1. Introduction
Natural products are important sources of new drug discoveries and play an important role in drug development [
1]. Synthetic molecules and natural products could practically resolve the long-standing medication design challenges. The development of active antimicrobials from natural products would be aided by modern chemistry and bioinformatics approaches [
2]. 35% of medications available in the market are reported to be derived from natural materials, with 25% of those accounted for by plants [
3]. Natural products, particularly antibiotics and therapeutic agents of cancer, with their distinctive benefits, are essential in the process of novel drug discovery [
4].
However, bacterial infections have a significant impact on public health. Treatment of bacterial infections is becoming more difficult as a result of an increase in the number of patients with complex underlying problems and microorganisms resistant to current antimicrobial medications [
5,
6,
7]. These issues, as well as drug cytotoxicity and a limited therapeutic spectrum, have prompted the development of new antimicrobial drugs [
5]. One of the main reasons to discover new antimicrobial agents or to improve the efficacy of already available antibiotics is antimicrobial resistance (AMR), which is one of the world’s most serious public health issues at present. Worldwide antibiotic resistance threatens our progress in healthcare, food production, and ultimately life expectancy. Therefore, the identification of new antimicrobial agents is of immense need to save lives and protect health systems.
β-Lap is a natural compound isolated from the heartwood of lapacho tree (
Tabebuia impetiginosa) found in different parts of South America [
8]. Numerous studies have demonstrated that β-lap has a variety of pharmacological actions, including anticancer [
9], anti-
Trypanosoma cruzi—the causative agent of Chagas disease—[
10], antibacterial [
11], and antifungal properties. Having antifungal activity against azole-resistant
Candida spp. isolates not only inhibits the biofilm formation but disrupts the already formed mature biofilms. Cristian Salas et al. used β-Lap to treat Chagas disease, one of the most serious endemic disorders caused by
Trypanosoma cruzi, and found that β-Lap inhibited epimastigote development in
Trypanosoma cruzi cells and trypomastigote vitality in vitro [
10].
β-Lap possesses antibacterial properties which act against a variety of bacteria species, including
M. tuberculosis, its multidrug resistant (MDR) clinical isolates and methicillin resistant
S. aureus (MRSA). β-Lap showed additive effect in combination with first line tuberculosis drugs and synergic effect with N-acetylcysteine against
M. tuberculosis H37Rv [
12]. In addition, it synergistically inhibited the growth of methicillin resistant
S. aureus (MRSA) in combination with fluoroquinolones, carbapenems, and other beta-lactam antibiotic [
13]. Generation of hydrogen peroxide and superoxide anions in bacterial cells has been demonstrated as one of the mechanisms for the action of β-Lap [
14].
Nutrient limitation, slow growth, adaptive stress responses, and formation of persistent cells are hypothesized to constitute a multi-layered biofilm, which leads to the poor antibiotic penetration [
15] and thus difficult eradication of biofilm producers [
16]. It is believed that the reduced antibiotic susceptibility of bacteria in biofilms is due to a combination of poor antibiotic penetration, an altered microenvironment, adaptive responses, and the presence of bacterial persisted cells. Therefore, there is a need to develop a novel antibiotic to inhibit the biofilm formation and to disrupt the formed biofilms, which would allow the cells to grow actively and thus might help the existing antibiotics to clear infections involving biofilms which are refractory to current treatments [
17].
2. Materials and Methods
2.1. Solvents, Media, and Chemicals Used in This Study
β-Lap was purchased from Solarbio Life Science (Beijing, China) and dissolved in dimethyl sulfoxide (DMSO). Gram’s crystal violet, a bacteriological stain, was obtained from TITAN BIOTECH. Phosphate buffered saline (PBS) tablets were obtained from Sigma Aldrich®, Dorset, UK. Methanol (≥99.8%) was purchased from Sigma-Aldrich® GmbH Chemie, Steinheim, France. Acetic acid (≥99.8%) was purchased from Sigma-Aldrich® GmbH Chemie, Steinheim, Germany. Six antibiotics belonging to different classes and potencies were purchased from different pharmaceutical companies. Gentamicin (10 µg) from Becton Dickinson, NJ, USA, vancomycin (30 µg), amikacin (30 µg), tetracycline (30 µg), ciprofloxacin (5 µg) from Bioanalyse® Ankara, Turkey, and amoxicillin (30 µg) from Oxoid™ Basingstoke, Hampshire, UK.
2.2. Bacterial Strains Used for Antimicrobial Susceptibility Testing
Laboratory bacterial strains of Staphylococcus epidermidis, Pseudomonas aeruginosa ATCC 27853, Staphylococcus saprophyticus, Staphylococcus aureus, Pseudomonas spp., Shigella, Escherichia coli, Klebsiella pneumonia, Streptococcus mutans, Mycobacterium smegmatis, and Enterococcus faecalis grown in trypticase soy broth were tested for antibacterial activity of β-Lap. All strains are preserved as glycerol stocks in the freezer at −80 °C.
