Genomic Analysis Reveals New Integrative Conjugal Elements and Transposons in GBS Conferring Antimicrobial Resistance
Abstract
1. Introduction
2. Results
2.1. Phenotypic and Genotypic AMR Profiling
2.2. Tetracycline Resistance and Mobile Genetic Elements
2.3. Macrolide Resistance and Mobile Genetic Elements
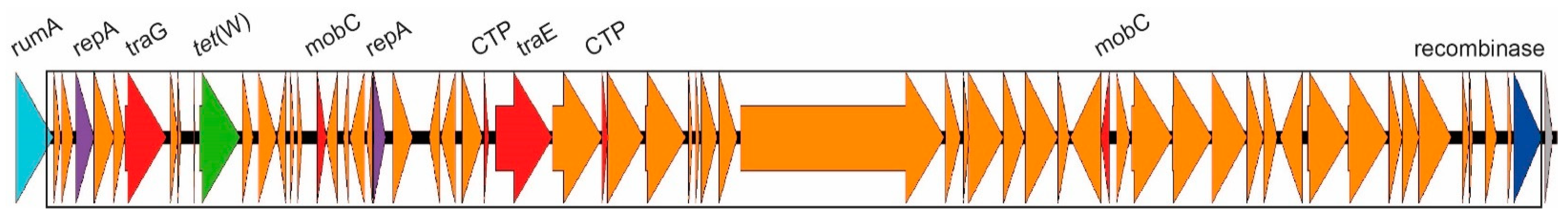
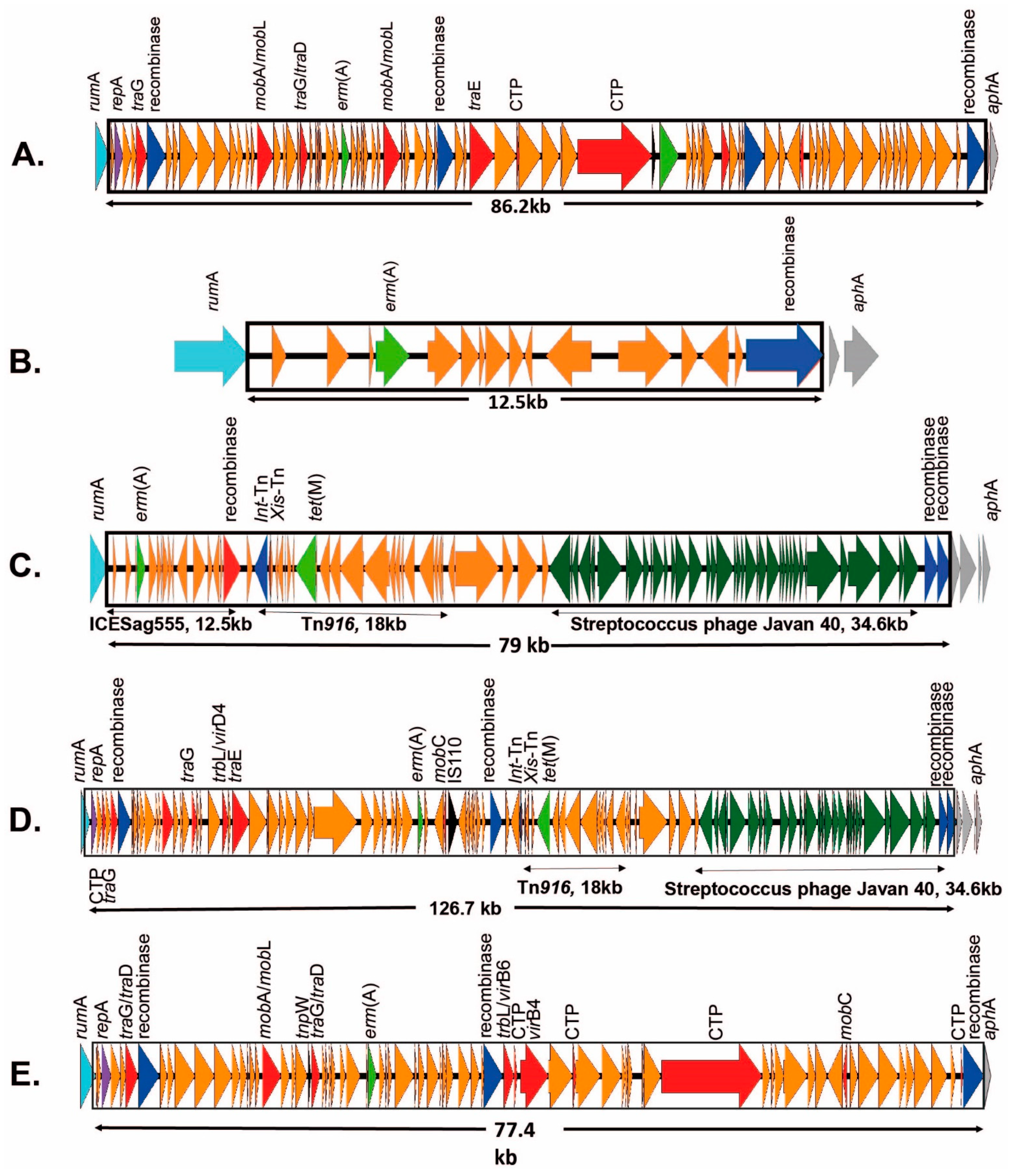
2.3.1. ICEs Carrying the erm(A) Gene
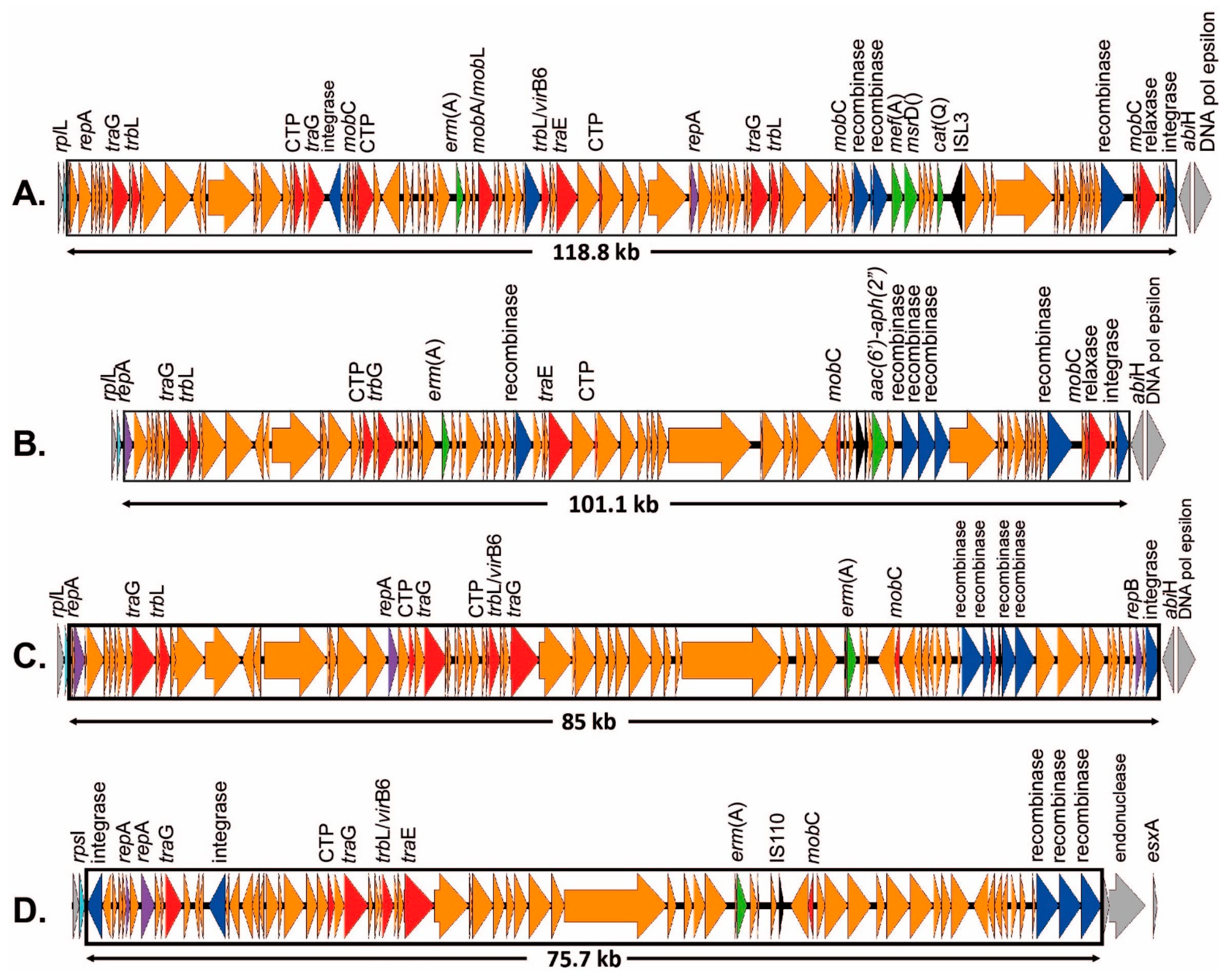
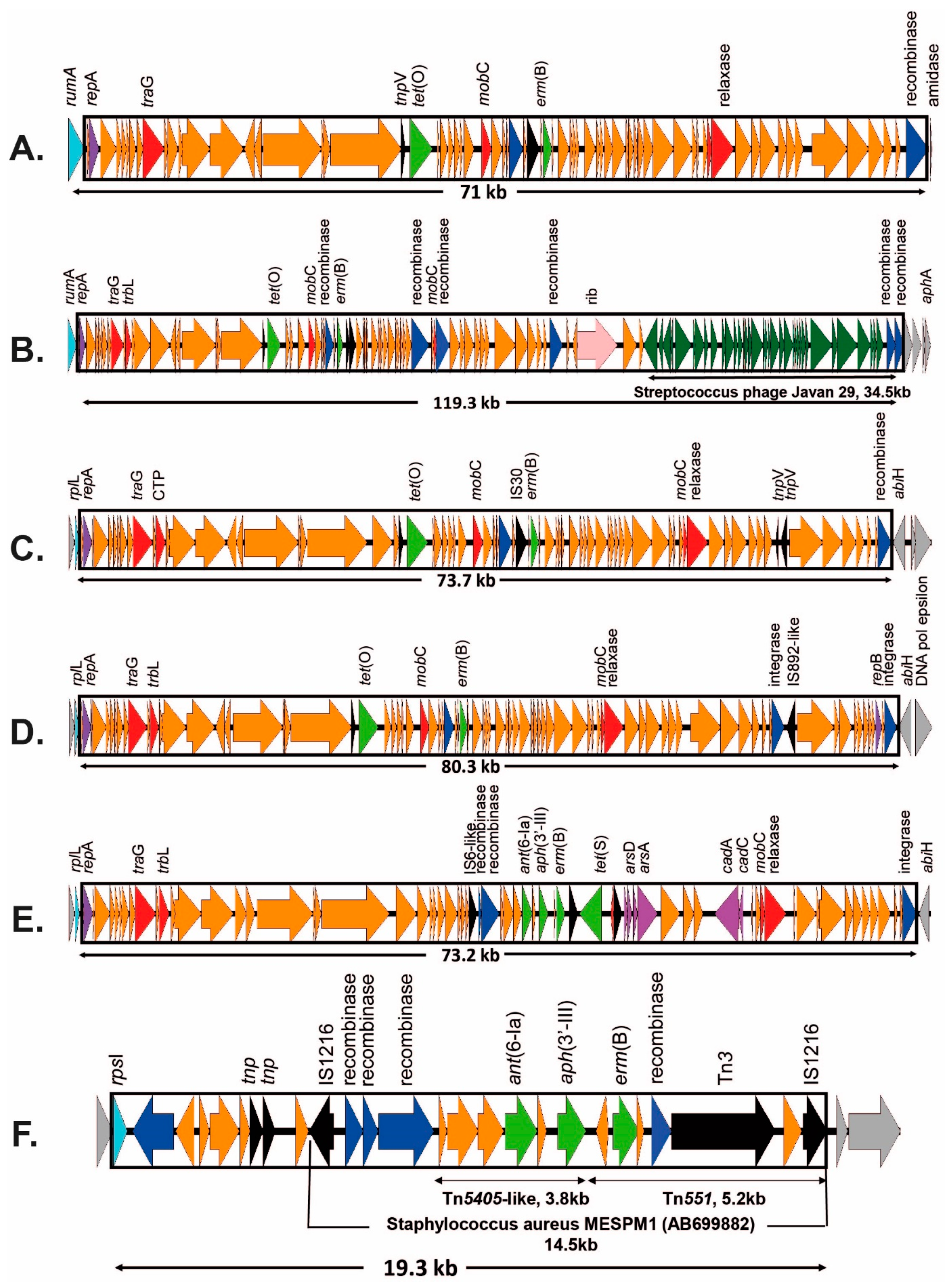
2.3.2. ICEs Carrying the erm(B) Gene
2.3.3. ICEs Carrying mef(A)/msr(D) Genes
2.3.4. ICEs Carrying the lsa(C) Gene
3. Discussion
4. Materials and Methods
4.1. Clinical GBS Strains and Their Whole Genome Sequences
4.2. Phenotypic Antibiotic Susceptibility Testing (AST)
4.3. Long-Read Whole Genome Sequencing
4.4. Resistome Profile Associated with the Mobile Genetic Elements
4.5. Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Navarro-Torné, A.; Curcio, D.; Moïsi, J.C.; Jodar, L. Burden of invasive group B Streptococcus disease in non-pregnant adults: A systematic review and meta-analysis. PLoS ONE 2021, 16, e0258030. [Google Scholar] [CrossRef]
- Rudman, J.; Carter, E.; Coehlo, J.; Hopkins, K.; Chalker, V.J.; Guy, R.; Lamagni, T.L. Laboratory surveillance of pyogenic and non-pyogenic streptococcal bacteraemia in England (2020). Health Prot. Rep. 2020, 15, 1–23. [Google Scholar]
- Healy, B. Public H.W. Group B Streptococcal Infection. Available online: https://bestpractice.bmj.com/topics/en-gb/924 (accessed on 3 December 2022).
- Hayes, K.; O’Halloran, F.; Cotter, L. A review of antibiotic resistance in Group B Streptococcus: The story so far. Crit. Rev. Microbiol. 2020, 46, 253–269. [Google Scholar] [CrossRef]
- CLSI. M100 Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022; ISBN 978-1-68440-135-2. [Google Scholar]
- Chu, Y.W.; Tse, C.; Tsang, G.K.L.; So, D.K.S.; Fung, J.T.L.; Lo, J.Y.C. Invasive group B Streptococcus isolates showing reduced susceptibility to penicillin in Hong Kong. J. Antimicrob. Chemother. 2007, 60, 1407–1409. [Google Scholar] [CrossRef]
