Discovery of Novel Resistance Mechanisms of Vibrio parahaemolyticus Biofilm against Aminoglycoside Antibiotics
Abstract
1. Introduction
2. Results
2.1. No Linear Correlation between Antibiotic Resistance of Planktonic Cells and Biofilm-Forming Ability
2.2. There Is a Significant Correlation between Antibiotic Resistance of Cells Growing in Biofilms and Biofilm-Forming Ability
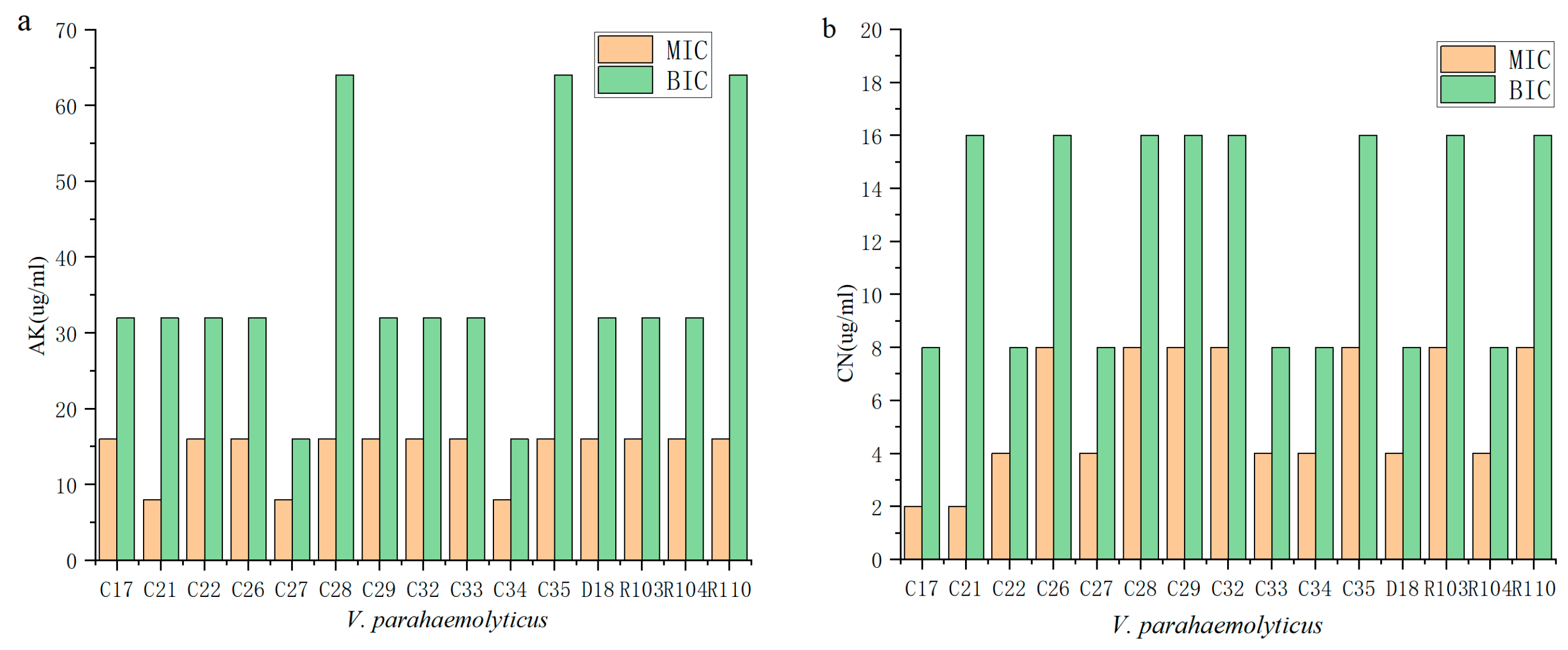
2.3. Biofilms Are More Protective against Antibiotics
2.4. Biofilm Enhances Antibiotic Resistance through Its Own Structural Characteristics
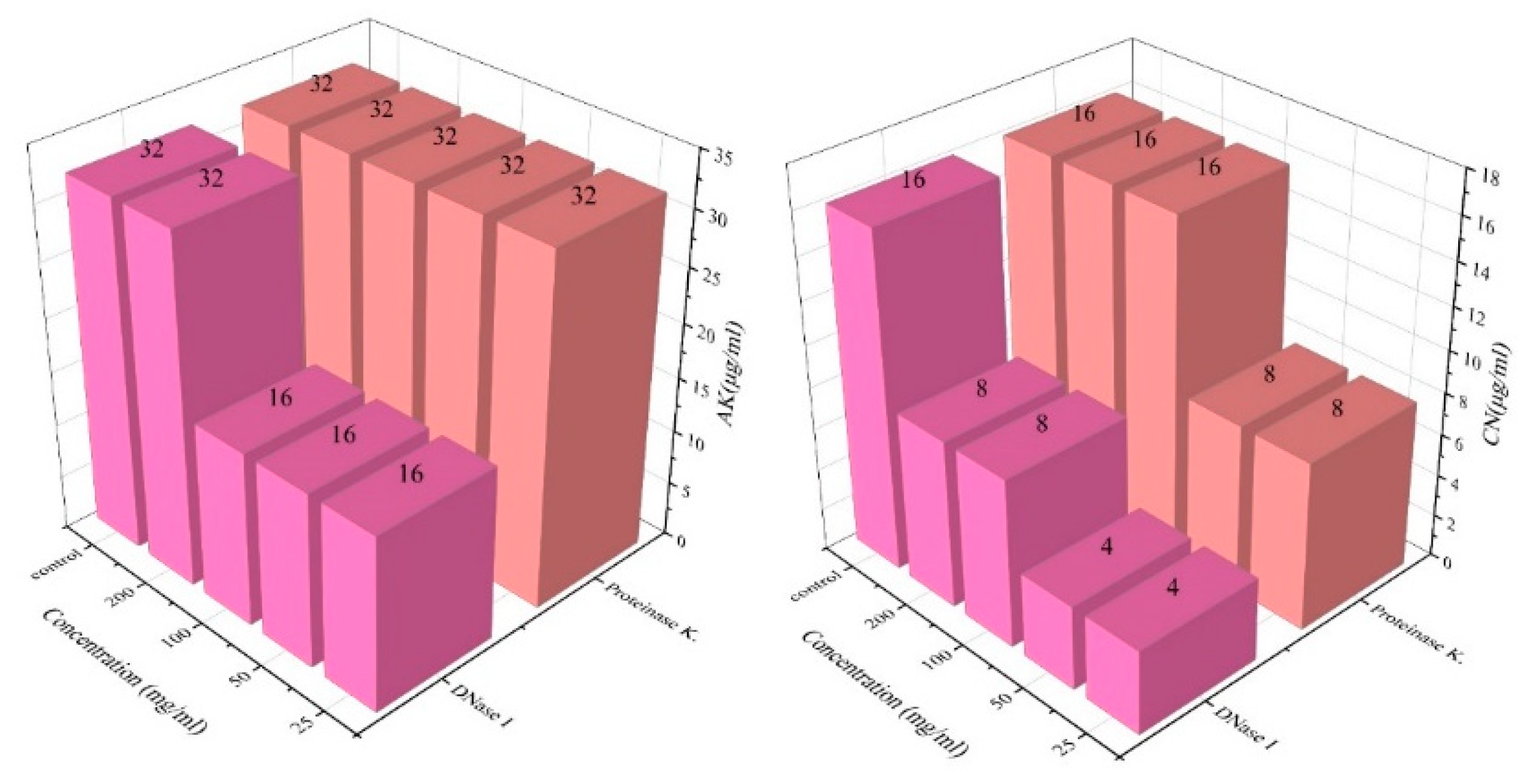
2.5. Enzyme-Treated Biofilms Are Less Resistant to Antibiotics
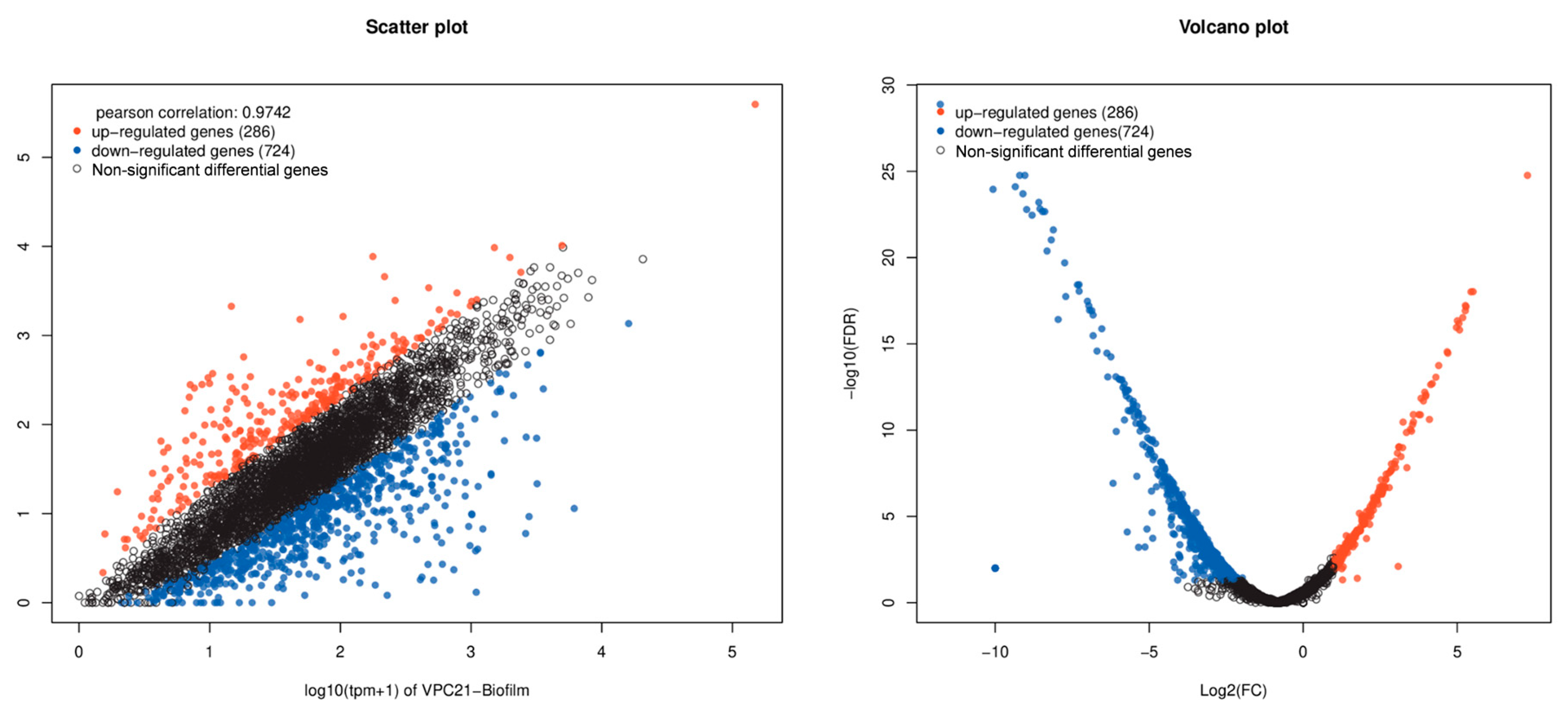
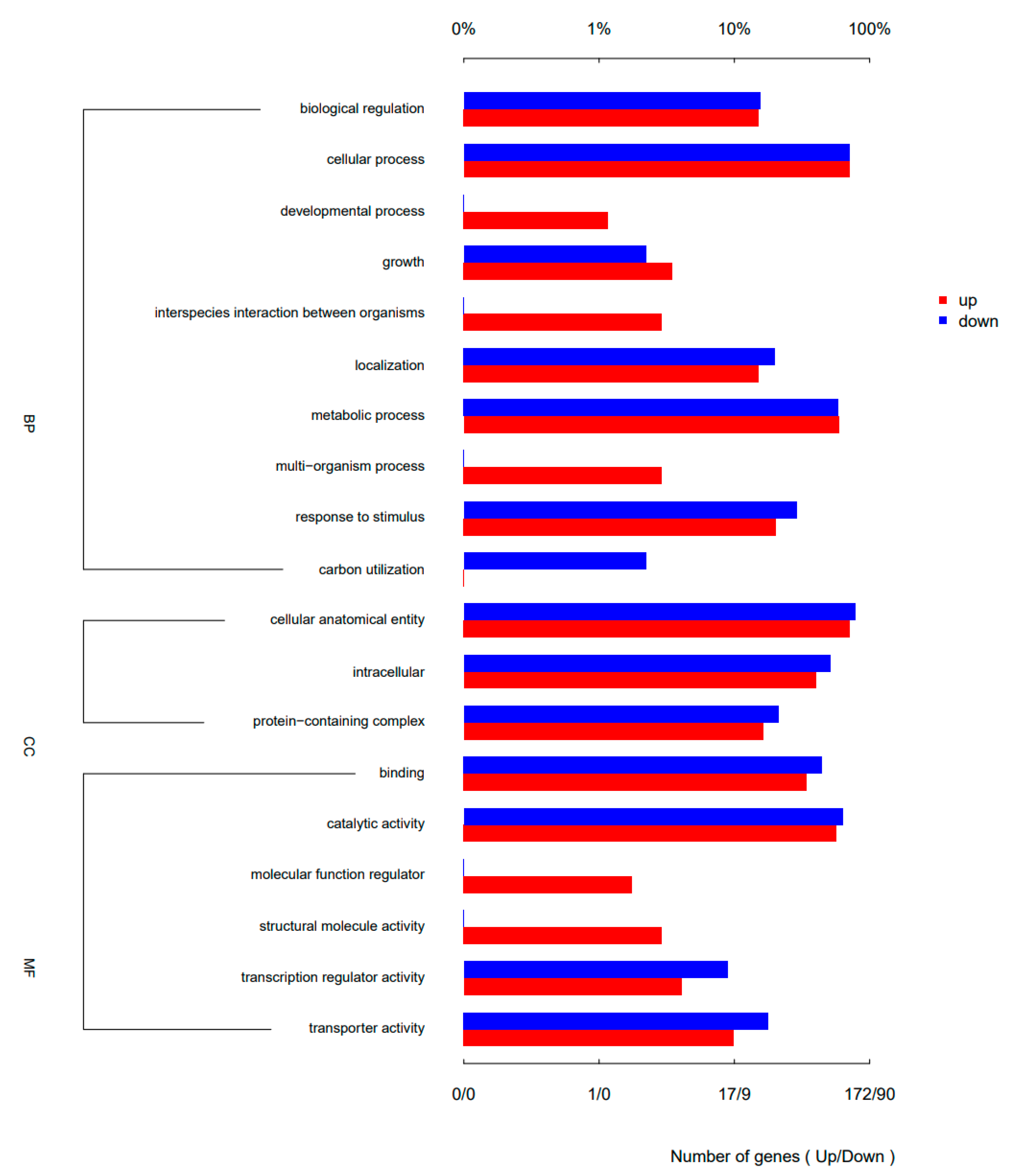
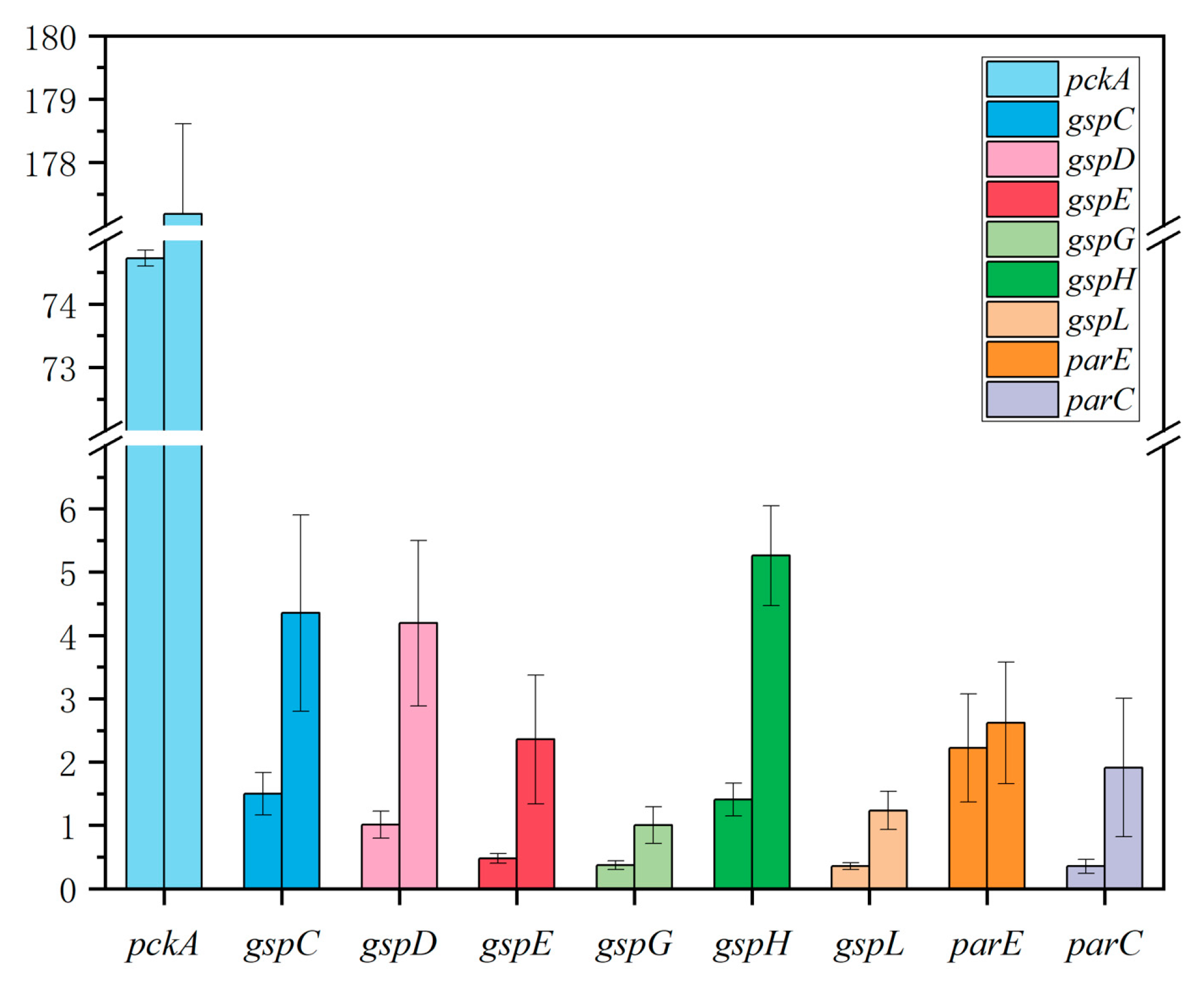
2.6. There Were Significant Differences in Gene Expression between Biofilm and Planktonic Cells
3. Discussion
3.1. “Enclosure” Type Resistance Mechanism of VP Biofilm against AGAs
3.2. “Neutralization” Type Resistance Mechanism of VP Biofilm against AGAs
3.3. “Regulatory” Type Resistance Mechanism of VP Biofilm against AGAs
4. Materials and Methods
4.1. Bacteria Solution Preparation
4.2. Preparation of Antibiotic Sensitive Testing Plate (96-Well Plate)
4.3. Determination of Planktonic Cells Minimum Inhibitory Concentration (MIC)
4.4. Determination of Biofilm Minimum Inhibitory Concentration (BIC)
4.5. Determination of Biofilm Biomass
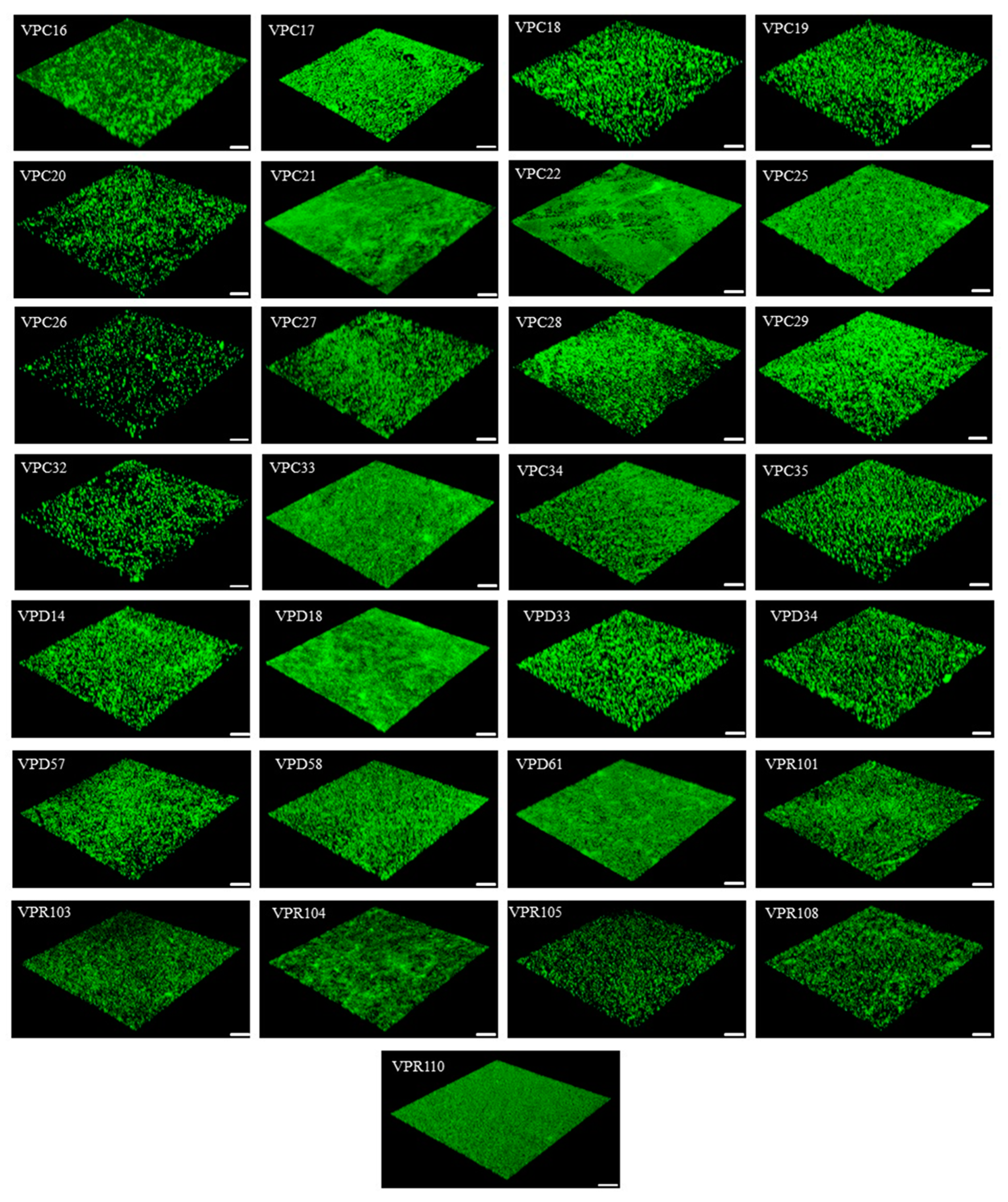
4.6. Confocal Laser Scanning Microscope (CLSM)
4.7. Enzyme Treatment of Biofilm (DNase I Enzyme and Protease K)
4.8. RNA-Sequencing
4.9. Quantitative Reverse Transcription-Polymerase Chain Reaction (q-PCR)
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007–2015; WHO: Geneva, Switzerland, 2015. [Google Scholar]
- Nicolaou, K.C.; Rigol, S. A brief history of antibiotics and select advances in their synthesis. J. Antibiot. 2018, 71, 153–184. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.F.; Zhong, C.Q.; Zhou, Y.P.; Lu, M.R.; Zong, G.L.; Zhang, P.P.; Cheng, M.T.; Cao, G.X. The integrative and conjugative element ICECspPOL2 contributes to the outbreak of Multi-Antibiotic-Resistant bacteria for Chryseobacterium spp. and Elizabethkingia spp. Microbiol. Spectr. 2022, 9, e02005-21. [Google Scholar]
- Suarez, S.A.; Martiny, A.C. Gene Amplification Uncovers Large Previously Unrecognized Cryptic Antibiotic Resistance Potential in E. coli. Microbiol. Spectr. 2022, 9, e00289-21. [Google Scholar] [CrossRef] [PubMed]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2015, 6, 34. [Google Scholar]
- Kirk, M.D.; Pires, S.M.; Black, R.E.; Caipo, M.; Crump, J.A.; Devleesschauwer, B.; Dopfer, D.; Fazil, A.; Fischer-Walker, C.L.; Hald, T.; et al. World health organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: A Data Synthesis. PLoS Med. 2015, 12, e1001921. [Google Scholar]
