Identification of Efflux Pump Mutations in Pseudomonas aeruginosa from Clinical Samples
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Identification of Bacterial Isolates
2.2. Extraction of Genomic DNA
2.3. Molecular Identification of Bacterial Isolates
2.4. Antibiotic Susceptibility Testing
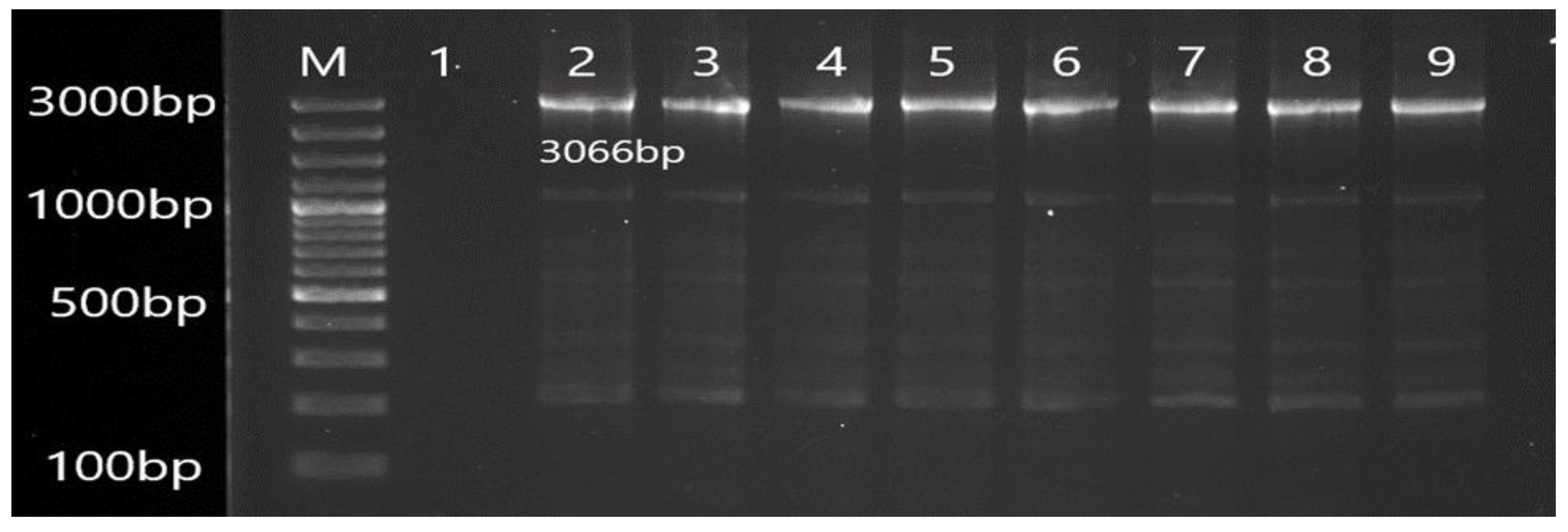
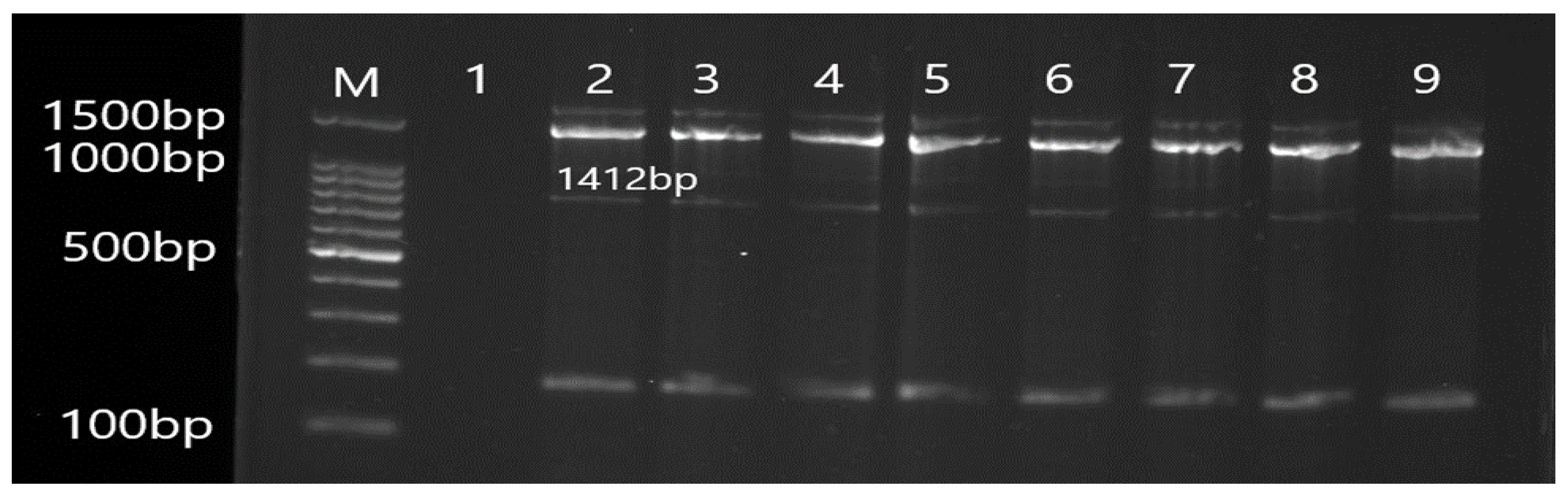
2.5. Molecular Detection of Efflux Pump Resistance Genes
2.6. Mutational Analysis of PCR Products
2.7. Computational Studies
2.8. Statistical Analysis
3. Results
3.1. Antibiotics Susceptibility Testing
3.2. Molecular Detection of Efflux Pump Resistance Genes in Isolates of P. aeruginosa