- Kimura, K.; Suzuki, S.; Wachino, J.I.; Kurokawa, H.; Yamane, K.; Shibata, N.; Nagano, N.; Kato, H.; Shibayama, K.; Arakawa, Y. First molecular characterization of group B streptococci with reduced penicillin susceptibility. Antimicrob. Agents Chemother. 2008, 52, 2890–2897. [Google Scholar] [CrossRef]
- Nagano, N.; Nagano, Y.; Toyama, M.; Kimura, K.; Tamura, T.; Shibayama, K.; Arakawa, Y. Nosocomial spread of multidrug-resistant group B streptococci with reduced penicillin susceptibility belonging to clonal complex 1. J. Antimicrob. Chemother. 2012, 67, 849–856. [Google Scholar] [CrossRef]
- Seki, T.; Kimura, K.; Reid, M.E.; Miyazaki, A.; Banno, H.; Jin, W.; Wachino, J.I.; Yamada, K.; Arakawa, Y. High isolation rate of MDR group B streptococci with reduced penicillin susceptibility in Japan. J. Antimicrob. Chemother. 2015, 70, 2725–2728. [Google Scholar] [CrossRef]
- Dahesh, S.; Hensler, M.E.; Van Sorge, N.M.; Gertz, R.E.; Schrag, S.; Nizet, V.; Beall, B.W. Point mutation in the group B streptococcal pbp2x gene conferring decreased susceptibility to β-lactam antibiotics. Antimicrob. Agents Chemother. 2008, 52, 2915–2918. [Google Scholar] [CrossRef]
- Metcalf, B.J.; Chochua, S.; Gertz, R.E.; Hawkins, P.A.; Ricaldi, J.; Li, Z.; Walker, H.; Tran, T.; Rivers, J.; Mathis, S.; et al. Short-read whole genome sequencing for determination of antimicrobial resistance mechanisms and capsular serotypes of current invasive Streptococcus agalactiae recovered in the USA. Clin. Microbiol. Infect. 2017, 23, 574.e7–574.e14. [Google Scholar] [CrossRef]
- Francois Watkins, L.K.; McGee, L.; Schrag, S.J.; Beall, B.; Jain, J.H.; Pondo, T.; Farley, M.M.; Harrison, L.H.; Zansky, S.M.; Baumbach, J.; et al. Epidemiology of Invasive Group B Streptococcal Infections among Nonpregnant Adults in the United States, 2008–2016. JAMA Intern. Med. 2019, 179, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Gaudreau, C.; Lecours, R.; Ismaïl, J.; Gagnon, S.; Jetté, L.; Roger, M. Prosthetic hip joint infection with a Streptococcus agalactiae isolate not susceptible to penicillin G and ceftriaxone. J. Antimicrob. Chemother. 2009, 65, 594–595. [Google Scholar] [CrossRef]