- Chen, C.X.; Liao, X.P.; Jiang, H.X.; Zhu, H.Q.; Yue, L.; Li, S.J.; Fang, B.H.; Liu, Y.H. Characteristics of Escherichia coli biofilm production, genetic typing, drug resistance pattern and gene expression under aminoglycoside pressures. Environ. Toxicol. Pharmacol. 2010, 30, 5–10. [Google Scholar] [CrossRef]
- Wang, N.Y.; Luo, J.; Deng, F.; Huang, Y.S.; Zhou, H. Antibiotic Combination Therapy: A Strategy to Overcome Bacterial Resistance to Aminoglycoside Antibiotics. Front. Pharmacol. 2022, 13, 839808. [Google Scholar] [CrossRef]
- Jennings, L.K.; Storek, K.M.; Ledvina, H.E.; Coulon, C.; Marmont, L.S.; Sadovskaya, I.; Secor, P.R.; Tseng, B.S.; Scian, M.; Filloux, A.; et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA 2015, 112, 11353–11358. [Google Scholar] [CrossRef]
- Karygianni, L.; Ren, Z.; Koo, H.; Thurnheer, T. Biofilm Matrixome: Extracellular Components in Structured Microbial Communities. Trends Microbiol. 2020, 28, 668–681. [Google Scholar] [CrossRef]
- Mccarty, S.; Woods, E.; Percival, S.L. Chapter Nine-Biofilms: From Concept to Reality; Elsevier Inc.: Amsterdam, The Netherlands, 2014; pp. 143–163. [Google Scholar]
- Yamaguchi-Kuroda, Y.; Kikuchi, Y.; Kokubu, E.; Ishihara, K. Porphyromonas gingivalis diffusible signaling molecules enhance Fusobacterium nucleatum biofilm formation via gene expression modulation. J. Oral Microbiol. 2023, 15, 2165001. [Google Scholar] [CrossRef]
- Mah, T.-F.C.; O’Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef] [PubMed]
- González, J.F.; Alberts, H.; Lee, J.; Doolittle, L.; Gunn, J.S. Biofilm Formation Protects Salmonella from the Antibiotic Ciprofloxacin In Vitro and In Vivo in the Mouse Model of chronic Carriage. Sci. Rep. 2018, 8, 222. [Google Scholar] [CrossRef] [PubMed]
- Shenkutie, A.M.; Yao, M.Z.; Siu, G.K.-H.; Wong, B.K.C.; Leung, P.H.-M. Biofilm-Induced Antibiotic Resistance in Clinical Acinetobacter baumannii Isolates. Antibiotics 2020, 9, 817. [Google Scholar] [CrossRef] [PubMed]
- Goodyear, M.C.; Garnier, N.E.; Levesque, R.C.; Khursigara, C.M. Liverpool Epidemic Strain Isolates of Pseudomonas aeruginosa Display High Levels of Antimicrobial Resistance during Both Planktonic and Biofilm Growth. Microbiol. Spectr. 2022, 10, e01024-22. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.M.; Zhao, Y.; Pan, Y.J.; Liu, H.Q. Comparison on the Growth Variability of Vibrio parahaemolyticus Coupled With Strain Sources and Genotypes Analyses in Simulated Gastric Digestion Fluids. Front. Microbiol. 2020, 11, 212. [Google Scholar] [CrossRef]
- Su, Y.-C.; Liu, C. Vibrio parahaemolyticus: A concern of seafood safety. Food Microbiol. 2007, 24, 549–558. [Google Scholar] [CrossRef]