2.3. Zone of Inhibition
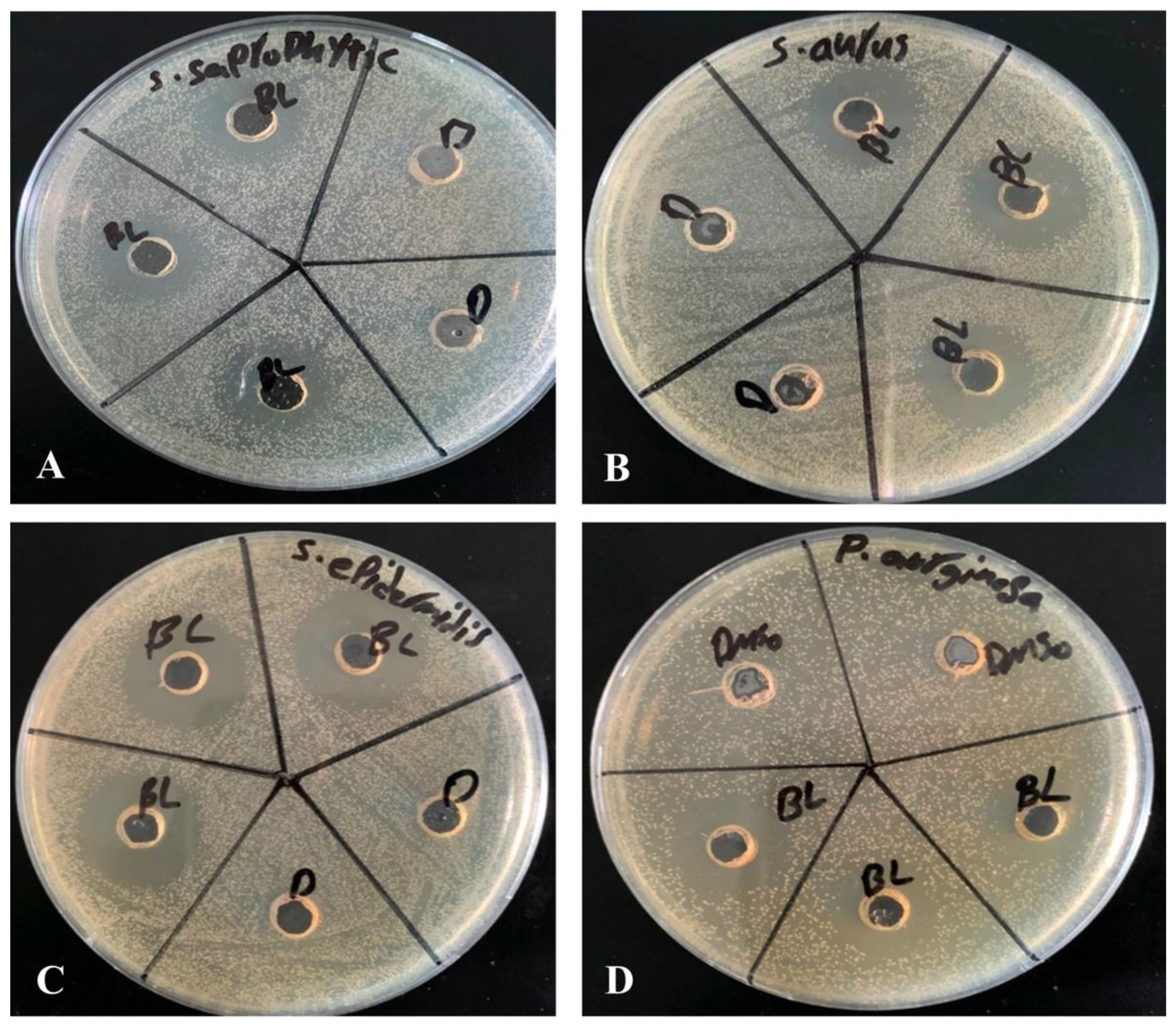
Agar well diffusion method is widely used to evaluate the antimicrobial activity of plants or microbial extracts. Wells of 6 mm diameter were punched aseptically with a sterile tip on trypticase soy agar (TSA) plates. Using aseptic techniques, inoculums of 0.5 McFarland were prepared by taking a colony from TSA plate into 2 mL of normal saline and mixing it thoroughly by vortexing. Subsequently, the bacterial suspension was diluted 100-fold by transferring 20 µL of the bacterial suspension in to 2 mL of sterile normal saline. Above diluted bacterial suspensions were spread over the agar plate’s surface using a sterile cotton swab to obtain a uniform microbial growth on plates. Then, 20 µL of 2.5 mM β-Lap dissolved in DMSO was dispensed into the wells of the plate, while, for control, 20 µL of DMSO was dispensed into separate wells. After 24-h incubation, a ruler was used to measure the diameter of a clear zone around the well.
2.4. Minimum Inhibitory Concentration (MIC) by Broth Dilution Method
MIC, the lowest concentration of antimicrobial agent which completely inhibits the growth of microorganism, was determined by broth dilution method as described [
18] with slight modification. In brief, some individual colonies of a bacterial strain were suspended into 2 mL of saline to an optical density of 0.5 McFarland. The culture was subsequently diluted 100-fold in trypticase soy broth. Then, 180 µL of diluted culture was dispensed into the wells of 96-well plate of having periphery wells filled with 200 µL of distilled water to avoid evaporation during incubation period. The 11th column was filled with 200 µL of sterile trypticase soy broth for the control of sterility of growth medium. Wells of the column 2 were reserved for control to which 20 µL of DMSO was added. The 20 μL of stock solutions of 10 mM, 5 mM, 2 mM, 0.4 mM, 0.08 mM, 0.016 mM, 0.0032 mM, and 0.00064 prepared in DMSO were added to the wells of a row from 3rd column to 10th column. For each bacterial strain, three wells per dilution of β-Lap were used. The plates were incubated for 24 h at 37 °C. The lowest concentration of the compound at which no visible growth was observed is considered as MIC for the bacterial strain tested.
2.5. Minimum Bactericidal Concentration by Streaking Method
The minimum bactericidal concentration (MBC) is the lowest concentration of an antimicrobial at which microorganism is completely killed. A sterilized loop was used to streak the cultures from the wells of MIC up to the wells of highest concentration of β-Lap on TSA plates. The concentration of β-Lap where no growth was detected is considered as MBC for the bacterial strain tested.
2.6. Microplate AlamarBlue Assay (MABA)
The Microplate AlamarBlue
® assay (MABA) is a sensitive, rapid, and nonradiometric method, which evaluates cellular health and offers the potential for screening large numbers of antimicrobial compounds. MABA was performed as described [
18] with a slight modification In brief, 180 µL of aforementioned 100-fold diluted culture of a bacterial strain in trypticase soy broth containing 1× alamarBlue were dispensed into the wells of 96-well plate. Then, 20 µL of β-Lap at its MIC and 2×MIC were dispensed in triplicate culture wells. For positive control and medium sterility 20 µL of DMSO were added to triplicate wells of culture and growth media, respectively. The periphery wells were filled with 200 µL of sterile water to avoid evaporation of the well content. The plate was sealed with breathable sealing film (Merck) to allow exchange of gasses. The plate was incubated in FLUOstar
® Omega microplate reader overnight. The fluorescent readings (excitation/emission maxima at 544/590 nm) were recorded after every 30 min. The average values of blank corrected fluorescence units were plotted against time using Microsoft Excel software. Standard deviations were calculated, and graphs were constructed by using Microsoft Excel software, version 16.66.1.