- Longtin, J.; Vermeiren, C.; Shahinas, D.; Tamber, G.S.; McGeer, A.; Low, D.E.; Katz, K.; Pillai, D.R. Novel mutations in a patient isolate of Streptococcus agalactiae with reduced penicillin susceptibility emerging after long-term oral suppressive therapy. Antimicrob. Agents Chemother. 2011, 55, 2983–2985. [Google Scholar] [CrossRef]
- Sigaúque, B.; Kobayashi, M.; Vubil, D.; Nhacolo, A.; Chaúque, A.; Moaine, B.; Massora, S.; Mandomando, I.; Nhampossa, T.; Bassat, Q.; et al. Invasive bacterial disease trends and characterization of group B streptococcal isolates among young infants in southern Mozambique, 2001–2015. PLoS ONE 2018, 13, e0191193. [Google Scholar] [CrossRef] [PubMed]
- Lamagni, T.L.; Keshishian, C.; Efstratiou, A.; Guy, R.; Henderson, K.L.; Broughton, K.; Sheridan, E. Emerging trends in the epidemiology of invasive group B streptococcal disease in England and Wales, 1991–2010. Clin. Infect. Dis. 2013, 57, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Varaldo, P.E.; Montanari, M.P.; Giovanetti, E. Genetic elements responsible for erythromycin resistance in streptococci. Antimicrob. Agents Chemother. 2009, 53, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Malbruny, B.; Werno, A.M.; Murdoch, D.R.; Leclercq, R.; Cattoir, V. Cross-resistance to lincosamides, streptogramins A, and pleuromutilins due to the lsa(C) Gene in Streptococcus agalactiae UCN70. Antimicrob. Agents Chemother. 2011, 55, 1470–1474. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, P.A.; Law, C.S.; Metcalf, B.J.; Chochua, S.; Jackson, D.M.; Westblade, L.F.; Jerris, R.; Beall, B.W.; McGee, L. Cross-resistance to lincosamides, streptogramins A and pleuromutilins in Streptococcus agalactiae isolates from the USA. J. Antimicrob. Chemother. 2017, 72, 1886–1892. [Google Scholar] [CrossRef]
