchanan, S.K. The beta-barrel membrane protein insertase machinery from Gram-negative bacteria. Curr. Opin. Struct. Biol. 2015, 31, 35–42. [Google Scholar] [CrossRef]
- Rouvière, P.E.; Gross, C.A. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 1996, 10, 3170–3182. [Google Scholar] [CrossRef] [PubMed]
- Lazar, S.W.; Kolter, R. SurA assists the folding of Escherichia coli outer membrane proteins. J. Bacteriol. 1996, 178, 1770–1773. [Google Scholar] [CrossRef] [PubMed]
- Behrens, S.; Maier, R.; de Cock, H.; Schmid, F.X.; Gross, C.A. The SurA periplasmic PPIase lacking its parvulin domains functions in vivo and has chaperone activity. EMBO J. 2001, 20, 285–294. [Google Scholar] [CrossRef]
- Bitto, E.; McKay, D.B. Crystallographic Structure of SurA, a Molecular Chaperone that Facilitates Folding of Outer Membrane Porins. Structure 2002, 10, 1489–1498. [Google Scholar] [CrossRef]
- Jiang, J.; Zhang, X.; Chen, Y.; Wu, Y.; Zhou, Z.H.; Chang, Z.; Sui, S.-F. Activation of DegP chaperone-protease via formation of large cage-like oligomers upon binding to substrate proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 11939–11944. [Google Scholar] [CrossRef]
- Ge, X.; Wang, R.; Ma, J.; Liu, Y.; Ezemaduka, A.N.; Chen, P.R.; Fu, X.; Chang, Z. DegP primarily functions as a protease for the biogenesis of beta-barrel outer membrane proteins in the Gram-negative bacterium Escherichia coli. FEBS J. 2014, 281, 1226–1240. [Google Scholar] [CrossRef]
- Rollauer, S.E.; Sooreshjani, M.A.; Noinaj, N.; Buchanan, S.K. Outer membrane protein biogenesis in Gram-negative bacteria. Philos Trans. R. Soc. Lond B. Biol. Sci. 2015, 370. [Google Scholar] [CrossRef]
- Driessen, A.J.; Nouwen, N. Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 2008, 77, 643–667. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Malinverni, J.; Ruiz, N.; Kim, S.; Silhavy, T.J.; Kahne, D. Identification of a Multicomponent Complex Required for Outer Membrane Biogenesis in Escherichia coli. Cell 2005, 121, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Bakelar, J.; Buchanan, S.K.; Noinaj, N. The structure of the beta-barrel assembly machinery complex. Science 2016, 351, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Li, H.; Dong, H.; Zeng, Y.; Zhang, Z.; Paterson, N.G.; Stansfeld, P.J.; Wang, Z.; Zhang, Y.; Wang, W.; et al. Structural basis of outer membrane protein insertion by the BAM complex. Nature 2016, 531, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Zheng, J.; Wang, Y.; Yang, X.; Liu, Y.; Sun, C.; Cao, B.; Zhou, H.; Ni, D.; Lou, J.; et al. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat. Struct. Mol. Biol. 2016, 23, 192–196. [Google Scholar] [CrossRef]
- Wu, R.; Bakelar, J.W.; Lundquist, K.; Zhang, Z.; Kuo, K.M.; Ryoo, D.; Pang, Y.T.; Sun, C.; White, T.; Klose, T.; et al. Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM. Nat. Commun. 2021, 12, 7131. [Google Scholar] [CrossRef]
- Tomasek, D.; Rawson, S.; Lee, J.; Wzorek, J.S.; Harrison, S.C.; Li, Z.; Kahne, D. Structure of a nascent membrane protein as it folds on the BAM complex. Nature 2020, 583, 473–478. [Google Scholar] [CrossRef]
- Iadanza, M.G.; Higgins, A.J.; Schiffrin, B.; Calabrese, A.N.; Brockwell, D.J.; Ashcroft, A.E.; Radford, S.E.; Ranson, N.A. Lateral opening in the intact beta-barrel assembly machinery captured by cryo-EM. Nat. Commun. 2016, 7, 12865. [Google Scholar] [CrossRef]
- Iadanza, M.G.; Schiffrin, B.; White, P.; Watson, M.A.; Horne, J.E.; Higgins, A.J.; Calabrese, A.N.; Brockwell, D.J.; Tuma, R.; Kalli, A.C.; et al. Distortion of the bilayer and dynamics of the BAM complex in lipid nanodiscs. Commun. Biol. 2020, 3, 766. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Han, L.; Li, B.; Zhang, M.; Zhou, H.; Luo, Q.; Zhang, X.; Huang, Y. Structures of the beta-barrel assembly machine recognizing outer membrane protein substrates. FASEB J. 2021, 35, e21207. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Capalash, N.; Sharma, P. Immunoprotective potential of BamA, the outer membrane protein assembly factor, against MDR Acinetobacter baumannii. Sci. Rep. 2017, 7, 12411. [Google Scholar] [CrossRef] [PubMed]