2.7. Antibiotic Activity Assay
To investigate the effect of the β-Lap compound for several commercial antibiotics, the 0.5 McFarland of bacterial suspension was diluted 100-fold in normal saline and the resulting bacterial suspension was streaked on TSA plates using a sterile cotton swab to achieve a uniform spread of microbial colonies. The antibiotic discs (diameter of 6 mm) were placed on the surface of inoculated plate to determine its inhibition zone size. For the effect on commercial antibiotics, 20 µL of β-Lap from 2.5 mM working stock were dispensed onto the antibiotic discs. The size of inhibition zone was measured in millimeters after 18–24 h of incubation at 37 °C.
2.8. Crystal Violet Biofilm Assay
First, 180 µL of 100-fold diluted bacterial suspension obtained from 0.5 McFarland were dispensed into the wells of 96-well plate. To determine the minimum biofilm inhibition concentration (MBIC), 20 µL of β-Lap compound at 5-fold dilutions ranging from 0.5 mM to 0.0016 mM were dispensed in triplicate wells. For the control 20 µL of DMSO were dispensed into the triplicate wells. After 24 h of incubation under static conditions of growth at 37 °C, the wells were rinsed thrice with sterile PBS (phosphate-buffered saline, pH 7.4) to remove the non-adherent bacteria. The cells on the walls of wells were fixed with 99% (v/v) methanol for 15 min. After discarding methanol wells were dried in laminar flow for 5 min. The attached biofilm cells were stained with 0.5% crystal violet (Sigma-Aldrich) for 5 min at room temperature. The excess stain was removed by washing thrice with distilled water and the crystal violet-bound cells were solubilized in 33% acetic acid. The absorbance readings at 570 nm were taken in FLUOstar® Omega microplate reader and the averages of the blank corrected values were plotted using Microsoft Excel. For mature biofilm formation, the cultures were allowed to grow statically for 24 or 48 h at 37 °C. The biofilms formed were washed thrice with PBS and incubated with sterile growth medium containing β-Lap. 24 h later the biofilms were washed, stained, and estimated as described above. The percentage of biofilm inhibition was calculated by the following formula: Biofilm inhibition (%) = [(Control OD570 nm − Treated OD570 nm)/Control OD570 nm] × 100.
2.9. Colony Forming Units (CFU)
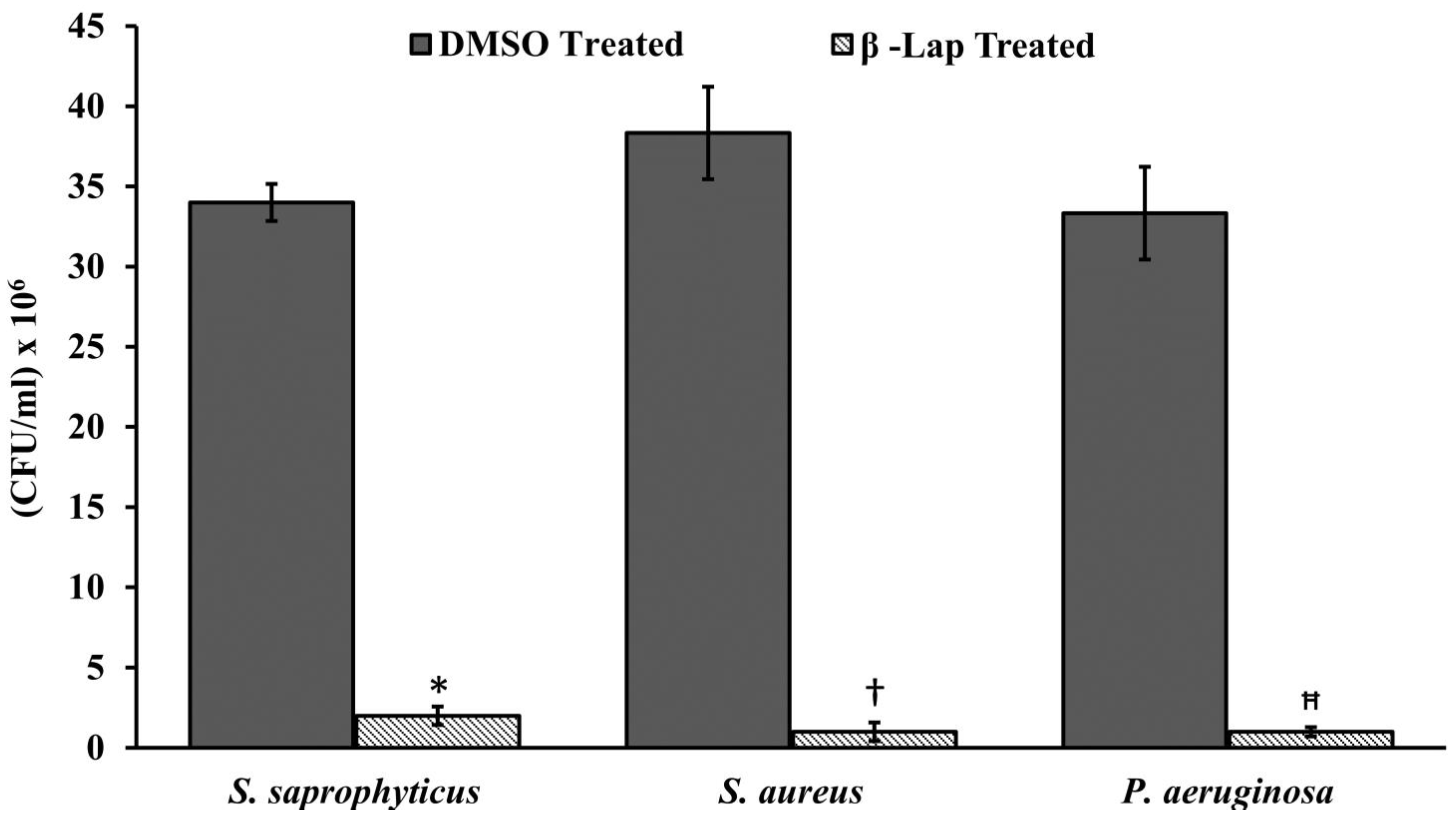
CFU of the cultures treated with DMSO or β-Lap were enumerated by plating 100 µL of 10-fold dilutions on nutrient agar plates. The colony count was calculated by the formula, CFU/mL = (Number of colonies in 0.1 mL of culture plated/0.1) × dilution factor. For hydrogen peroxide sensitivity assay, cells treated overnight with 1/5xMIC were pelleted down, washed thrice with 1xPBS, and re-suspended in 1xPBS containing 20 mM H2O2. After one hour of incubation at 37 °C, the cell suspensions were serially diluted and 100 µL were plated on nutrient agar plates. CFUs/mL ere enumerated from DMSO treated and β-Lap treated samples for each strain.