- Zhou, K.; Xie, L.; Han, L.; Guo, X.; Wang, Y.; Sun, J. ICESag37, a novel integrative and conjugative element carrying antimicrobial resistance genes and potential virulence factors in Streptococcus agalactiae. Front. Microbiol. 2017, 8, 1921. [Google Scholar] [CrossRef]
- Cunha, V.D.; Davies, M.R.; Douarre, P.; Rosinski-, I. Streptococcus agalactiae clones infecting humans were selected and fixed through the extensive use of tetracycline. Nat. Commun. 2015, 5, 4544. [Google Scholar] [CrossRef]
- Brochet, M.; Couvé, E.; Glaser, P.; Guédon, G.; Payot, S. Integrative conjugative elements and related elements are major contributors to the genome diversity of Streptococcus agalactiae. J. Bacteriol. 2008, 190, 6913–6917. [Google Scholar] [CrossRef]
- Ambroset, C.; Coluzzi, C.; Guédon, G.; Devignes, M.-D.; Loux, V.; Lacroix, T.; Payot, S.; Leblond-Bourget, N. New Insights into the Classification and Integration Specificity of Streptococcus Integrative Conjugative Elements through Extensive Genome Exploration. Front. Microbiol. 2015, 6, 1483. [Google Scholar] [CrossRef]
- Morici, E.; Simoni, S.; Brenciani, A.; Giovanetti, E.; Varaldo, P.E.; Mingoia, M. A new mosaic integrative and conjugative element from Streptococcus agalactiae carrying resistance genes for chloramphenicol (cat(Q)) and macrolides [mef(I) and erm(TR)]. J. Antimicrob. Chemother. 2017, 72, 64–67. [Google Scholar] [CrossRef]
- Grosso, M.D.; Camilli, R.; Rizzi, E.; Pietrelli, A.; Bellis, G. De ICE Spy 009, a Conjugative Genetic Element Carrying mef (E) in Streptococcus pyogenes. Antimicrob. Agents Chemother 2012, 60, 3906–3912. [Google Scholar] [CrossRef]
- Achard, A.; Villers, C.; Pichereau, V.; Leclercq, R. New lnu (C) gene conferring resistance to lincomycin by nucleotidylation in Streptococcus agalactiae UCN36. Antimicrob. Agents Chemother. 2005, 49, 2716–2719. [Google Scholar] [CrossRef]
- Douarre, P.E.; Sauvage, E.; Poyart, C.; Glaser, P. Host specificity in the diversity and transfer of lsa resistance genes in group B Streptococcus. J. Antimicrob. Chemother. 2015, 70, 3205–3213. [Google Scholar] [CrossRef]