- Noinaj, N.; Kuszak, A.J.; Gumbart, J.C.; Lukacik, P.; Chang, H.; Easley, N.C.; Lithgow, T.; Buchanan, S.K. Structural insight into the biogenesis of beta-barrel membrane proteins. Nature 2013, 501, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.; Hsu, S.C.; Bedard, J.; Inoue, K.; Jarvis, P. The Omp85-related chloroplast outer envelope protein OEP80 is essential for viability in Arabidopsis. Plant Physiol. 2008, 148, 235–245. [Google Scholar] [CrossRef]
- Hsu, S.C.; Patel, R.; Bedard, J.; Jarvis, P.; Inoue, K. Two distinct Omp85 paralogs in the chloroplast outer envelope membrane are essential for embryogenesis in Arabidopsis thaliana. Plant Signal Behav. 2008, 3, 1134–1135. [Google Scholar] [CrossRef]
- Topel, M.; Ling, Q.; Jarvis, P. Neofunctionalization within the Omp85 protein superfamily during chloroplast evolution. Plant Signal Behav. 2012, 7, 161–164. [Google Scholar] [CrossRef]
- Noinaj, N.; Gumbart, J.C.; Buchanan, S.K. The beta-barrel assembly machinery in motion. Nat. Rev. Microbiol. 2017, 15, 197–204. [Google Scholar] [CrossRef]
- Bakelar, J.; Buchanan, S.K.; Noinaj, N. Structural snapshots of the beta-barrel assembly machinery. FEBS J. 2017, 284, 1778–1786. [Google Scholar] [CrossRef]
- Wu, R.; Stephenson, R.; Gichaba, A.; Noinaj, N. The big BAM theory: An open and closed case? Biochim. Et Biophys. Acta. Biomembr. 2020, 1862, 183062. [Google Scholar] [CrossRef]
- Noinaj, N.; Kuszak, A.J.; Balusek, C.; Gumbart, J.C.; Buchanan, S.K. Lateral Opening and Exit Pore Formation Are Required for BamA Function. Structure 2014, 22, 1055–1062. [Google Scholar] [CrossRef] [PubMed]
- Webb, C.T.; Selkrig, J.; Perry, A.J.; Noinaj, N.; Buchanan, S.K.; Lithgow, T. Dynamic association of BAM complex modules includes surface exposure of the lipoprotein BamC. J. Mol. Biol. 2012, 422, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Rassam, P.; Copeland, N.A.; Birkholz, O.; Toth, C.; Chavent, M.; Duncan, A.L.; Cross, S.J.; Housden, N.G.; Kaminska, R.; Seger, U.; et al. Supramolecular assemblies underpin turnover of outer membrane proteins in bacteria. Nature 2015, 523, 333–336. [Google Scholar] [CrossRef] [PubMed]
- Sikora, A.E.; Wierzbicki, I.H.; Zielke, R.A.; Ryner, R.F.; Korotkov, K.V.; Buchanan, S.K.; Noinaj, N. Structural and functional insights into the role of BamD and BamE within the beta-barrel assembly machinery in Neisseria gonorrhoeae. J. Biol. Chem. 2018, 293, 1106–1119. [Google Scholar] [CrossRef]
- Bennion, D.; Charlson, E.S.; Coon, E.; Misra, R. Dissection of beta-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol. Microbiol. 2010, 77, 1153–1171. [Google Scholar] [CrossRef] [PubMed]
- Browning, D.F.; Matthews, S.A.; Rossiter, A.E.; Sevastsyanovich, Y.R.; Jeeves, M.; Mason, J.L.; Wells, T.J.; Wardius, C.A.; Knowles, T.J.; Cunningham, A.F.; et al. Mutational and topological analysis of the Escherichia coli BamA protein. PLoS ONE 2013, 8, e84512. [Google Scholar] [CrossRef] [PubMed]
- Ghequire, M.G.K.; Swings, T.; Michiels, J.; Buchanan, S.K.; De Mot, R. Hitting with a BAM: Selective Killing by Lectin-Like Bacteriocins. mBio 2018, 9, e02138-17. [Google Scholar] [CrossRef]
- Hart, E.M.; Mitchell, A.M.; Konovalova, A.; Grabowicz, M.; Sheng, J.; Han, X.; Rodriguez-Rivera, F.P.; Schwaid, A.G.; Malinverni, J.C.; Balibar, C.J.; et al. A small-molecule inhibitor of BamA impervious to efflux and the outer membrane permeability barrier. Proc. Natl. Acad. Sci. USA 2019, 116, 21748–21757. [Google Scholar] [CrossRef]
- Imai, Y.; Meyer, K.J.; Iinishi, A.; Favre-Godal, Q.; Green, R.; Manuse, S.; Caboni, M.; Mori, M.; Niles, S.; Ghiglieri, M.; et al. A new antibiotic selectively kills Gram-negative pathogens. Nature 2019, 576, 459–464. [Google Scholar] [CrossRef]