3.3. Mutational Analysis of Antibiotic-Resistant Efflux Pump Genes
3.4. Mutation Impact on Structure Stability
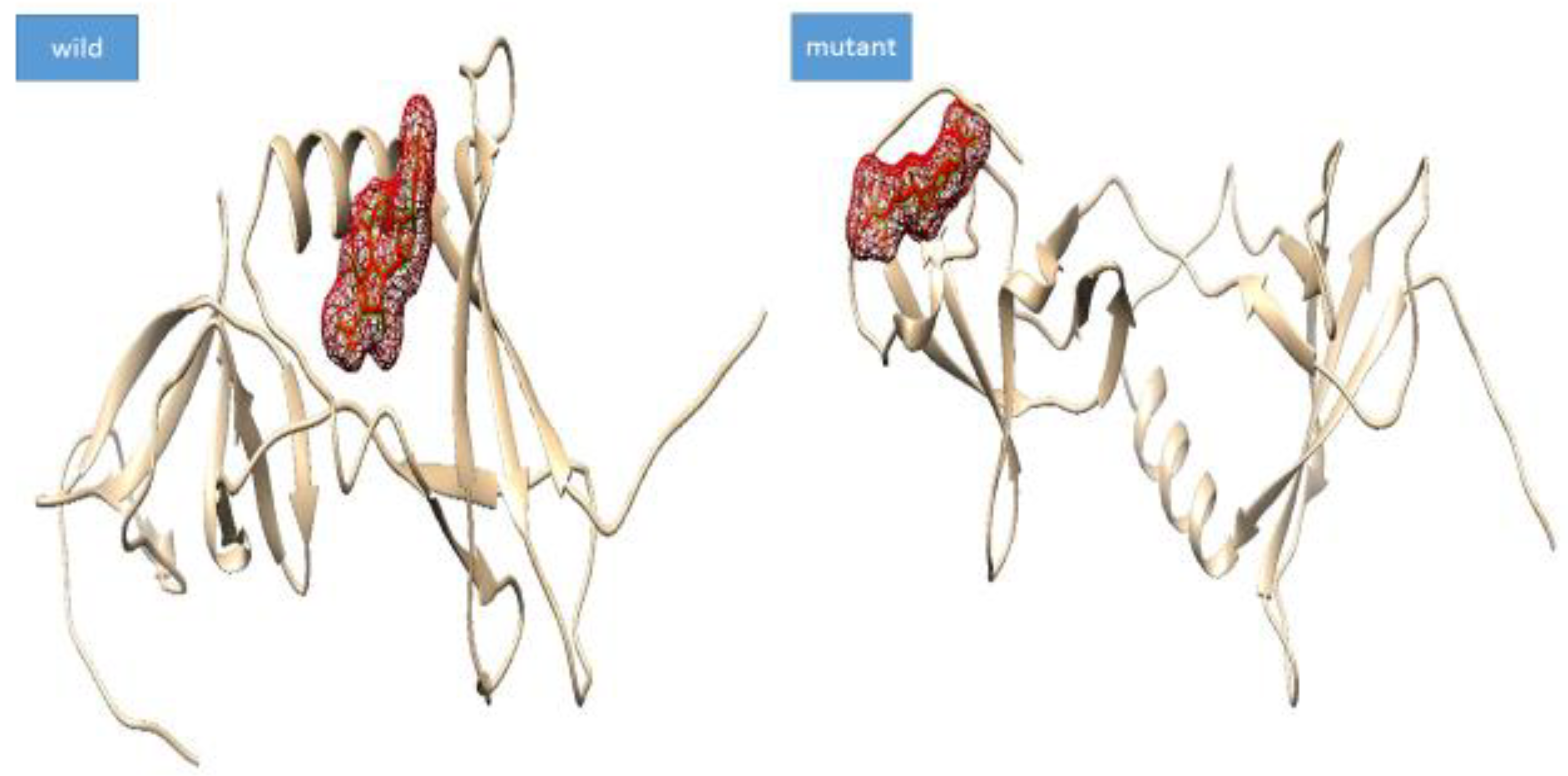
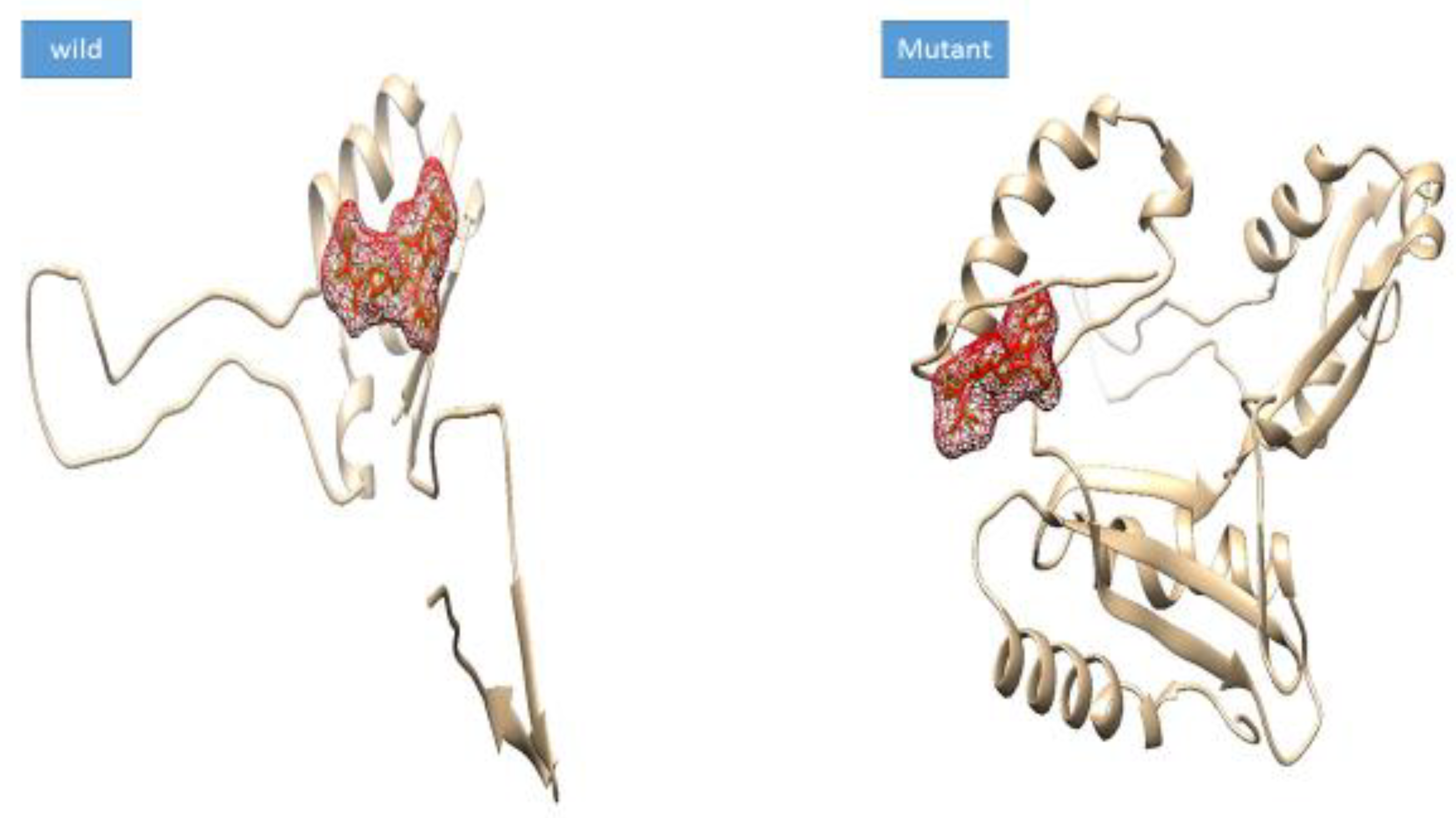
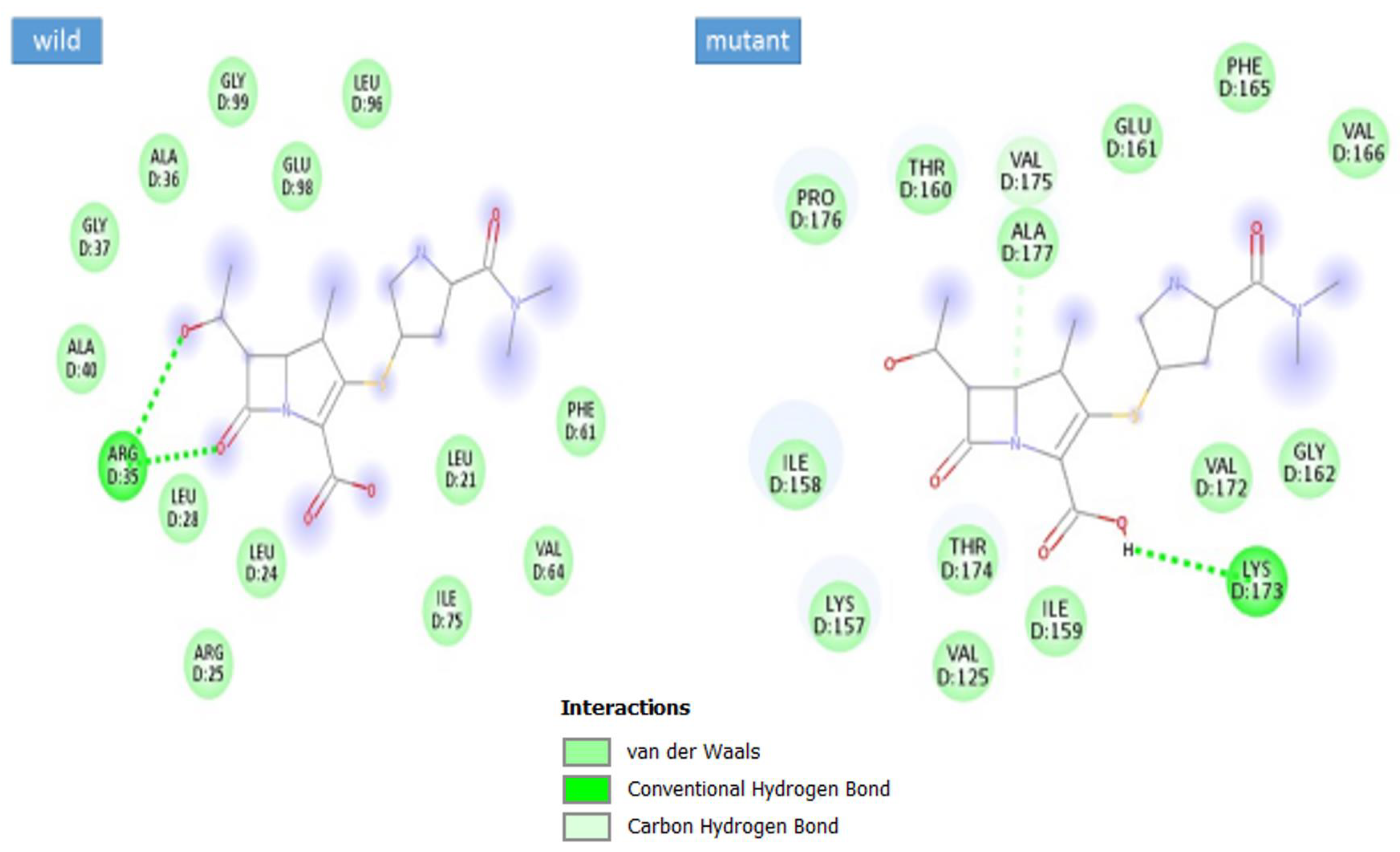
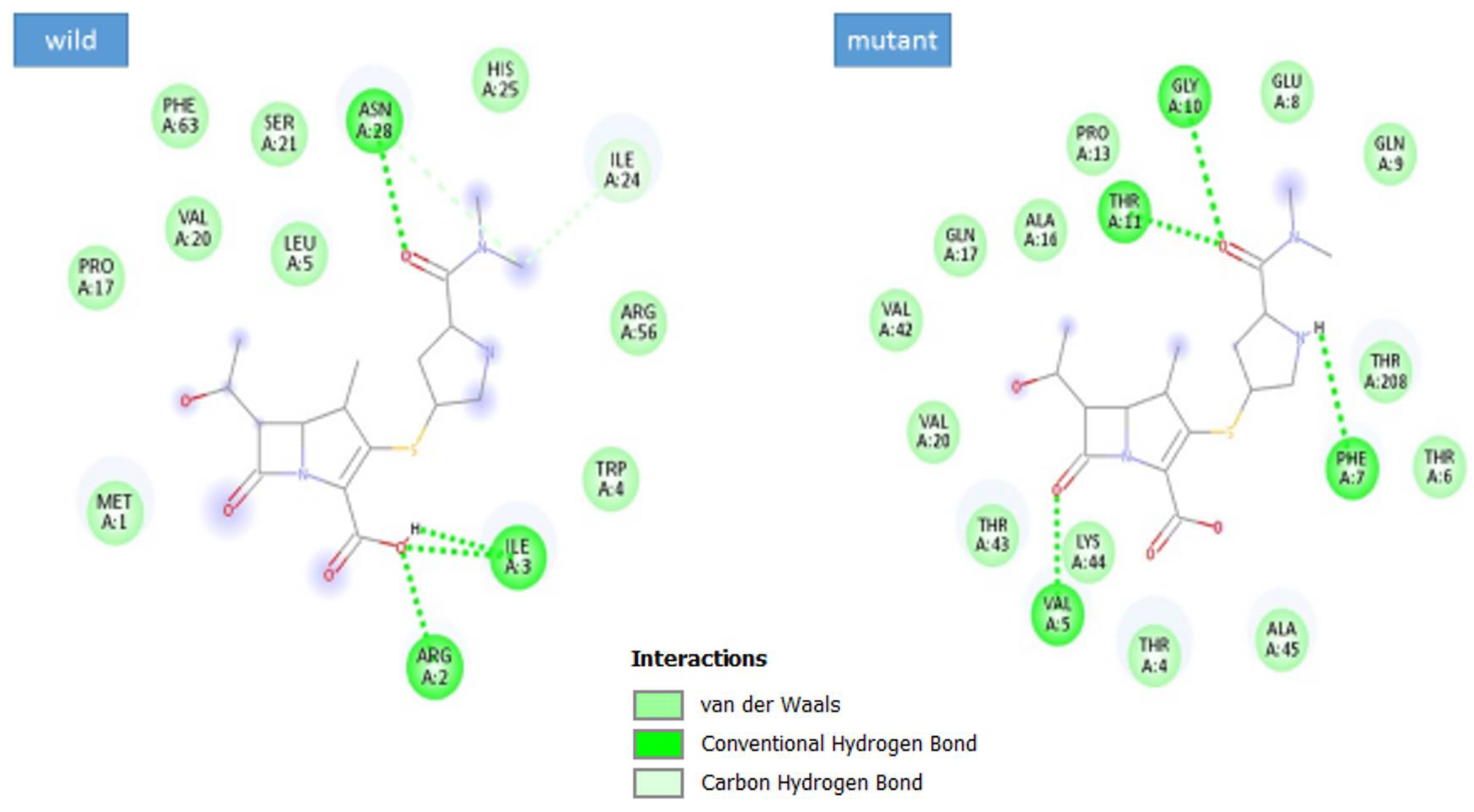
3.5. Docking Analysis
4. Discussion
5. Conclusions
6. Future Recommendations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peix, A.; Martha-Helena, B.; Encarna, V. Historical evolution and current status of the taxonomy of genus Pseudomonas. Infect. Genet. Evol. 2009, 9, 1132–1147. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.M. Pyoverdines: Pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species. Arch. Microbiol. 2000, 174, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Meliani, A. The multifactorial resistance of Pseudomonas aeruginosa. EXCLI J. 2020, 19, 813–816. [Google Scholar] [PubMed]
- Streeter, K.; Katouli, M. Pseudomonas aeruginosa: A review of their Pathogenesis and Prevalence in Clinical Settings and the Environment. Epidemiol. Infect. 2016, 2, 25–32. [Google Scholar] [CrossRef]
- El Solh, A.A.; Alhajhusain, A. Update on the treatment of Pseudomonas aeruginosa pneumonia. J. Antimicrob. Chemother. 2009, 64, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, E.B.; Vincent, T.H. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev. Pharmacoecon. Outcomes Res. 2010, 10, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lina, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.R.; Jane, L.B.; Stephen, L.; Kim, L. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. Res. 2010, 192, 6191–6199. [Google Scholar] [CrossRef] [PubMed]
- Breidenstein, E.B.; Elena, B.M.; de la Fuente-Núñez, C.; Robert, E.H. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.; Vassiliy, N.B.; Vito, R.; Niraj, M.; Pierpaolo, C.; Ulrich, K.; Paolo, R.; Vargiu, A.; Baylay, A.; Smith, H.; et al. AcrB drug-binding