- Niu, L.; Ba, Y.B.; Bai, F.J.; Liu, H.Q.; Pan, Y.J.; Zhao, Y. Comparison of antimicrobial resistance of Vibrio parahaemolyticus isolated from different sources. J. Food Sci. 2019, 38, 9–16. [Google Scholar]
- Han, N.; Mizan, F.R.; Jahid, I.K.; Ha, S.-D. Biofilm formation by Vibrio parahaemolyticus on food and food contact surfaces increases with rise in temperature. Food Control 2016, 70, 161–166. [Google Scholar] [CrossRef]
- Li, H.; Tang, R.; Lou, Y.; Cui, Z.L.; Chen, W.J.; Hong, Q.; Zhang, Z.H.; Malakar, P.K.; Pan, Y.J.; Zhao, Y. A Comprehensive Epidemiological Research for Clinical Vibrio parahaemolyticus in Shanghai. Front. Microbiol. 2017, 8, 1043. [Google Scholar] [CrossRef]
- Ahmed, H.A.; El Bayomi, R.M.; Hussein, M.A.; Khedr, M.H.; Remela, E.M.A.; El-Ashram, A.M. Molecular characterization, antibiotic resistance pattern and biofilm formation of Vibrio parahaemolyticus and V. cholerae isolated from crustaceans and humans. Int. J. Food Microbiol. 2018, 274, 31–37. [Google Scholar] [CrossRef]
- Carvalho, G.; Fouchet, D.; Danesh, G.; Godeux, A.-S.; Laaberki, M.-H.; Pontier, D.; Charpentier, X.; Venner, S. Bacterial Transformation Buffers Environmental Fluctuations through the Reversible Integration of Mobile Genetic Elements. mBio 2020, 11, e02443-19. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.G.; Murray, S.; Pascoe, B.; Bray, J.; Sheppard, S.K. Biofilm morphotypes and population structure among staphylococcus epidermidis from commensal and clinical samples. PLoS ONE 2016, 11, e0151240. [Google Scholar]
- Pauter, K.; Railean-Plugaru, V.; Złoch, M.; Pomastowski, P.; Szultka-Młyńska, M.; Buszewski, B. Identification, Structure and Characterization of Bacillus tequilensis Biofilm with the Use of Electrophoresis and Complementary Approaches. J. Clin. Med. 2022, 11, 722. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.W.; Mah, T.-F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Gupta, P.; Mankere, B.; Keloth, S.C.; Tuteja, U.; Pandey, P.; Chelvam, K.T. Increased antibiotic resistance exhibited by the biofilm of Vibrio cholerae O139. J. Antimicrob. Chemother. 2018, 73, 1841–1847. [Google Scholar] [CrossRef]
- Yonezawa, H.; Osaki, T.; Hanawa, T.; Kurata, S.; Ochiai, K.; Kamiya, S. Impact of Helicobacter pylori Biofilm Formation on Clarithromycin Susceptibility and Generation of Resistance Mutations. PLoS ONE 2013, 8, e73301. [Google Scholar] [CrossRef]
- Garousi, M.; Tabar, S.M.; Mirazi, H.; Asgari, P.; Sabeghi, P.; Salehi, A.; Khaledi, A.; Sanani, M.G.P.; Mirzahosseini, H.K. A global systematic review and meta-analysis on correlation between biofilm producers and non-biofilm producers with antibiotic resistance in Uropathogenic Escherichia coli. Microb. Pathog. 2022, 164, 105412. [Google Scholar] [CrossRef]