- Kaur, H.; Jakob, R.P.; Marzinek, J.K.; Green, R.; Imai, Y.; Bolla, J.R.; Agustoni, E.; Robinson, C.V.; Bond, P.J.; Lewis, K.; et al. The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase. Nature 2021, 593, 125–129. [Google Scholar] [CrossRef]
- Lee, J.; Sutterlin, H.A.; Wzorek, J.S.; Mandler, M.D.; Hagan, C.L.; Grabowicz, M.; Tomasek, D.; May, M.D.; Hart, E.M.; Silhavy, T.J.; et al. Substrate binding to BamD triggers a conformational change in BamA to control membrane insertion. Proc. Natl. Acad. Sci. USA 2018, 115, 2359–2364. [Google Scholar] [CrossRef] [PubMed]
- Leonard-Rivera, M.; Misra, R. Conserved residues of the putative L6 loop of Escherichia coli BamA play a critical role in the assembly of beta-barrel outer membrane proteins, including that of BamA itself. J. Bacteriol. 2012, 194, 4662–4668. [Google Scholar] [CrossRef] [PubMed]
- Lundquist, K.; Bakelar, J.; Noinaj, N.; Gumbart, J.C. C-terminal kink formation is required for lateral gating in BamA. Proc. Natl. Acad. Sci. USA 2018, 115, E7942–E7949. [Google Scholar] [CrossRef] [PubMed]
- Luther, A.; Urfer, M.; Zahn, M.; Muller, M.; Wang, S.Y.; Mondal, M.; Vitale, A.; Hartmann, J.B.; Sharpe, T.; Monte, F.L.; et al. Chimeric peptidomimetic antibiotics against Gram-negative bacteria. Nature 2019, 576, 452–458. [Google Scholar] [CrossRef]
- Storek, K.M.; Auerbach, M.R.; Shi, H.; Garcia, N.K.; Sun, D.; Nickerson, N.N.; Vij, R.; Lin, Z.; Chiang, N.; Schneider, K.; et al. Monoclonal antibody targeting the beta-barrel assembly machine of Escherichia coli is bactericidal. Proc. Natl. Acad. Sci. USA 2018, 115, 3692–3697. [Google Scholar] [CrossRef]
- Wzorek, J.S.; Lee, J.; Tomasek, D.; Hagan, C.L.; Kahne, D.E. Membrane integration of an essential beta-barrel protein prerequires burial of an extracellular loop. Proc. Natl. Acad. Sci. USA 2017, 114, 2598–2603. [Google Scholar] [CrossRef]
- Miller, R.D.; Iinishi, A.; Modaresi, S.M.; Yoo, B.K.; Curtis, T.D.; Lariviere, P.J.; Liang, L.; Son, S.; Nicolau, S.; Bargabos, R.; et al. Computational identification of a systemic antibiotic for gram-negative bacteria. Nat. Microbiol. 2022, 7, 1661–1672. [Google Scholar] [CrossRef]
- Namdari, F.; Hurtado-Escobar, G.A.; Abed, N.; Trotereau, J.; Fardini, Y.; Giraud, E.; Velge, P.; Virlogeux-Payant, I. Deciphering the roles of BamB and its interaction with BamA in outer membrane biogenesis, T3SS expression and virulence in Salmonella. PLoS ONE 2012, 7, e46050. [Google Scholar] [CrossRef]
- Ruiz, N.; Falcone, B.; Kahne, D.; Silhavy, T.J. Chemical conditionality: A genetic strategy to probe organelle assembly. Cell 2005, 121, 307–317. [Google Scholar] [CrossRef]
- Steenhuis, M.; Abdallah, A.M.; de Munnik, S.M.; Kuhne, S.; Sterk, G.J.; van den Berg van Saparoea, B.; Westerhausen, S.; Wagner, S.; van der Wel, N.N.; Wijtmans, M.; et al. Inhibition of autotransporter biogenesis by small molecules. Mol. Microbiol. 2019, 112, 81–98. [Google Scholar] [CrossRef]
- Vuong, P.; Bennion, D.; Mantei, J.; Frost, D.; Misra, R. Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry. J. Bacteriol. 2008, 190, 1507–1517. [Google Scholar] [CrossRef]
- Fuangthong, M.; Sallabhan, R.; Atichartpongkul, S.; Rangkadilok, N.; Sriprang, R.; Satayavivad, J.; Mongkolsuk, S. The omlA gene is involved in multidrug resistance and its expression is inhibited by coumarins in Xanthomonas campestris pv. phaseoli. Arch Microbiol. 2008, 189, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.J.; Browning, D.F.; Jeeves, M.; Maderbocus, R.; Rajesh, S.; Sridhar, P.; Manoli, E.; Emery, D.; Sommer, U.; Spencer, A.; et al. Structure and function of BamE within the outer membrane and the beta-barrel assembly machine. EMBO Rep. 2011, 12, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Ochsner, U.A.; Vasil, A.I.; Johnson, Z.; Vasil, M.L. Pseudomonas aeruginosa fur overlaps with a gene encoding a novel outer membrane lipoprotein, OmlA. J. Bacteriol. 1999, 181, 1099–1109. [Google Scholar] [CrossRef] [PubMed]
- Sklar, J.G.; Wu, T.; Gronenberg, L.S.; Malinverni, J.C.; Kahne, D.; Silhavy, T.J. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. USA 2007, 104, 6400–6405. [Google Scholar] [CrossRef] [PubMed]