2.10. Catalase Activity Assay
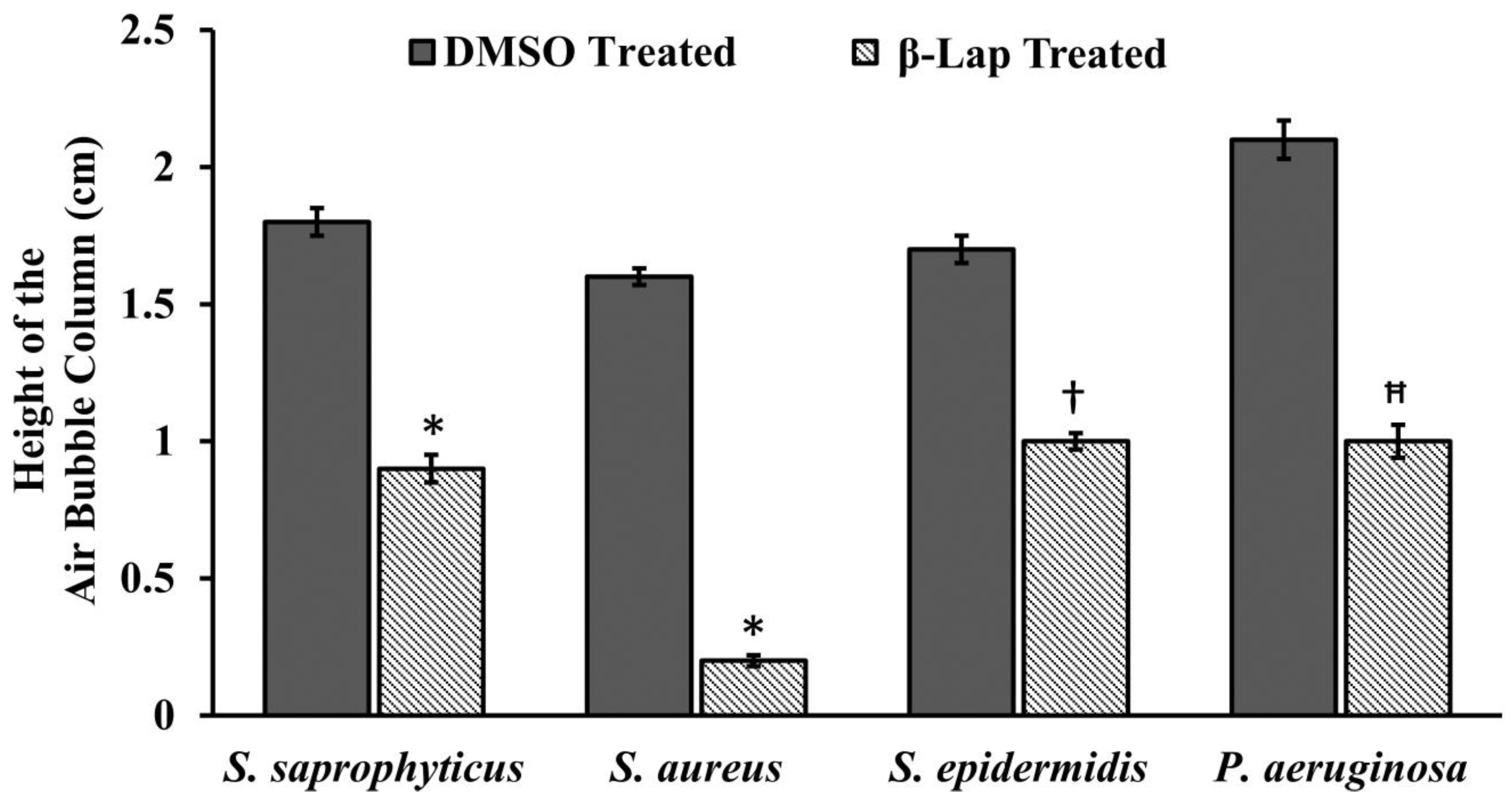
For catalase activity, the bacterial cultures containing β-Lap at its 1/100xMIC were incubated overnight in a shaking incubator at 200 RPM. For control, bacterial cultures in presence of DMSO were incubated under similar conditions. The 200 µL of 3% H2O2 were dispensed into one ml of the overnight culture in a glass tube and mixed immediately. The height of air bubble column formed above the culture was measured using a ruler. The experiment was setup in triplicates for each strain and the average length of the air bubble column was plotted using Microsoft Excel.