- Fauzia, K.A.; Miftahussurur, M.; Syam, A.F.; Waskito, L.A.; Doohan, D.; Rezkitha, Y.A.A.; Matsumoto, T.; Tuan, V.P.; Akada, J.; Yonezawa, H.; et al. Biofilm Formation and Antibiotic Resistance Phenotype of Helicobacter pylori Clinical Isolates. Toxins 2020, 12, 473. [Google Scholar] [CrossRef]
- Cordeiro, R.D.A.; Evangelista, A.J.D.J.; Serpa, R.; De Andrade, A.R.C.; Mendes, P.B.L.; Oliveira, J.; De Alencar, L.P.; Pereira, V.S.; Lima-Neto, R.; Brilhante, R.N.; et al. Cefepime and Amoxicillin Increase Metabolism and Enhance Caspofungin Tolerance of Candida albicans Biofilms. Front. Microbiol. 2019, 10, 1337. [Google Scholar] [CrossRef]
- Schilcher, K.; Andreoni, F.; Haunreiter, V.D.; Seidl, K.; Hasse, B.; Zinkernagel, A.S. Modulation of Staphylococcus aureus Biofilm Matrix by Subinhibitory Concentrations of Clindamycin. Antimicrob. Agents Chemother. 2016, 60, 5957–5967. [Google Scholar] [CrossRef]
- Van, A.H.; Van, D.P.; Coenye, T. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol. 2014, 22, 326–333. [Google Scholar]
- Solihin, J.; Waturangi, D.E.; Purwadaria, T. Induction of amylase and protease as antibiofilm agents by starch, casein, and yeast extract in Arthrobacter sp. CW01. BMC Microbiol. 2021, 21, 232. [Google Scholar] [CrossRef] [PubMed]
- Secchi, E.; Savorana, G.; Vitale, A.; Eberl, L.; Stocker, R.; Rusconi, R. The structural role of bacterial eDNA in the formation of biofilm streamers. Proc. Natl. Acad. Sci. USA 2022, 119, e2113723119. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, H.; Charron-Mazenod, L.; Lewenza, S. Extracellular DNA Chelates Cations and Induces Antibiotic Resistance in Pseudomonas aeruginosa Biofilms. PLoS Pathog. 2008, 4, e1000213. [Google Scholar] [CrossRef]
- Tseng, B.S.; Zhang, W.; Harrison, J.J.; Quach, T.P.; Song, J.L.; Penterman, J.; Singh, P.K.; Chopp, D.L.; Packman, A.I.; Parsek, M.R. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ. Microbiol. 2013, 15, 2865–2878. [Google Scholar]
- Chiang, W.-C.; Nilsson, M.; Jensen, P.Ø.; Høiby, N.; Nielsen, T.E.; Givskov, M.; Tolker-Nielsen, T. Extracellular DNA Shields against Aminoglycosides in Pseudomonas aeruginosa Biofilms. Antimicrob. Agents Chemother. 2013, 57, 2352–2361. [Google Scholar] [CrossRef]
- Brown, H.L.; Hanman, K.; Reuter, M.; Betts, R.P.; Van Vliet, A.H.M. Campylobacter jejuni biofilms contain extracellular DNA and are sensitive to DNase I treatment. Front. Microbiol. 2015, 6, 699. [Google Scholar] [CrossRef]
- Shukla, S.K.; Rao, T.S. Dispersal of Bap-mediated Staphylococcus aureus biofilm by proteinase K. J. Antibiot. 2013, 66, 55–60. [Google Scholar] [CrossRef]
- Gordon, C.A.; Hodges, N.A.; Marriott, C. Antibiotic interaction and diffusion through alginate and exopolysaccharide of cystic fibrosis-derived Pseudomonas aeruginosa. J. Antimicrob. Chemother. 1988, 22, 667–674. [Google Scholar] [CrossRef]