- Volokhina, E.B.; Beckers, F.; Tommassen, J.; Bos, M.P. The beta-barrel outer membrane protein assembly complex of Neisseria meningitidis. J. Bacteriol. 2009, 191, 7074–7085. [Google Scholar] [CrossRef]
- Anwari, K.; Webb, C.T.; Poggio, S.; Perry, A.J.; Belousoff, M.; Celik, N.; Ramm, G.; Lovering, A.; Sockett, R.E.; Smit, J.; et al. The evolution of new lipoprotein subunits of the bacterial outer membrane BAM complex. Mol. Microbiol. 2012, 84, 832–844. [Google Scholar] [CrossRef]
- Storek, K.M.; Vij, R.; Sun, D.; Smith, P.A.; Koerber, J.T.; Rutherford, S.T. The Escherichia coli beta-Barrel Assembly Machinery Is Sensitized to Perturbations under High Membrane Fluidity. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef]
- Konovalova, A.; Grabowicz, M.; Balibar, C.J.; Malinverni, J.C.; Painter, R.E.; Riley, D.; Mann, P.A.; Wang, H.; Garlisi, C.G.; Sherborne, B.; et al. Inhibitor of intramembrane protease RseP blocks the sigma(E) response causing lethal accumulation of unfolded outer membrane proteins. Proc. Natl. Acad. Sci. USA 2018, 115, E6614–E6621. [Google Scholar] [CrossRef]
- Mahoney, T.F.; Ricci, D.P.; Silhavy, T.J. Classifying beta-Barrel Assembly Substrates by Manipulating Essential Bam Complex Members. J. Bacteriol. 2016, 198, 1984–1992. [Google Scholar] [CrossRef]
- Onufryk, C.; Crouch, M.-L.; Fang, F.C.; Gross, C.A. Characterization of Six Lipoproteins in the σE Regulon. J. Bacteriol. 2005, 187, 4552–4561. [Google Scholar] [CrossRef]
- Rigel, N.W.; Schwalm, J.; Ricci, D.P.; Silhavy, T.J. BamE modulates the Escherichia coli beta-barrel assembly machine component BamA. J. Bacteriol. 2012, 194, 1002–1008. [Google Scholar] [CrossRef] [PubMed]
- Malinverni, J.C.; Werner, J.; Kim, S.; Sklar, J.G.; Kahne, D.; Misra, R.; Silhavy, T.J. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 2006, 61, 151–164. [Google Scholar] [CrossRef]
- Genevrois, S.; Steeghs, L.; Roholl, P.; Letesson, J.J.; van der Ley, P. The Omp85 protein of Neisseria meningitidis is required for lipid export to the outer membrane. EMBO J. 2003, 22, 1780–1789. [Google Scholar] [CrossRef] [PubMed]
- Voulhoux, R.; Tommassen, J. Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly. Res. Microbiol. 2004, 155, 129–135. [Google Scholar] [CrossRef]
- Gentle, I.; Gabriel, K.; Beech, P.; Waller, R.; Lithgow, T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 2004, 164, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Psonis, J.J.; Chahales, P.; Henderson, N.S.; Rigel, N.W.; Hoffman, P.S.; Thanassi, D.G. The small molecule nitazoxanide selectively disrupts BAM-mediated folding of the outer membrane usher protein. J. Biol. Chem. 2019, 294, 14357–14369. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, X.; Zhang, J.; Lin, Y.; You, X.; Chen, M.; Wang, Y.; Zhu, N.; Si, S. Identification of a Compound That Inhibits the Growth of Gram-Negative Bacteria by Blocking BamA-BamD Interaction. Front. Microbiol. 2020, 11, 1252. [Google Scholar] [CrossRef]
- Vij, R.; Lin, Z.; Chiang, N.; Vernes, J.M.; Storek, K.M.; Park, S.; Chan, J.; Meng, Y.G.; Comps-Agrar, L.; Luan, P.; et al. A targeted boost-and-sort immunization strategy using Escherichia coli BamA identifies rare growth inhibitory antibodies. Sci. Rep. 2018, 8, 7136. [Google Scholar] [CrossRef] [PubMed]
- White, P.; Haysom, S.F.; Iadanza, M.G.; Higgins, A.J.; Machin, J.M.; Whitehouse, J.M.; Horne, J.E.; Schiffrin, B.; Carpenter-Platt, C.; Calabrese, A.N.; et al. The role of membrane destabilisation and protein dynamics in BAM catalysed OMP folding. Nat. Commun. 2021, 12, 4174. [Google Scholar] [CrossRef]
- Kaur, H.; Hartmann, J.B.; Jakob, R.P.; Zahn, M.; Zimmermann, I.; Maier, T.; Seeger, M.A.; Hiller, S. Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach. J. Biomol. NMR 2019, 73, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Urfer, M.; Bogdanovic, J.; Lo Monte, F.; Moehle, K.; Zerbe, K.; Omasits, U.; Ahrens, C.H.; Pessi, G.; Eberl, L.; Robinson, J.A. A Peptidomimetic Antibiotic Targets Outer Membrane Proteins and Disrupts Selectively the Outer Membrane in Escherichia coli. J. Biol. Chem. 2016, 291, 1921–1932. [Google Scholar] [CrossRef]