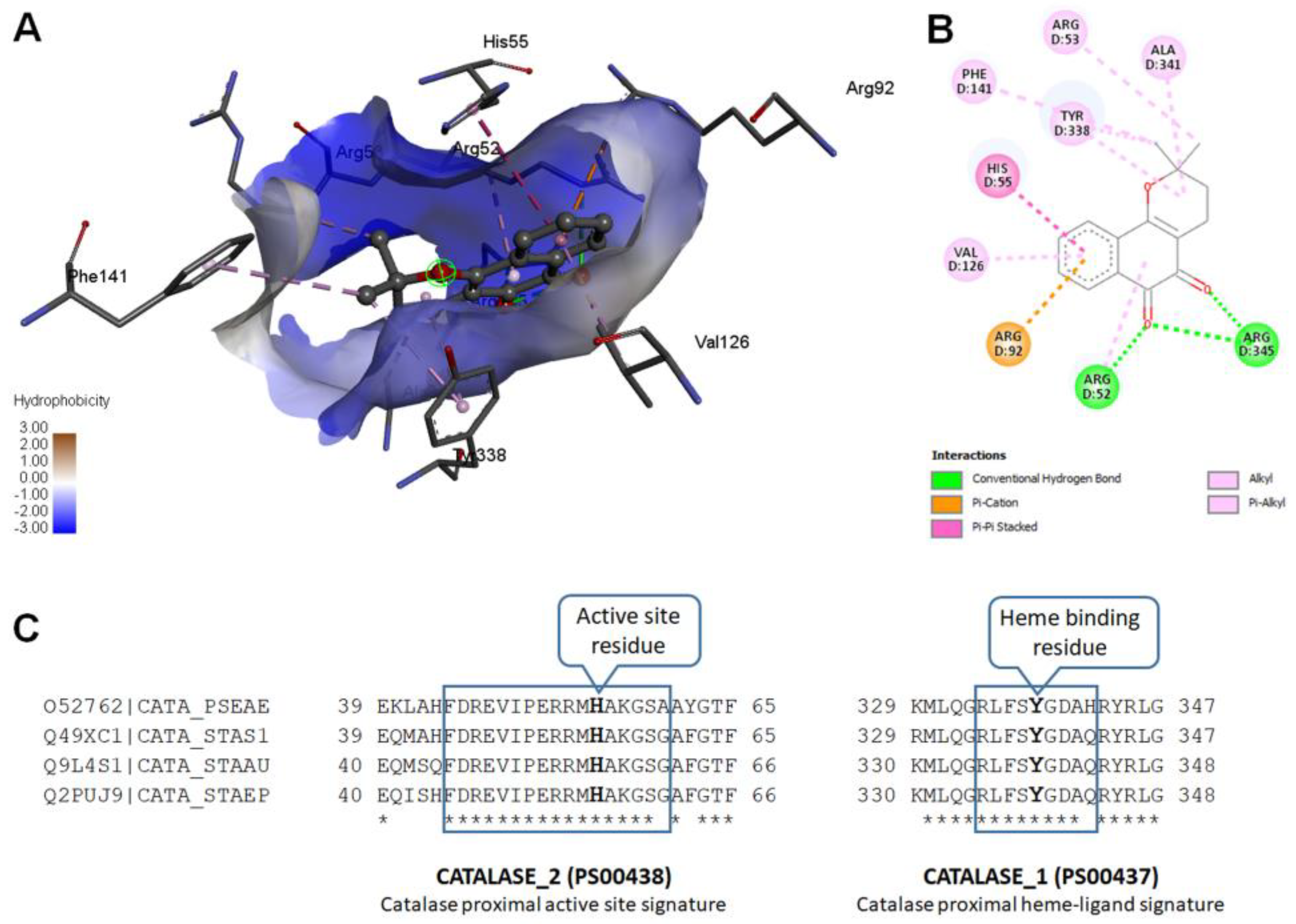
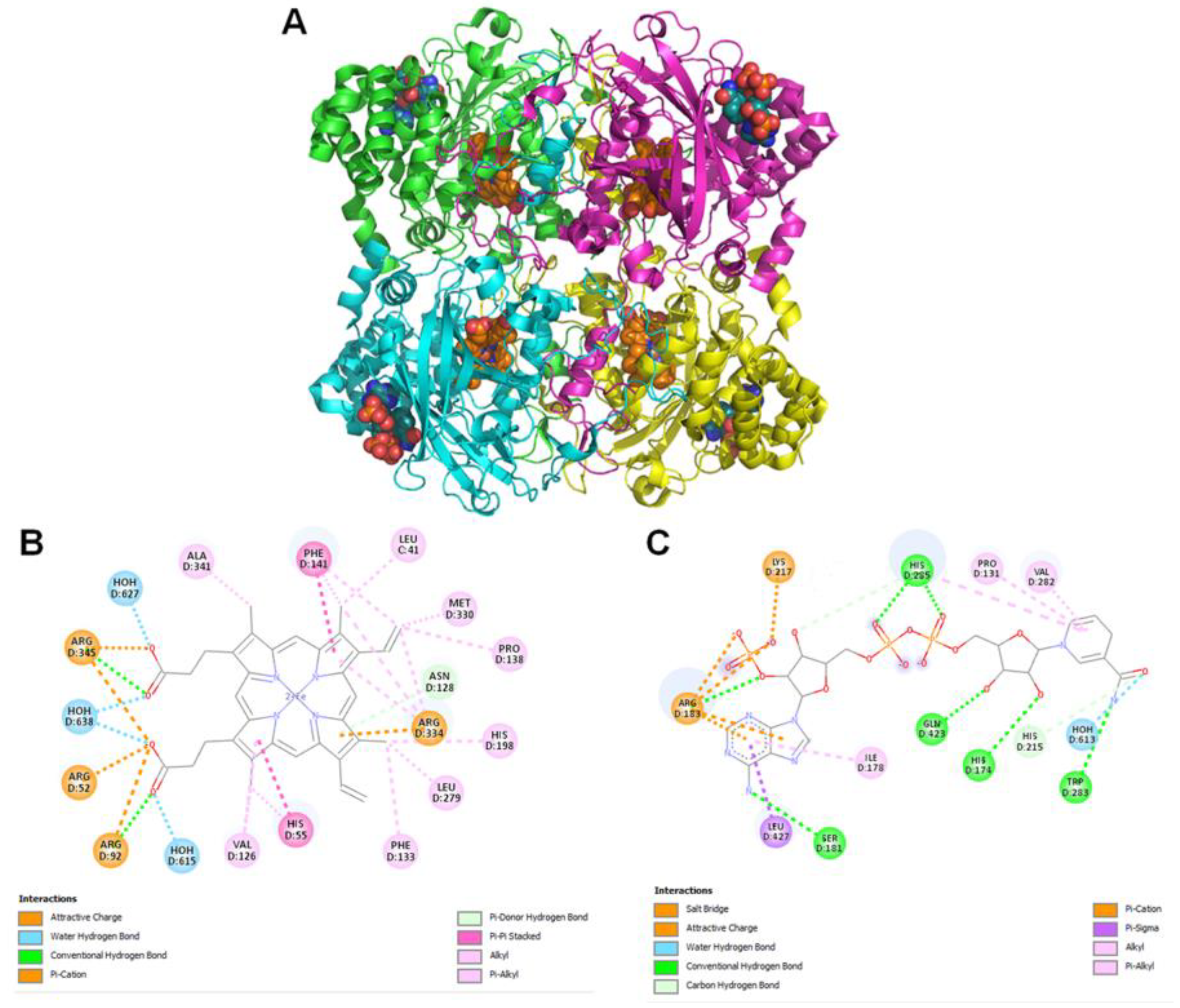
2.11. Molecular Modeling
Modeled
Pseudomonas aeruginosa catalase complexed with β-Lapachone using molecular docking approach. Coordinates of crystal structure of
P. aeruginosa catalase (KatA tetramer, PDB code: 4e37, resolution 2.53 Å) with heme and NADPH bound was downloaded from Protein Data Bank (
https://www.rcsb.org/structure/4E37) on 20 December 2022. Bound ligands and water molecules were removed for docking studies. β-Lap structure (.sdf file, PubChem CID: 3885) were downloaded from PubChem database [
19] (
https://pubchem.ncbi.nlm.nih.gov/compound/3885, accessed on 20 December 2022). To generate KatA-ligand complexes CB-Dock2 [
20], cavity guided blind docking, web server available at
https://cadd.labshare.cn/cb-dock2/ (accessed on 20 December 2022) was used. Modeled KatA-ligand complexes were analyzed using BIOVIA Discovery Studio Visualizer, v21, 2021, Protein-Ligand Interaction Profiler [
21] available at
https://plip-tool.biotec.tu-dresden.de/plip-web/plip/index (accessed on 20 December 2022) and PyMOL available at
https://pymol.org (accessed on 20 December 2022) to identify biologically relevant KatA-ligand complexes.