- Nagano, N.; Nagano, Y.; Kimura, K.; Tamai, K.; Yanagisawa, H.; Arakawa, Y. Genetic heterogeneity in pbp genes among clinically isolated group B streptococci with reduced penicillin susceptibility. Antimicrob. Agents Chemother. 2008, 52, 4258–4267. [Google Scholar] [CrossRef]
- Africa, C.W.J.; Kaambo, E. Group B Streptococcus serotypes in pregnant women from the Western Cape region of South Africa. Front. Public Health 2018, 6, 356. [Google Scholar] [CrossRef] [PubMed]
- Yi, A.; Kim, C.K.; Kimura, K.; Arakawa, Y.; Hur, M.; Yun, Y.M.; Moon, H.W. First case in Korea of group B Streptococcus with reduced penicillin susceptibility harboring amino acid substitutions in penicillin-binding protein 2x. Ann. Lab. Med. 2019, 39, 414–416. [Google Scholar] [CrossRef]
- Buu-Hoi, A.; Le Bouguenec, C.; Horaud, T. High-level chromosomal gentamicin resistance in Streptococcus agalactiae (Group B). Antimicrob. Agents Chemother. 1990, 34, 985–988. [Google Scholar] [CrossRef] [PubMed]
- Mingoia, M.; Morici, E.; Tili, E.; Giovanetti, E.; Montanari, M.P.; Varaldo, P.E. Characterization of Tn5801.Sag, a variant of Staphylococcus aureus Tn916 family transposon Tn5801 that is widespread in clinical isolates of Streptococcus agalactiae. Antimicrob. Agents Chemother. 2013, 57, 4570–4574. [Google Scholar] [CrossRef] [PubMed]
- Teatero, S.; Ramoutar, E.; McGeer, A.; Li, A.; Melano, R.G.; Wasserscheid, J.; Dewar, K.; Fittipaldi, N. Clonal Complex 17 Group B Streptococcus strains causing invasive disease in neonates and adults originate from the same genetic pool. Sci. Rep. 2016, 6, 20047. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Gao, C.; Huang, L.; Guan, X.; Ji, W.; Chang, C.Y.; McIver, D.J.; Deng, Q.; Zhong, H.; Xie, Y.; et al. Predominance of III/ST19 and Ib/ST10 Lineages with High Multidrug Resistance in Fluoroquinolone-Resistant Group B Streptococci Isolates in Which a New Integrative and Conjugative Element Was Identified. Front. Microbiol. 2021, 11, 609526. [Google Scholar] [CrossRef] [PubMed]