- Mori, N.; Ishii, Y.; Tateda, K.; Kimura, S.; Kouyama, Y.; Inoko, H.; Mitsunaga, S.; Yamaguchi, K.; Yoshihara, E. A peptide based on homologous sequences of the beta-barrel assembly machinery component BamD potentiates antibiotic susceptibility of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2012, 67, 2173–2181. [Google Scholar] [CrossRef]
- Hagan, C.L.; Wzorek, J.S.; Kahne, D. Inhibition of the beta-barrel assembly machine by a peptide that binds BamD. Proc. Natl. Acad. Sci. USA 2015, 112, 2011–2016. [Google Scholar] [CrossRef] [PubMed]
- Vimala, A.; Ramakrishnan, C.; Gromiha, M.M. Identifying a potential receptor for the antibacterial peptide of sponge Axinella donnani endosymbiont. Gene 2015, 566, 166–174. [Google Scholar] [CrossRef]
- Ghequire, M.G.K.; De Mot, R. LlpB represents a second subclass of lectin-like bacteriocins. Microb. Biotechnol. 2019, 12, 567–573. [Google Scholar] [CrossRef]
- Shamir, E.R.; Warthan, M.; Brown, S.P.; Nataro, J.P.; Guerrant, R.L.; Hoffman, P.S. Nitazoxanide inhibits biofilm production and hemagglutination by enteroaggregative Escherichia coli strains by blocking assembly of AafA fimbriae. Antimicrob. Agents Chemother. 2010, 54, 1526–1533. [Google Scholar] [CrossRef] [PubMed]
- Zav’yalov, V.; Zavialov, A.; Zav’yalova, G.; Korpela, T. Adhesive organelles of Gram-negative pathogens assembled with the classical chaperone/usher machinery: Structure and function from a clinical standpoint. FEMS Microbiol. Rev. 2010, 34, 317–378. [Google Scholar] [CrossRef]
- Srinivas, N.; Jetter, P.; Ueberbacher, B.J.; Werneburg, M.; Zerbe, K.; Steinmann, J.; Van der Meijden, B.; Bernardini, F.; Lederer, A.; Dias, R.L.; et al. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science 2010, 327, 1010–1013. [Google Scholar] [CrossRef] [PubMed]
- Hagan, C.L.; Westwood, D.B.; Kahne, D. bam Lipoproteins Assemble BamA in vitro. Biochemistry 2013, 52, 6108–6113. [Google Scholar] [CrossRef]
- Diederichs, K.A.; Ni, X.; Rollauer, S.E.; Botos, I.; Tan, X.; King, M.S.; Kunji, E.R.S.; Jiang, J.; Buchanan, S.K. Structural insight into mitochondrial beta-barrel outer membrane protein biogenesis. Nat. Commun. 2020, 11, 3290. [Google Scholar] [CrossRef] [PubMed]
- Lundquist, K.; Billings, E.; Bi, M.; Wellnitz, J.; Noinaj, N. The assembly of beta-barrel membrane proteins by BAM and SAM. Mol. Microbiol. 2021, 115, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Paschen, S.A.; Neupert, W.; Rapaport, D. Biogenesis of beta-barrel membrane proteins of mitochondria. Trends Biochem. Sci. 2005, 30, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Kutik, S.; Stojanovski, D.; Becker, L.; Becker, T.; Meinecke, M.; Kruger, V.; Prinz, C.; Meisinger, C.; Guiard, B.; Wagner, R.; et al. Dissecting membrane insertion of mitochondrial beta-barrel proteins. Cell 2008, 132, 1011–1024. [Google Scholar] [CrossRef] [PubMed]
- Misra, R. Assembly of the beta-Barrel Outer Membrane Proteins in Gram-Negative Bacteria, Mitochondria, and Chloroplasts. ISRN Mol. Biol. 2012, 2012, 708203. [Google Scholar] [CrossRef]
- Richardson, L.G.; Paila, Y.D.; Siman, S.R.; Chen, Y.; Smith, M.D.; Schnell, D.J. Targeting and assembly of components of the TOC protein import complex at the chloroplast outer envelope membrane. Front. Plant Sci. 2014, 5, 269. [Google Scholar] [CrossRef]
- Schwenkert, S.; Dittmer, S.; Soll, J. Structural components involved in plastid protein import. Essays Biochem. 2018, 62, 65–75. [Google Scholar] [CrossRef]
- Kim, D.E.; Chivian, D.; Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004, 32, W526–W531. [Google Scholar] [CrossRef]
- Beard, H.; Cholleti, A.; Pearlman, D.; Sherman, W.; Loving, K.A. Applying physics-based scoring to calculate free energies of binding for single amino acid mutations in protein-protein complexes. PLoS ONE 2013, 8, e82849. [Google Scholar] [CrossRef]