2.12. Statistical Analysis
IBM SPSS statistical software version 21 was used to perform the statistical analysis of data. All the data sets were obtained in triplicates from more than one biological replicate and the values were interpreted as means ± standard deviation (SD). One way ANOVA with Tukey’s post-hoc test was used to assess the differences between treated and untreated samples. The p < 0.05 was considered statistically significant.
4. Discussion
In recent decades the increase in infectious cases caused by bacterial species and the resulting disproportionate use of antimicrobials has led to the development of resistance to antimicrobial drugs [
23]. To overcome such hurdles, scientists gained interest in recent decades in identifying new antimicrobials, for which plants and their secondary metabolites are a prospective source of bioactive molecules as potential therapeutic agents [
24]. Naphthoquinones, a group of secondary metabolites commonly found in higher plants, have great structural diversity and biological activity.
In the present study, we evaluated the ability of β-Lap, a naphthoquinone, to inhibit the growth of several pathogenic bacteria. Out of eleven bacterial strains screened, only four
S. saprophyticus, S. aureus,
S. epidermidis, and
P. aeruginosa were susceptible to the tested concentration (2.5 mM) of β-Lap with zone of inhibition ranging from 17.7 mm to 26.7 mm (
Table 1). Among them
S. epidermidis,
S. saprophyticus, and
P. aeruginosa showed the highest zone of inhibition, while
S. aureus comparatively showed lower zone of inhibition. As expected, a positive correlation was observed between inhibition zone sizes and corresponding MIC values. To further evaluate the bactericidal activity of β-Lap, cells were treated with increasing concentration of β-Lap (1/2xMIC, MIC, 2xMIC and 4xMIC) and as expected CFUs enumerated decreased in a dose dependent matter, suggesting that the β-Lap compound was bactericidal in nature (
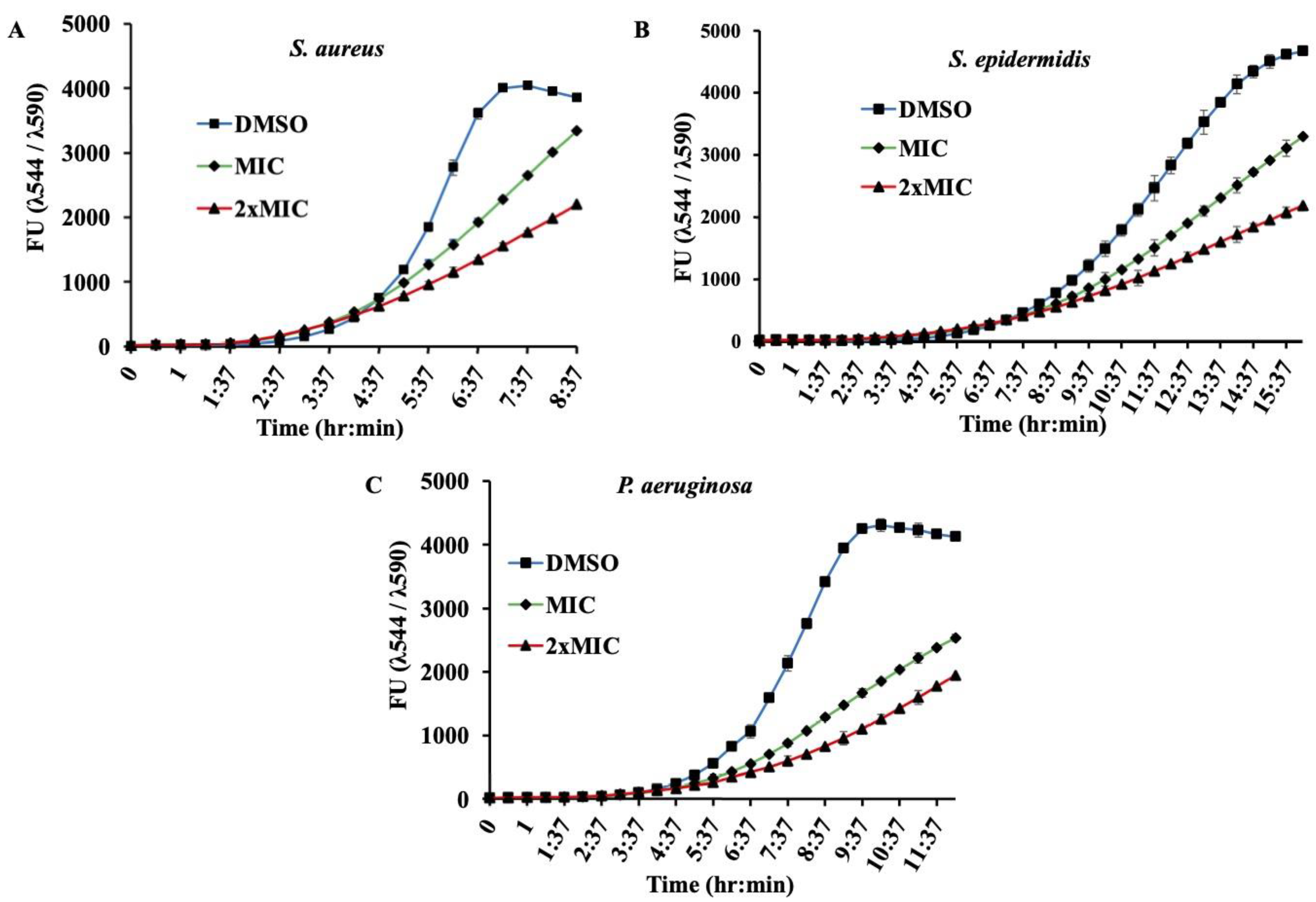
Supplementary Figure S1). In addition to this endpoint analysis, growth kinetics of bacterial strains in presence of MIC and 2xMIC of β-Lap confirmed the decrease in growth rate in a dose-depended manner (
Figure 2). Though antibacterial activity of the β-Lap has not been studied in detail, our data were consistent with the other previous observations. In one of the studies, β-Lap encapsulated in liposomes inhibited different MRSA isolates [
11]. In another study it inhibited the methicillin sensitive and resistant strains of
S. aureus and methicillin resistant strains of
S. epidermidis and
S. haemolyticus [
25]. Among the naphthoquinones (lapachol, α-lapachone, and β-lapachone) tested against several strains of MRSA, β-Lap exhibited not only antibacterial activity but synergic activity in association with commercial antibiotics [
13]. Since in the present study all the β-Lap susceptible strains are aerobes and Gram positive, we suspected that the activity of catalases might be compromised in such strains. In support of this notion, we assayed the sensitivity of β-Lap treated cells towards H
2O
2 treatment. It is evident from the
Figure 3 and
Figure 4 that the cells treated with β-Lap were very sensitive to H
2O
2 treatment due to the large reduction in their catalase activity. The cause of reduction of the catalase activity needs to be investigated, which is our future goal of research. It could be either due to the reduction in the level of catalase or inhibition of its catalytic activity. Molecular modelling predicted the inhibition of catalase activity by binding to the proximal active site and heme binding sites of the enzyme.