pocket substitution confers clinically relevant resistance and altered substrate specificity. Proc. Natl. Acad. Sci. USA 2015, 112, 3511–3516. [Google Scholar] [CrossRef] [PubMed]
- Suresh, M.; Narayanan, N.; Vimal, K.; Jayasree, P.; Kumar, P. Mutational and Phylogenetic Analysis of nfxB Gene in Multidrug-Resistant Clinical Isolates of Pseudomonas aeruginosa Hyperexpressing MexCD-OprJ Efflux Pump. Adv. Microbiol. 2019, 9, 993. [Google Scholar] [CrossRef]
- Kohler, T.; Michéa-Hamzehpour, M.; Henze, U.; Gotoh, N.; Kocjancic, C.L.; Pechère, J. Characterization of MexE–MexF–OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 1997, 23, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Bialvaei, A.Z.; Rahbar, M.; Hamidi-Farahani, R.; Asgari, A.; Esmailkhani, A.; Soleiman-Meigooni, S. Expression of RND efflux pumps mediated antibiotic resistance in Pseudomonas aeruginosa clinical strains. Microb. Pathog. 2021, 153, 104789. [Google Scholar] [CrossRef] [PubMed]
- Younes, K.; Memar, M.Y.; Farajnia, S.; Adibkia, K.; Kafil, H.S.; Ghotaslou, R. Molecular epidemiology and carbapenem resistance of Pseudomonas aeruginosa isolated from patients with burns. J. Wound Care 2021, 30, 2. [Google Scholar]