- Diekema, D.J.; Andrews, J.I.; Huynh, H.; Rhomberg, P.R.; Doktor, S.R.; Beyer, J.; Shortridge, V.D.; Flamm, R.K.; Jones, R.N.; Pfaller, M.A. Molecular epidemiology of macrolide resistance in neonatal bloodstream isolates of group B streptococci. J. Clin. Microbiol. 2003, 41, 2659–2661. [Google Scholar] [CrossRef]
- Von Both, U.; Ruess, M.; Mueller, U.; Fluegge, K.; Sander, A.; Berner, R. A serotype V clone is predominant among erythromycin-resistant Streptococcus agalactiae isolates in a southwestern region of Germ(A)ny. J. Clin. Microbiol. 2003, 41, 2166–2169. [Google Scholar] [CrossRef]
- Tsai, M.H.; Hsu, J.F.; Lai, M.Y.; Lin, L.C.; Chu, S.M.; Huang, H.R.; Chiang, M.C.; Fu, R.H.; Lu, J.J. Molecular characteristics and antimicrobial resistance of group B Streptococcus strains causing invasive disease in neonates and adults. Front. Microbiol. 2019, 10, 264. [Google Scholar] [CrossRef] [PubMed]
- Lopes, E.; Fernandes, T.; Machado, M.P.; Carriço, J.A.; Melo-Cristino, J.; Ramirez, M.; Martins, E.R.; Oliveira, H.; Vaz, T.; Gião, M.; et al. Increasing macrolide resistance among Streptococcus agalactiae causing invasive disease in non-pregnant adults was driven by a single capsular-transformed lineage, Portugal, 2009 to 2015. Eurosurveillance 2018, 23, 1700473. [Google Scholar] [CrossRef]
- Khan, U.B.; Jauneikaite, E.; Andrews, R.; Chalker, V.J.; Spiller, O.B. Identifying large-scale recombination and capsular switching events in Streptococcus agalactiae strains causing disease in adults in the UK between 2014 and 2015. Microb. Genom. 2022, 8, 000783. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, S.M.; Nasaj, M.; Hosseini, S.M.; Arabestani, M.R. Survey of strain distribution and antibiotic resistance pattern of group B streptococci (Streptococcus agalactiae) isolated from clinical specimens. GMS Hyg. Infect. Control 2016, 11, Doc18. [Google Scholar] [CrossRef]
- Sands, K.; Carvalho, M.J.; Portal, E.; Thomson, K.; Dyer, C.; Akpulu, C.; Andrews, R.; Ferreira, A.; Gillespie, D.; Hender, T.; et al. Characterization of antimicrobial-resistant Gram-negative bacteria that cause neonatal sepsis in seven low- and middle-income countries. Nat. Microbiol. 2021, 6, 512–523. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Tansirichaiya, S.; Rahman, M.A.; Roberts, A.P. The Transposon Registry. Mob. DNA 2019, 10, 40. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Kaas, R.S.; Seyfarth, A.M.; Agersø, Y.; Lund, O.; Larsen, M.V.; Aarestrup, F.M. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J. Antimicrob. Chemother. 2013, 68, 771–777. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.; Harris, S.R.; Berriman, M.; Parkhill, J.; McQuillan, J.A. Artemis: An integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 2012, 28, 464–469. [Google Scholar] [CrossRef]

| Clonal Complex | Serotype | Insertion Site | Number of Isolates | ARGs | Mobile Genetic Element | Novel | Accession Number |
|---|---|---|---|---|---|---|---|
| CC1 | Ib | random A | 1 | erm(B) + tet(M) | Tn3872 | N | OP715845 |
| V | random | 3 | erm(B) + tet(M) | Tn3872 | N | OP715845 | |
| V | rpsI | 1 | erm(A) | ICESag100414 | Y | OP715842 | |
| V | rumA | 2 | erm(A) + tet(M) erm(A) + tet(M) | ICESag082 ICESag098 | Y Y | OP715838 OP715840 | |
| VI | rplL | 1 | erm(B) + ant(6-Ia) + aph(3′-III) + tet(S) | ICESag662 | Y | OP508062 | |
| CC12 | Ib | rumA | 2 | erm(A) erm(A) | ICESag428 ICESag555 | Y in ICESag082 | OP715841 OP715841 |
| Ib | unknown B | 1 | erm(B) + tet(M) | undefined | ND | none | |
| II | rumA | 4 | erm(A) + tet(O) | ICESag509 | Y | OP715843 | |
| CC17 | III | rumA | 1 | erm(A) | ICESag066 | Y | OP715837 |
| III | rumA | 2 | erm(B) + ant(6-Ia) + aph(3′-III) + aad(E) + tet(O) | ICESag37 | N | OP508056 | |
| III | rumA + rplL | 2 | erm(B) + ant(6-Ia) + aph(3′-III) + aad(E) + tet(O) + mef(A)/msr(D) C | ICESag37 + ICESpy009 | N | OP508056 + OP508057 | |
| CC19 | II | rumA | 1 | erm(B) + tet(O) | ICESag624 | Y | OP715844 |
| III | rumA (random) | 1 | erm(B) + tet(M) (+ lsa(C)) | ICESag071 (+Tn7589) | Y Y | OQ054582 OQ269476 | |
| Ia | rplL | 1 | erm(B) + tet(O) | ICESag195 | Y | OP508060 | |
| Ib | unknown B | 1 | lsa(C) D | ICE2603tRNAlys | N | none | |
| III | unknown B | 1 | lsa(C) D | ICE2603tRNAlys | N | none | |