- Billings, N.; Millan, M.R.; Caldara, M.; Rusconi, R.; Tarasova, Y.; Stocker, R.; Ribbeck, K.; Parsek, M.R. The Extracellular Matrix Component Psl Provides Fast-Acting Antibiotic Defense in Pseudomonas aeruginosa Biofilms. PLoS Pathog. 2013, 9, e1003526. [Google Scholar] [CrossRef]
- Rees, D.C.; Johnson, E.; Lewinson, O. ABC transporters: The power to change. Nat. Rev. Mol. Cell Biol. 2009, 10, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Rees, D.C. Structural and Mechanistic Diversity of ABC Transporters. Biophys. J. 2014, 106, 434a. [Google Scholar] [CrossRef]
- Li, B.; Zhang, Y.J.; Guo, Q.; He, S.Y.; Fan, J.S.; Xu, L.Y.; Zhang, Z.M.; Wu, W.Y.; Chu, H.Q. Antibacterial peptide RP557 increases the antibiotic sensitivity of Mycobacterium abscessus by inhibiting biofilm formation. Sci. Total Environ. 2022, 807, 151855. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.C.; Cai, Y.Y.; Chen, Z.H.; Li, H.J.; Xu, Z.Z.; Li, W.J.; Jia, J.H.; Sun, Y.D. Spot-mediated NapA upregulation promotes oxidative stress-induced Helicobacter pylori biofilm formation and confers multidrug resistance. Antimicrob. Agents Chemother. 2021, 65, e00152-21. [Google Scholar] [CrossRef] [PubMed]
- Parker, A.E.; Walker, D.K.; Goeres, D.M.; Allan, N.; Olson, M.E. Ruggedness and reproducibility of the MBEC biofilm disinfectant efficacy test. J. Microbiol. Methods 2014, 102, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Song, X.Y.; Zhang, Z.H.; Fu, J.J.; Wang, X.; Malakar, P.K.; Liu, H.Q.; Pan, Y.J.; Zhao, Y. Removal of Foodborne Pathogen Biofilms by Acidic Electrolyzed Water. Front. Microbiol. 2017, 8, 988. [Google Scholar] [CrossRef]
- Antoniani, D.; Bocci, P.; Maciąg, A.; Raffaelli, N.; Landini, P. Monitoring of diguanylate cyclase activity and of cyclic-di-GMP biosynthesis by whole-cell assays suitable for high-throughput screening of biofilm inhibitors. Appl. Microbiol. Biotechnol. 2010, 85, 1095–1104. [Google Scholar] [CrossRef]
- Michler-Kozma, D.N.; Neu, T.R.; Gabel, F. Environmental conditions affect the food quality of plastic associated biofilms for the benthic grazer Physa fontinalis. Sci. Total Environ. 2022, 816, 151663. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Chang, H.Y.; Tong, C.B.S. Using edgeR software to analyze complex RNA-Seq data. Acta Hortic. 2021, 1307, 171–176. [Google Scholar] [CrossRef]
- Zou, Q.; Wei, H.; Chen, Z.L.; Ye, P.; Zhang, J.Q.; Sun, M.Q.; Huang, L.; Li, J. Soil particle size fractions affect arsenic (As) release and speciation: Insights into dissolved organic matter and functional genes. J. Hazard. Mater. 2022, 443, 130100. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Guo, L.X.; Chen, B.W.; Zhang, Z.H.; Wang, J.J.; Liu, H.Q.; Pan, Y.J.; Zhao, Y. Quantitative analysis of Vibrio parahaemolyticus biofilm structure based on high-throughput confocal laser scanning microscopy. Acta Microbiol. Sin. 2020, 60, 715–726. [Google Scholar]