- Gross, S.; Panter, F.; Pogorevc, D.; Seyfert, C.E.; Deckarm, S.; Bader, C.D.; Herrmann, J.; Muller, R. Improved broad-spectrum antibiotics against Gram-negative pathogens via darobactin biosynthetic pathway engineering. Chem. Sci. 2021, 12, 11882–11893. [Google Scholar] [CrossRef]
- Willett, J.L.; Gucinski, G.C.; Fatherree, J.P.; Low, D.A.; Hayes, C.S. Contact-dependent growth inhibition toxins exploit multiple independent cell-entry pathways. Proc. Natl. Acad. Sci. UAS 2015, 112, 11341–11346. [Google Scholar] [CrossRef]
- Aoki, S.K.; Malinverni, J.C.; Jacoby, K.; Thomas, B.; Pamma, R.; Trinh, B.N.; Remers, S.; Webb, J.; Braaten, B.A.; Silhavy, T.J.; et al. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol. Microbiol. 2008, 70, 323–340. [Google Scholar] [CrossRef] [PubMed]
- Ruhe, Z.C.; Wallace, A.B.; Low, D.A.; Hayes, C.S. Receptor polymorphism restricts contact-dependent growth inhibition to members of the same species. mBio 2013, 4, e00480-13. [Google Scholar] [CrossRef]
- Webb, C.T.; Heinz, E.; Lithgow, T. Evolution of the beta-barrel assembly machinery. Trends Microbiol. 2012, 20, 612–620. [Google Scholar] [CrossRef]
- Heinz, E.; Lithgow, T. A comprehensive analysis of the Omp85/TpsB protein superfamily structural diversity, taxonomic occurrence, and evolution. Front. Microbiol. 2014, 5, 370. [Google Scholar] [CrossRef]
- Howard, A.; O’Donoghue, M.; Feeney, A.; Sleator, R.D. Acinetobacter baumannii: An emerging opportunistic pathogen. Virulence 2012, 3, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Havenga, B.; Ndlovu, T.; Clements, T.; Reyneke, B.; Waso, M.; Khan, W. Exploring the antimicrobial resistance profiles of WHO critical priority list bacterial strains. BMC Microbiol. 2019, 19, 303. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Ranaghan, K.E.; Azam, S.S. Combating tigecycline resistant Acinetobacter baumannii: A leap forward towards multi-epitope based vaccine discovery. Eur. J. Pharm. Sci. 2019, 132, 1–17. [Google Scholar] [CrossRef] [PubMed]
- El-Rami, F.E.; Zielke, R.A.; Wi, T.; Sikora, A.E.; Unemo, M. Quantitative Proteomics of the 2016 WHO Neisseria gonorrhoeae Reference Strains Surveys Vaccine Candidates and Antimicrobial Resistance Determinants. Mol. Cell Proteom. 2019, 18, 127–150. [Google Scholar] [CrossRef]
- Baarda, B.I.; Zielke, R.A.; Nicholas, R.A.; Sikora, A.E. PubMLST for Antigen Allele Mining to Inform Development of Gonorrhea Protein-Based Vaccines. Front. Microbiol. 2018, 9, 2971. [Google Scholar] [CrossRef]
- Zielke, R.A.; Wierzbicki, I.H.; Baarda, B.I.; Gafken, P.R.; Soge, O.O.; Holmes, K.K.; Jerse, A.E.; Unemo, M.; Sikora, A.E. Proteomics-driven Antigen Discovery for Development of Vaccines Against Gonorrhea. Mol. Cell Proteom. 2016, 15, 2338–2355. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.; Wang, X.; Wang, X.; Teng, D.; Wang, J. In silico analysis and recombinant expression of BamA protein as a universal vaccine against Escherichia coli in mice. Appl. Microbiol. Biotechnol. 2016, 100, 5089–5098. [Google Scholar] [CrossRef] [PubMed]
- Grassmann, A.A.; Kremer, F.S.; Dos Santos, J.C.; Souza, J.D.; Pinto, L.D.S.; McBride, A.J.A. Discovery of Novel Leptospirosis Vaccine Candidates Using Reverse and Structural Vaccinology. Front. Immunol. 2017, 8, 463. [Google Scholar] [CrossRef] [PubMed]
- Baliga, P.; Shekar, M.; Venugopal, M.N. Potential Outer Membrane Protein Candidates for Vaccine Development Against the Pathogen Vibrio anguillarum: A Reverse Vaccinology Based Identification. Curr. Microbiol. 2018, 75, 368–377. [Google Scholar] [CrossRef] [PubMed]
| Bam Mutant | Mutation | Effect | Publication | |
|---|---|---|---|---|
| 1 | BamA | BamAΔR64 | Slight sensitivity to vancomycin and rifampin | Bennion et al., 2010. [36] |
| 2 | BamA | 46 N-terminal insertions into POTRA domains and linker, 41 insertions into β-barrel | Variable vancomycin sensitivity | Browning et al., 2013. [37] |
| 3 | BamA | BamA G667V, T671A, R666C | Resistance to LlpA | Ghequire et al., 2018. [38] |