The combinational therapy of the new drugs with already existing commercial antimicrobials is an important strategy in the search of more effective antimicrobial agents for treating microbial infections. This can potentially increase efficacy, reduce toxicity, cure faster, prevent the emergence of resistance, and provide a broader spectrum of activity than mono therapy regimens. Combinational effect of β-Lap with other antibiotics on the bacterial strains tested (
Table 2) showed that it has a potential to increase the activity of some drugs in vitro. These results are in agreement with the synergistic effects between β-Lap and conventional antimicrobials against methicillin resistant
Staphylococcus aureus (MRSA) strains [
13]. It remains to be seen whether the β-Lap will have the same effect in combination with antibiotics in vivo.
Bacterial cells, being substantially more resistant to drugs and host immune responses in biofilms, pose a great challenge to chemotherapy of the infectious diseases. There is poor antibiotic penetration and slow growth due to nutrient limitation, which makes cells less prone to antibiotics. Hence, there is a need to develop new drugs that could penetrate well and kill the bacterial cells protected by multi-layered extracellular polymeric matrix.
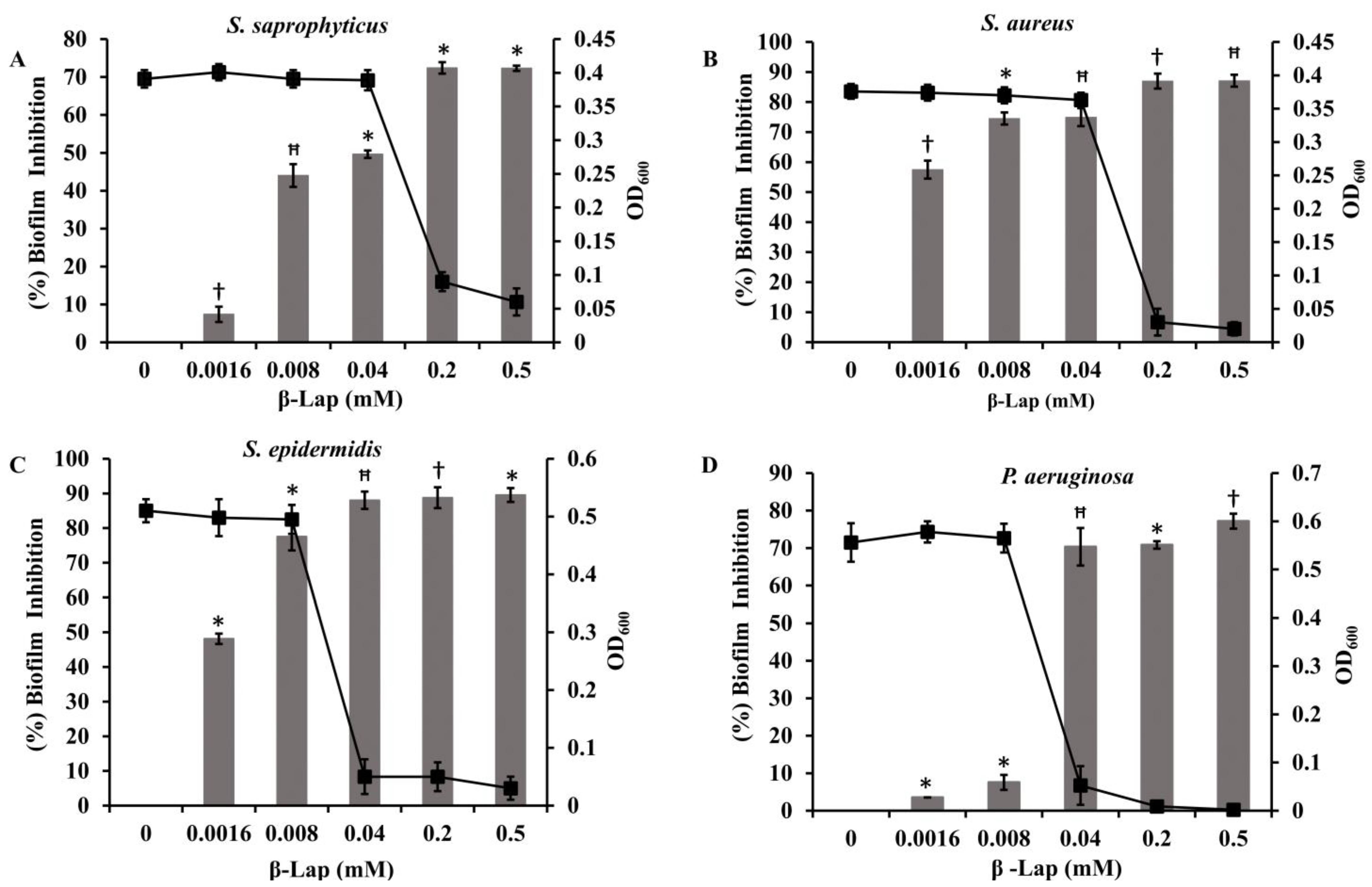
Four bacterial strains
S. saprophyticus, S. aureus, S. epidermidis, and
P. aeruginosa (ATCC 27853) were tested to investigate the effect of β-Lap on their biofilm formation. It was found that the biofilm formation in the tested strains significantly inhibited (50–80% biofilm inhibition) at MBIC, except
P. aeruginosa. Among the strains
S. saprophyticus, S. aureus, and
S. epidermidis, the biofilm formation of
S. aureus and
S. epidermidis was strongly inhibited. The inhibition was 75% for
S. aureus and 78% for