| III | unknown B | 1 | erm(B) + ant(6-Ia) + aph(3′-III) | undefined | ND | none | |
| V | rplL | 1 | erm(A) + aac(6′)-aph(2″) | ICESag139 | Y | OP508059 | |
| V | rplL | 2 | erm(A) + cat(Q) + mef(A) + msr(D) | ICESag236 | N | OP508058 | |
| V | rpsI | 1 | erm(B) + ant(6-Ia) + aph(3′-III) | ICESag738 | Y | OP508063 | |
| V | rumA | 1 | mef(A)/msr(D) | not apparent E | ND | none | |
| CC23 | Ia | rpsI | 1 | erm(A) | incomplete F | ND | none |
| Ia | Tn5801 | 4 | mef(A)/msr(D) adjacent to tet(M) | not apparent | ND | none | |
| Ia | plasmid | 1 | erm(T) | plasmid | N | NZ_JF308630 | |
| CC459 | IV | unknown | 1 | erm(A) + tet(M) | undefined | ND | none |
| IV | rplL | 2 | erm(A) erm(B) + tet(O) | ICESag084 ICESag206 | Y Y | OP715839 OP508061 | |
| CC452 | Ia | rplL | 1 | mef(A)/msr(D) | ICESpy009 | N | OP508061 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, U.B.; Portal, E.A.R.; Sands, K.; Lo, S.; Chalker, V.J.; Jauneikaite, E.; Spiller, O.B. Genomic Analysis Reveals New Integrative Conjugal Elements and Transposons in GBS Conferring Antimicrobial Resistance. Antibiotics 2023, 12, 544. https://doi.org/10.3390/antibiotics12030544
Khan UB, Portal EAR, Sands K, Lo S, Chalker VJ, Jauneikaite E, Spiller OB. Genomic Analysis Reveals New Integrative Conjugal Elements and Transposons in GBS Conferring Antimicrobial Resistance. Antibiotics. 2023; 12(3):544. https://doi.org/10.3390/antibiotics12030544
Chicago/Turabian StyleKhan, Uzma Basit, Edward A. R. Portal, Kirsty Sands, Stephanie Lo, Victoria J. Chalker, Elita Jauneikaite, and Owen B. Spiller. 2023. "Genomic Analysis Reveals New Integrative Conjugal Elements and Transposons in GBS Conferring Antimicrobial Resistance" Antibiotics 12, no. 3: 544. https://doi.org/10.3390/antibiotics12030544
APA StyleKhan, U. B., Portal, E. A. R., Sands, K., Lo, S., Chalker, V. J., Jauneikaite, E., & Spiller, O. B. (2023). Genomic Analysis Reveals New Integrative Conjugal Elements and Transposons in GBS Conferring Antimicrobial Resistance. Antibiotics, 12(3), 544. https://doi.org/10.3390/antibiotics12030544