| 4 | BamA | BamA E470K | Resistance to MRL-494 | Hart et al., 2019. [39] |
| 5 | BamA | BamA G429R, G429V, T434A, Q445P, A705T, G433D/E435K/F394V | Resistance to darobactin | Imai et al., 2019. [40] |
| 6 | BamA | G429V, G807V, E435K, Q445P, Q445P/T434A | Increased sensitivity to darobactin | Kaur et al., 2021 [41] |
| 7 | BamA | BamA F494L | With LptD Y721D, decreased vancomycin sensitivity; in WT, increased vancomycin sensitivity in nutrient-depleting conditions | Lee et al., 2018. [42] |
| 8 | BamA | R641E, Δ641RGF643, R641A/G642A/F643A, R641A/G642A, R641A/F643A, G642A/F643A | Increased sensitivity to vancomycin and rifampin | Leonard-Rivera et al., 2012. [43] |
| 9 | BamA | G807A, G807V, G807F | G807A, G807V, G807F: increased sensitivity to rifampicin; G807V, G807F: increased sensitivity to vancomycin | Lundquist et al., 2018. [44] |
| 10 | BamA | BamA D703Y | Decreased sensitivity to colistin | Luther et al., 2019. [45] |
| 11 | BamA | E554Q, H555Y, E554Q/H555Y, L6 deletion | E554Q, H555Y, E554Q/H555Y: decreased sensitivity to MAB1; L6 deletion: decreased sensitivity to MAB2 | Storek et al., 2018. [46] |
| 12 | BamA | BamA G771A, F738, V660A/R661A, V660A/R661A-LL (loop-to-lumen disulfide bond) | A strain lacking BamA: G771A: hypersensitivity to rifampicin; Strain lacking DegP-BamA G771A, F738: resistant to rifampicin, V660A/R661A: sensitive to rifampicin; (BamA V660A/R661A)-LL: decreased sensitivity to rifampicin | Wzorek et al., 2017. [47] |
| 13 | BamA | BamA G429, G809, L501Q, P782, G429V/G807V | Resistance to dynobactin | Miller et al., 2022. [48] |
| 14 | BamB | ΔBamB, BamB D227A, D229A, L173S/L175S/R176A | ΔBamB: increased sensitivity to amoxicillin; ΔBamB, D277A, and L173S/L175S/R176A: increased sensitivity to vancomycin, erythromycin, and bacitracin; ΔBamB, D277A, D229A, and L173S/L175S/R176A: increased sensitivity to rifampin, flumequine, and enrofloxacin | Namdari et al., 2012. [49] |
| 15 | BamB | recessive LOF mutations in yfgML locus via independent element insertions | yfgML: resistance to bile salts, chlorobiphenyl vancomycin (CBPV) | Ruiz et al., 2005. [50] |
| 16 | BamB | ΔBamB | Increased sensitivity to VUF15259 | Steenhuis et al., 2019. [51] |
| 17 | BamB | S172-A180 amino acid substitutions (scramble 1 & 2), L173S, L175S, R176A, L173S/L175S, L173S/R176A, L175S/R176A, L173S/L175S/R176A, YfgL(D227A)-His6 | Scramble 1 and 2: vancomycin hypersensitivity; R176A and either L173S or L175S: vancomycin sensitivity; L173S & L175S & R176A: vancomycin hypersensitivity; YfgL(D227A)-His6: slight increase in vancomycin sensitivity | Vuong et al., 2008. [52] |
| 18 | BamE | omlA: 170 bp insertion mutation via single recombination | Increased sensitivity to novobiocin, coumermycinA1, chloramphenicol, SDS, and menadione | Fuangthong et al., 2008. [53] |
| 19 | BamE | C20G, I32G, Q34G/C, G35C, N36G/C, Y37G, L38G, I46G, V55G, L59G, M64G/C, D66G, F68G/C, W73G, F74G, Y75G/C, V76G, R78G, Q88C, L91G, L93G, F95G/C, L101G | Increased sensitivity to vancomycin | Knowles et al., 2011. [54] |
| 20 | BamE | mutant strains: 6B- producing lesser amounts of OmlA (BamE) protein, 3A- lacking a functional omlA gene | 6B & 3A: increased sensitivity to SDS, deoxycholate 3A: increased sensitivity to nalidixic acid, rifampin, novobiocin, and chloramphenicol | Ochsner et al., 1999. [55] |
| 21 | BamE | smpA (strain lacking BamE) | Increased sensitivity (4-fold) to rifampin and cholate (2-fold); lethality on media with 0.5% SDS and 1 mM EDTA | Sklar et al., 2007. [56] |
| 22 | BamE | BamE deletion | Increased sensitivity to vancomycin | Volokhina et al., 2009. [57] |
| 23 | BamF | ΔBamF | Increased sensitivity to TritonX-100, SDS, nalidixic acid, rifampicin, vancomycin, and erythromycin | Anwari et al., 2012. [58] |
| 24 | BamA, BamB, BamC, BamE | ΔBamB, ΔBamC, ΔBamE, bamA101, BamA H555Y, V322A, P518L, T571M, G575D, G575S | ΔBamB, ΔBamC, ΔBamE, bamA101: increased sensitivity to MAB1; BamA H555Y, V322A, P518L, T571M, G575D, G575S: resistance to MAB1 | Storek et al., 2019. [59] |