| Biofilm-Forming Ability | Biofilm-Forming Ability | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Strong (+++) | Moderate (++) | Weak (+) | No Biofilm (−) | Strong (+++) | Moderate (++) | Weak (+) | No Biofilm (−) | ||
| VPC16 | + | VPD8 | − | ||||||
| VPC17 | +++ | VPD14 | + | ||||||
| VPC18 | + | VPD18 | +++ | ||||||
| VPC19 | + | VPD33 | + | ||||||
| VPC20 | +++ | VPD34 | + | ||||||
| VPC21 | +++ | VPD57 | + | ||||||
| VPC22 | +++ | VPD58 | + | ||||||
| VPC25 | ++ | VPD61 | +++ | ||||||
| VPC26 | ++ | VPR101 | ++ | ||||||
| VPC27 | + | VPR103 | ++ | ||||||
| VPC28 | +++ | VPR104 | +++ | ||||||
| VPC29 | +++ | VPR105 | + | ||||||
| VPC32 | +++ | VPR106 | − | ||||||
| VPC33 | + | VPR108 | + | ||||||
| VPC34 | ++ | VPR110 | + | ||||||
| VPC35 | +++ | VPR111 | − | ||||||
| AMP | CFPM | CAZ | AK | CN | TE | CIP | LEV | |
|---|---|---|---|---|---|---|---|---|
| VPC16 | >128 | 0.25 | 0.25 | 8 | 4 | 0.5 | 0.125 | 0.5 |
| VPC17 | 128 | 0.5 | 0.25 | 16 | 2 | 0.5 | 0.5 | 0.5 |
| VPC18 | >128 | 0.5 | 0.25 | 16 | 4 | 0.5 | 0.25 | 0.25 |
| VPC19 | >128 | 0.5 | 0.25 | 16 | 4 | 0.5 | 0.25 | 0.25 |
| VPC20 | >128 | 0.5 | 0.5 | 8 | 4 | 0.5 | 0.25 | 0.25 |
| VPC21 | 128 | 0.25 | 0.25 | 8 | 2 | 0.5 | 0.125 | 0.125 |
| VPC22 | >128 | 0.5 | 0.25 | 16 | 4 | 0.5 | 0.25 | 0.125 |
| VPC25 | >128 | 0.5 | 0.25 | 16 | 2 | 0.5 | 0.25 | 0.125 |
| VPC26 | 128 | 0.5 | 0.25 | 16 | 8 | 0.25 | 0.25 | 0.125 |
| VPC27 | 32 | 0.5 | 0.25 | 8 | 4 | 0.25 | 0.125 | 0.125 |
| VPC28 | >128 | 0.5 | 0.5 | 16 | 8 | 0.25 | 0.25 | 0.125 |
| VPC29 | 128 | 0.5 | 0.5 | 16 | 8 | 0.25 | 0.25 | 0.25 |
| VPC32 | 64 | 0.5 | 0.25 | 16 | 8 | 0.5 | 0.25 | 0.125 |
| VPC33 | >128 | 0.25 | 0.25 | 16 | 4 | 0.25 | 0.25 | 0.125 |
| VPC34 | 16 | 0.5 | 0.25 | 8 | 4 | 0.25 | 0.125 | 0.125 |
| VPC35 | 64 | 0.5 | 0.25 | 16 | 8 | 0.5 | 0.25 | 0.125 |
| VPD8 | >128 | 0.125 | 0.5 | 16 | 8 | 0.25 | 0.25 | 0.25 |
| VPD14 | 128 | 1 | 0.25 | 16 | 8 | 0.5 | 0.25 | 0.25 |
| VPD18 | >128 | 0.5 | 8 | 16 | 4 | 0.5 | 16 | 8 |
| VPD33 | >128 | 0.125 | 0.25 | 16 | 8 | 0.25 | 0.25 | 0.25 |
| VPD34 | >128 | 1 | 0.5 | 16 | 8 | 0.5 | 0.25 | 0.25 |
| VPD57 | 128 | 0.5 | 0.125 | 16 | 4 | 0.5 | 0.5 | 0.25 |
| VPD58 | 128 | 0.25 | 0.25 | 8 | 4 | 0.25 | 8 | 0.25 |
| VPD61 | >128 | 1 | 0.5 | 16 | 8 | 0.5 | 0.25 | 0.25 |
| VPR101 | 128 | 0.125 | 0.5 | 32 | 4 | 0.25 | 0.25 | 0.25 |
| VPR103 | >128 | 0.5 | 0.5 | 16 | 8 | 0.5 | 0.25 | 0.125 |
| VPR104 | 64 | 0.5 | 0.25 | 16 | 4 | 0.5 | 0.125 | 0.25 |
| VPR105 | >128 | 0.5 | 0.25 | 16 | 4 | 0.25 | 0.25 | 0.125 |
| VPR106 | >128 | 0.5 | 0.25 | 16 | 8 | 0.25 | 0.25 | 0.25 |
| VPR108 | >128 | 0.5 | 0.25 | 16 | 4 | 0.5 | 0.25 | 0.25 |
| VPR110 | >128 | 0.5 | 0.5 | 16 | 8 | 0.5 | 0.25 | 0.25 |
| VPR111 | 16 | 0.25 | 0.125 | 8 | 4 | 0.25 | 0.125 | 0.125 |