S. epidermidis at their respective MBIC. More importantly, no inhibition of growth occurred in the strains at MBIC (
Figure 7), suggesting that β-Lap targets the processes or factors required for biofilm formation without effecting the biological processes required for normal growth of the bacteria. These results suggested that the β-Lap could be potential antibiofilm agent against
S. aureus and
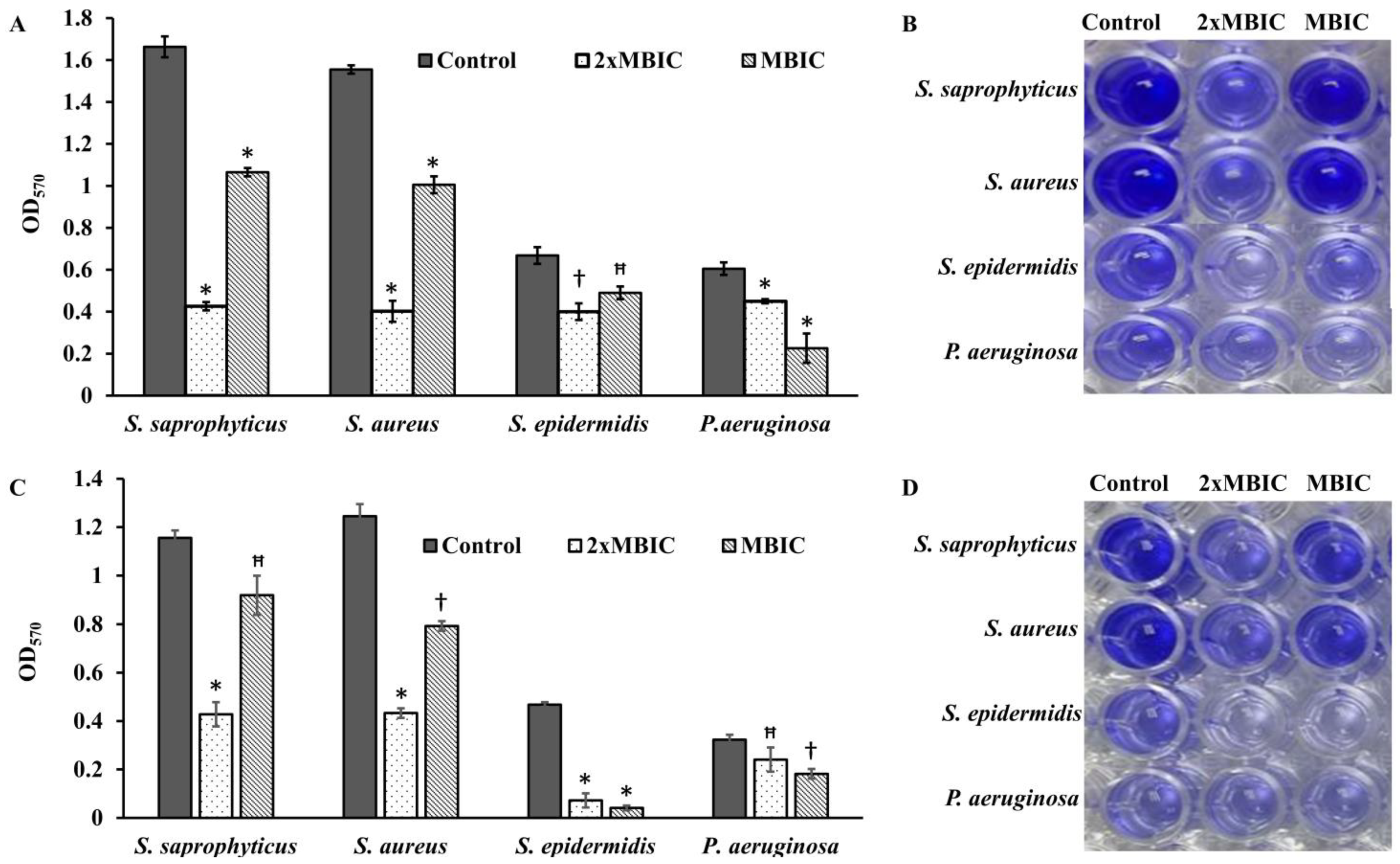
S. epidermidis. Not only did β-Lap inhibit biofilm formation, but it disrupted the mature biofilms of these bacteria. It was found that 48-h mature biofilms of
S. epidermidis were more sensitive to β-Lap as the biofilms were eradicated 11.5-fold by β-Lap treatment than that of control where the biofilms were treated with DMSO alone. Though biofilm formation of
P. aeruginosa was not inhibited by β-Lap, their 24-h mature biofilms were eradicated 2.7-fold more than that of control, where biofilms were treated with DMSO only. The mechanism behind the inhibition of biofilm formation and eradication of already formed biofilms needs to be investigated. Our results of antibacterial and antibiofilm activities of β-Lap corroborate well with the study of Fernandes AWC, et al., wherein the authors showed that β-Lap and Lapachol Oxime (semi-synthetic derivatives of Lapachol) exhibited antimicrobial and anti-biofilm potential against clinical isolates of
S. aureus [
26].
The results altogether certainly confirm that the β-Lap exhibited strong antibacterial and antibiofilm activities as well as enhanced the efficacy of commercial antibiotics tested. Therefore, β-Lap could be a drug of choice to be used in treating bacterial infections, but its activity in vivo needs to be investigated, which demands further research.
Conclusion: Taken together, our data suggest that β-Lap is a promising novel antibacterial agent that inhibits the bacterial catalase.