| 25 | BamA, BamB, BamD, BamE | mutant strain bamA101, ΔBamB, ΔBamC, BamD L13P | Mutant strain bamA101, BamD L13P: significantly increased sensitivity to batimastat; ΔBamB, ΔBamC: slightly increased sensitivity to batimastat | Konovalova et al., 2018. [60] |
| 26 | BamA, BamD | bamA101 (mutant strain with lower BamA expression), BamDRBS, BamDSS | Sensitivity to bile salts and SDS that is increased at temperatures lower than 37 °C | Mahoney et al., 2016. [61] |
| 27 | BamB, BamC | ΔyfgL (BamB deletion) ΔnlpB (BamC deletion) | ΔyfgL eliminated by kanamycin, increased sensitivity to SDS and novobiocin; ΔnlpB increased sensitivity to kanamycin | Onufryk et al., 2005. [62] |
| 28 | BamB, BamC, BamE | bamB::kan, ΔBamC/ΔBamE | bamB::kan, ΔBamC/ΔBamE: increased sensitivity to bacitracin, erythromycin, novobiocin, rifampin, and vancomycin | Rigel et al., 2012. [63] |
| 29 | BamC, BamD | BamC: insertion at codon 41 (nlpB::kan); BamD: insertion at codon 227 (yfiO::kan) | yfiO::kan allele caused lethality on a BamB LOF allele yfgl8 background; nlpB::dan yflG8 double mutants had irregular colony morphology when exposed to kanamycin | Wu et al., 2005. [15] |
| Class of Antimicrobial | Name | Source | Cellular Target | MIC | Ref. |
|---|---|---|---|---|---|
| Small Molecule | VUF 15259 | Autotransporter (AT) pathway | N/A | Steenhuis et al., 2019. [51] | |
| Nitazoxanide (NTZ) | BamB, BamE, BamD | N/A | Psonis et al., 2019. [68] | ||
| MRL-494 | BamA (Gram-negatives); Cytoplasmic membrane integrity (Gram-positives) | 25 μM (E. coli JCM320) | Hart et al., 2019. [39] | ||
| IMB-H4 | BamA, BamD | 4 μg/mL (E. coli ATCC 25922) | Li et al., 2020. [69] | ||
| Peptide/Protein | |||||
| Antibodies | MAB1 (monoclonal antibody) | Mouse/rat | BamA | N/A | Storek et al., 2018. [46] |
| anti-BamA monoclonal antibodies | Rat | BamA | N/A | Vij et al., 2018. [70] | |
| Fabs/Nanobodies | Fab1 | BamA | N/A | White et al., 2021. [71] | |
| nanoE6 | BamA | N/A | Kaur et al., 2019. [72] | ||
| nanoE7 | BamA | N/A | Kaur et al., 2019. [72] | ||
| Peptides | JB-95 (β-hairpin peptidomimetic) | possibly BamA or LptD; active against Gram-positives | 0.15 μg/mL E. coli (E. coli ATCC 25922) | Urfer et al., 2016. [73] | |
| FIRL (BamD mimic) | BamD | No solo antimicrobial activity; synergizes with existing drugs to lower MIC | Mori et al., 2012. [74] | ||
| Chimeric peptidomimetic antibiotics (peptides 3, 4, 7, 8) | BamA, LPS | Luther et al., 2019. [45] | |||
| Peptide 2 (BamA mimic) | E. coli | BamD | N/A | Hagan et al., 2015. [75] | |
| Antibacterial peptide | Axinella donnani | BamA | N/A | Vimala et al., 2015. [76] | |
| Darobactin A | Photorhabdus khanii | BamA | 4 μg/mL (E. coli MG1655) 2 μg/mL (E. coli ATCC 25922) | Imai et al., 2019. [40] | |
| Dynobactin A | Photorhabdus australis | BamA | 16 μg/mL (E. coli MG1655) 8 μg/mL (E. coli ATCC 25922) | Miller et al., 2022. [48] | |
| Lectin-like bacteriocins | LlpA | BamA | N/A | Ghequire et al., 2018; Ghequire et al., 2019. [38,77] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Overly Cottom, C.; Stephenson, R.; Wilson, L.; Noinaj, N. Targeting BAM for Novel Therapeutics against Pathogenic Gram-Negative Bacteria. Antibiotics 2023, 12, 679. https://doi.org/10.3390/antibiotics12040679
Overly Cottom C, Stephenson R, Wilson L, Noinaj N. Targeting BAM for Novel Therapeutics against Pathogenic Gram-Negative Bacteria. Antibiotics. 2023; 12(4):679. https://doi.org/10.3390/antibiotics12040679
Chicago/Turabian StyleOverly Cottom, Claire, Robert Stephenson, Lindsey Wilson, and Nicholas Noinaj. 2023. "Targeting BAM for Novel Therapeutics against Pathogenic Gram-Negative Bacteria" Antibiotics 12, no. 4: 679. https://doi.org/10.3390/antibiotics12040679
APA StyleOverly Cottom, C., Stephenson, R., Wilson, L., & Noinaj, N. (2023). Targeting BAM for Novel Therapeutics against Pathogenic Gram-Negative Bacteria. Antibiotics, 12(4), 679. https://doi.org/10.3390/antibiotics12040679