| AMP | CFPM | CAZ | AK | CN | TE | CIP | LEV | |
|---|---|---|---|---|---|---|---|---|
| SCC | −0.016 | 0.169 | 0.153 | 0.064 | −0.012 | 0.130 | 0.002 | −0.117 |
| p-value | 0.935 | 0.38 | 0.428 | 0.741 | 0.95 | 0.501 | 0.990 | 0.547 |
| AMP | CFPM | CAZ | AK | CN | TE | CIP | LEV | |
|---|---|---|---|---|---|---|---|---|
| VPC16 | 64 | 2 | 0.5 | 8 | 8 | 0.5 | 0.25 | 0.25 |
| VPC17 | >128 | 0.5 | 2 | 32 | 8 | 0.5 | 0.5 | 0.5 |
| VPC18 | >128 | 0.25 | 0.25 | 16 | 8 | 0.25 | 1 | 0.125 |
| VPC19 | >128 | 0.5 | 0.5 | 32 | 4 | 0.5 | 0.25 | 0.25 |
| VPC20 | >128 | 0.5 | 0.25 | 32 | 4 | 0.25 | 0.25 | 0.5 |
| VPC21 | >128 | 0.25 | 1 | 32 | 16 | 0.25 | 0.5 | 0.25 |
| VPC22 | >128 | 0.5 | 0.25 | 32 | 8 | 0.5 | 0.25 | 0.5 |
| VPC25 | >128 | 0.25 | 0.25 | 32 | 2 | 0 | 0.25 | 0.125 |
| VPC26 | >128 | 8 | 8 | 32 | 16 | 1 | 1 | 0.5 |
| VPC27 | 64 | 0.5 | 0.25 | 16 | 8 | 0.25 | 0.25 | 0.25 |
| VPC28 | >128 | 0.5 | 0.5 | 64 | 16 | 1 | 2 | 2 |
| VPC29 | 128 | 0.25 | 0.5 | 32 | 16 | 0.25 | 0.25 | 0.25 |
| VPC32 | 64 | 1 | 4 | 32 | 16 | 1 | 2 | 2 |
| VPC33 | >128 | 8 | 2 | 32 | 8 | 1 | 0.5 | 0.25 |
| VPC34 | 64 | 0.5 | 0.25 | 16 | 8 | 1 | 0.25 | 0.25 |
| VPC35 | 64 | 1 | 4 | 64 | 16 | 1 | 0.25 | 0.5 |
| VPD14 | 128 | 4 | 4 | 32 | 8 | 0.5 | 0.25 | 1 |
| VPD18 | >128 | 0.5 | 8 | 32 | 8 | 0.5 | 8 | 8 |
| VPD33 | >128 | 0 | 8 | 16 | 8 | 0.25 | 0.25 | 0.125 |
| VPD34 | >128 | 1 | 2 | 16 | 8 | 0.5 | 1 | 0.25 |
| VPD57 | 128 | 1 | 1 | 16 | 8 | 1 | 0.5 | 0.125 |
| VPD58 | 128 | 0 | 1 | 8 | 4 | 0.125 | 0.125 | 0.125 |
| VPD61 | >128 | 1 | 1 | 32 | 8 | 0.25 | 0.25 | 0.125 |
| VPR101 | >128 | 0 | 2 | 32 | 8 | 0.25 | 0.25 | 0.25 |
| VPR103 | >128 | 16 | 2 | 32 | 16 | 16 | 2 | 1 |
| VPR104 | 64 | 8 | 8 | 32 | 8 | 4 | 1 | 1 |
| VPR105 | 64 | 2 | 4 | 16 | 8 | 2 | 1 | 0.5 |
| VPR108 | 64 | 8 | 8 | 16 | 16 | 1 | 0.5 | 0.125 |
| VPR110 | 128 | 8 | 8 | 64 | 16 | 4 | 0.5 | 0.25 |
| AMP | CFPM | CAZ | AK | CN | TE | CIP | LEV | |
|---|---|---|---|---|---|---|---|---|
| SCC | 0.231 | −0.271 | −0.081 | 0.530 * | −0.083 | −0.254 | 0.018 | 0.352 |
| p-value | 0.229 | 0.154 | 0.677 | 0.003 | 0.668 | 0.184 | 0.927 | 0.061 |
| Strains | BV (×105 µm3) | AT (µm) | BR |
|---|---|---|---|
| VPC16 | 3.67 ± 0.51 | 3.10 ± 0.79 | 1.64 ± 0.04 |
| VPC17 | 12.68 ± 1.79 | 12.35 ± 1.78 | 1.62 ± 0.24 |
| VPC18 | 1.19 ± 0.52 | 1.25 ± 0.49 | 1.88 ± 0.05 |
| VPC19 | 1.08 ± 0.32 | 0.91 ± 0.15 | 1.89 ± 0.04 |
| VPC20 | 11.18 ± 0.47 | 11.16 ± 0.41 | 1.86 ± 0.06 |
| VPC21 | 17.18 ± 6.15 | 10.15 ± 9.84 | 1.16 ± 0.73 |
| VPC22 | 15.32 ± 4.90 | 17.78 ± 4.90 | 1.10 ± 0.32 |
| VPC25 | 10.38 ± 1.13 | 10.00 ± 0.83 | 1.00 ± 0.04 |
| VPC26 | 2.13 ± 0.04 | 0.15 ± 0.07 | 1.98 ± 0.01 |
| VPC27 | 3.45 ± 1.91 | 3.54 ± 2.77 | 1.65 ± 0.91 |
| VPC28 | 11.97 ± 1.01 | 12.64 ± 1.44 | 1.67 ± 0.15 |
| VPC29 | 13.69 ± 1.56 | 3.63 ± 1.29 | 1.55 ± 0.17 |
| VPC32 | 14.79 ± 0.12 | 0.88 ± 0.12 | 1.91 ± 0.01 |
| VPC33 | 11.35 ± 4.68 | 11.22 ± 2.90 | 0.89 ± 0.15 |
| VPC34 | 8.72 ± 5.25 | 8.02 ± 5.63 | 1.26 ± 0.42 |
| VPC35 | 22.04 ± 0.44 | 12.00 ± 0.31 | 1.79 ± 0.04 |
| VPD14 | 1.65 ± 0.56 | 1.58 ± 0.68 | 1.80 ± 0.07 |
| VPD18 | 17.40 ± 2.32 | 18.62 ± 2.01 | 0.96 ± 0.16 |
| VPD33 | 1.47 ± 0.94 | 1.39 ± 0.94 | 1.86 ± 0.10 |
| VPD34 | 1.68 ± 1.07 | 1.71 ± 1.05 | 1.83 ± 0.09 |
| VPD57 | 1.48 ± 0.90 | 1.55 ± 0.67 | 1.81 ± 0.1 |
| VPD58 | 4.82 ± 3.83 | 4.71 ± 4.08 | 1.48 ± 0.38 |
| VPD61 | 17.24 ± 2.03 | 13.90 ± 2.31 | 0.78 ± 0.04 |
| VPR101 | 16.05 ± 3.42 | 15.95 ± 2.59 | 1.27 ± 0.37 |
| VPR103 | 14.64 ± 3.37 | 15.29 ± 3.06 | 1.43 ± 0.27 |
| VPR104 | 17.38 ± 3.05 | 17.71 ± 2.04 | 1.18 ± 0.24 |
| VPR105 | 0.74 ± 0.51 | 0.99 ± 0.50 | 1.88 ± 0.06 |
| VPR108 | 3.39 ± 0.31 | 3.06 ± 0.52 | 1.63 ± 0.04 |
| VPR110 | 17.22 ± 6.36 | 17.05 ± 5.65 | 1.29 ± 0.52 |
| BV (×105 µm3) | AT (µm) | BR | ||||
|---|---|---|---|---|---|---|
| SCC | p-Value | SCC | p-Value | SCC | p-Value | |
| Ampicillins | 0.018 | 0.924 | 0.101 | 0.601 | −0.143 | 0.459 |
| Cefepime | 0.016 | 0.933 | 0.11 | 0.957 | 0.053 | 0.784 |
| Ceftazidime | 0.066 | 0.733 | 0.021 | 0.914 | 0.094 | 0.628 |
| Amikacin | 0.813 ** | 0.000 | 0.642 ** | 0.000 | 0.029 | 0.883 |
| Gentamicin | 0.411 * | 0.027 | 0.130 | 0.501 | −0.283 | 0.137 |
| Tetracycline | 0.136 | 0.483 | 0.088 | 0.649 | 0.115 | 0.553 |
| Ciprofloxacin | 0.117 | 0.547 | −0.039 | 0.841 | 0.115 | 0.422 |
| Levofloxacin | 0.430 * | 0.03 | 0.272 | 0.153 | 0.096 | 0.62 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, C.; Yuan, M.; Tao, Q.; Xu, T.; Liu, J.; Huang, Z.; Wu, Q.; Pan, Y.; Zhao, Y.; Zhang, Z. Discovery of Novel Resistance Mechanisms of Vibrio parahaemolyticus Biofilm against Aminoglycoside Antibiotics. Antibiotics 2023, 12, 638. https://doi.org/10.3390/antibiotics12040638
Tian C, Yuan M, Tao Q, Xu T, Liu J, Huang Z, Wu Q, Pan Y, Zhao Y, Zhang Z. Discovery of Novel Resistance Mechanisms of Vibrio parahaemolyticus Biofilm against Aminoglycoside Antibiotics. Antibiotics. 2023; 12(4):638. https://doi.org/10.3390/antibiotics12040638
Chicago/Turabian StyleTian, Cuifang, Mengqi Yuan, Qian Tao, Tianming Xu, Jing Liu, Zhenhua Huang, Qian Wu, Yingjie Pan, Yong Zhao, and Zhaohuan Zhang. 2023. "Discovery of Novel Resistance Mechanisms of Vibrio parahaemolyticus Biofilm against Aminoglycoside Antibiotics" Antibiotics 12, no. 4: 638. https://doi.org/10.3390/antibiotics12040638
APA StyleTian, C., Yuan, M., Tao, Q., Xu, T., Liu, J., Huang, Z., Wu, Q., Pan, Y., Zhao, Y., & Zhang, Z. (2023). Discovery of Novel Resistance Mechanisms of Vibrio parahaemolyticus Biofilm against Aminoglycoside Antibiotics. Antibiotics, 12(4), 638. https://doi.org/10.3390/antibiotics12040638