- Mohseni, N.; Rad, M.; Khorramian Toosi, B.; Mokhtari, A.R.; Yahyaraeyat, R.; Salehi, T.Z. Evaluation of MexAB-OprM efflux pump and determination of antimicrobial susceptibility in Pseudomonas aeruginosa human and veterinary isolates. Bulg. J. Vet. Med. 2019, 24, 200–207. [Google Scholar] [CrossRef]
- Ehiagbe, I.; Ehiagbe, F.; Akinshipe, B.; Ilobanafor, R. Fluoroquinolone efflux pump of Pseudomonas aeruginosa. NISEB J. 2019, 11, 4. [Google Scholar]
- Rudy, M.A.; Dolatabadi, S.; Zare, H.; Ghazvini, K. Characterization of efflux pump-mediated resistance against fluoro-quinolones among clinical isolates of Pseudomonas aeruginosa in the northeast of Iran and its association with mortality of infected patients. Cell 2019, 98, 1248938. [Google Scholar]
- Ali, Z.; Mumtaz, N.; Naz, S.A.; Jabeen, N.; Shafique, M. Multi-Drug Resistant Pseudomonas Aeruginosa: A threat of nosocomial infections in tertiary care hospitals. J. Pak. Med. Assoc. 2015, 65, 12. [Google Scholar] [PubMed]
- Kishk, R.; Abdalla, M.; Hashish, A.; Nemr, N.; El Nahhas, N.; Alkahtani, S.; Abdel-Daim, M.; Kishk, S. Efflux MexAB-mediated resistance in P. aeruginosa isolated from patients with healthcare associated infections. Pathogens 2020, 9, 471. [Google Scholar] [CrossRef] [PubMed]
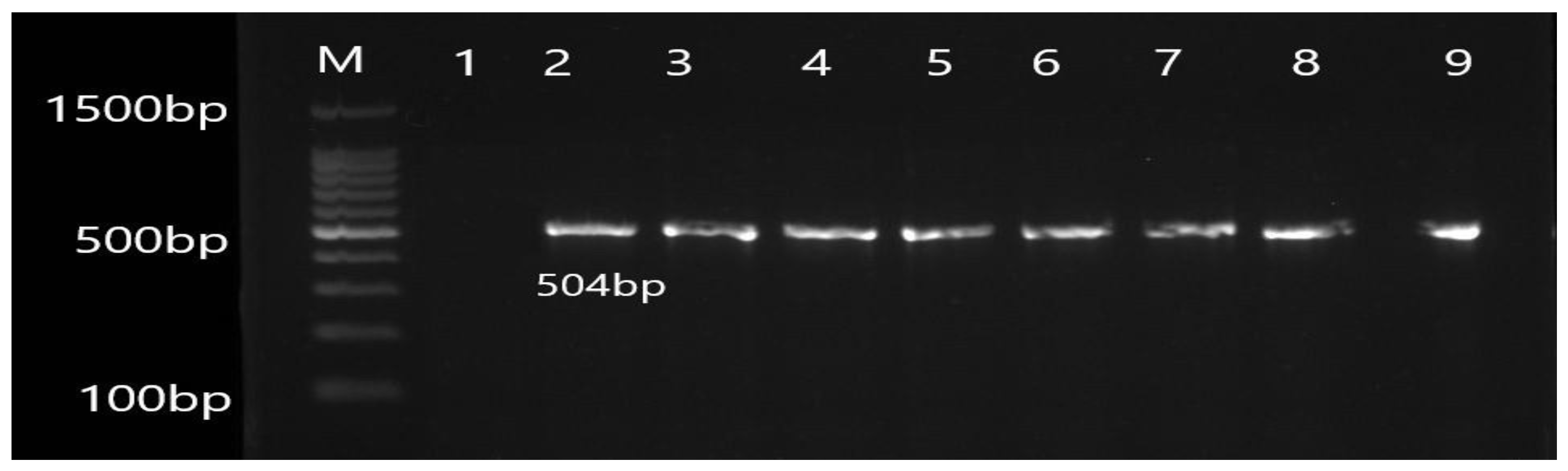
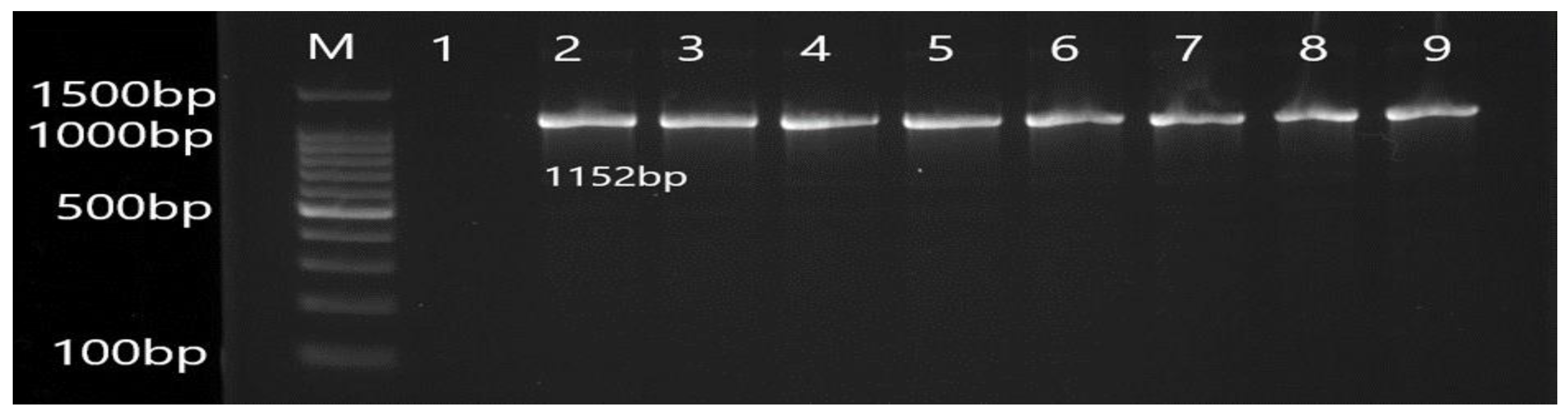
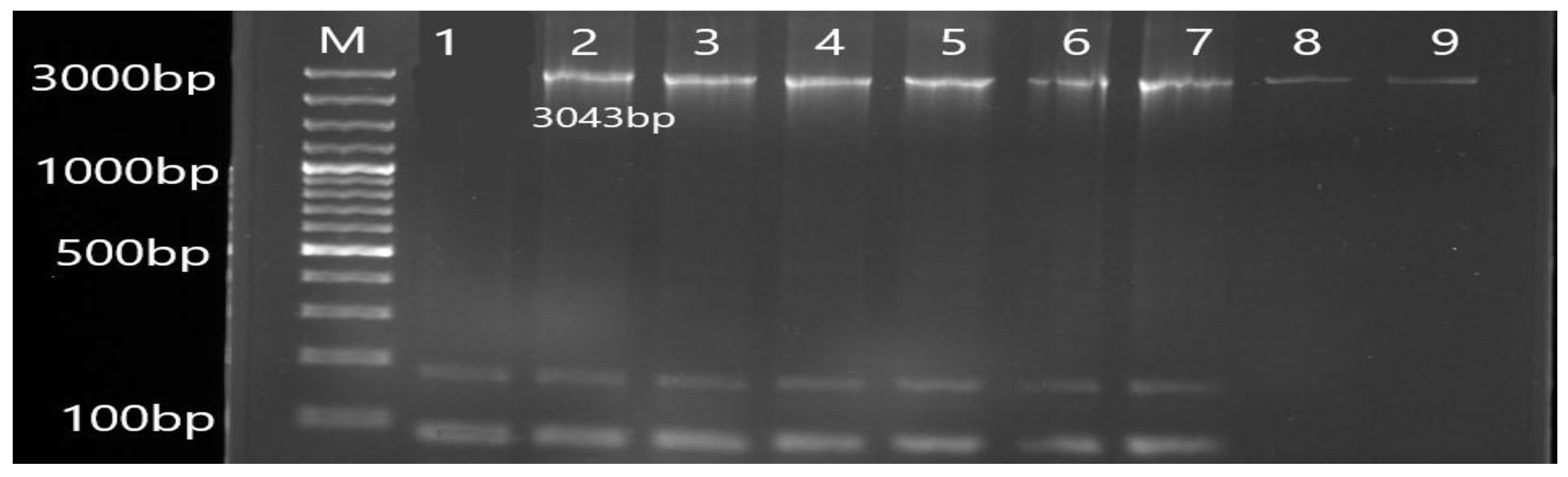
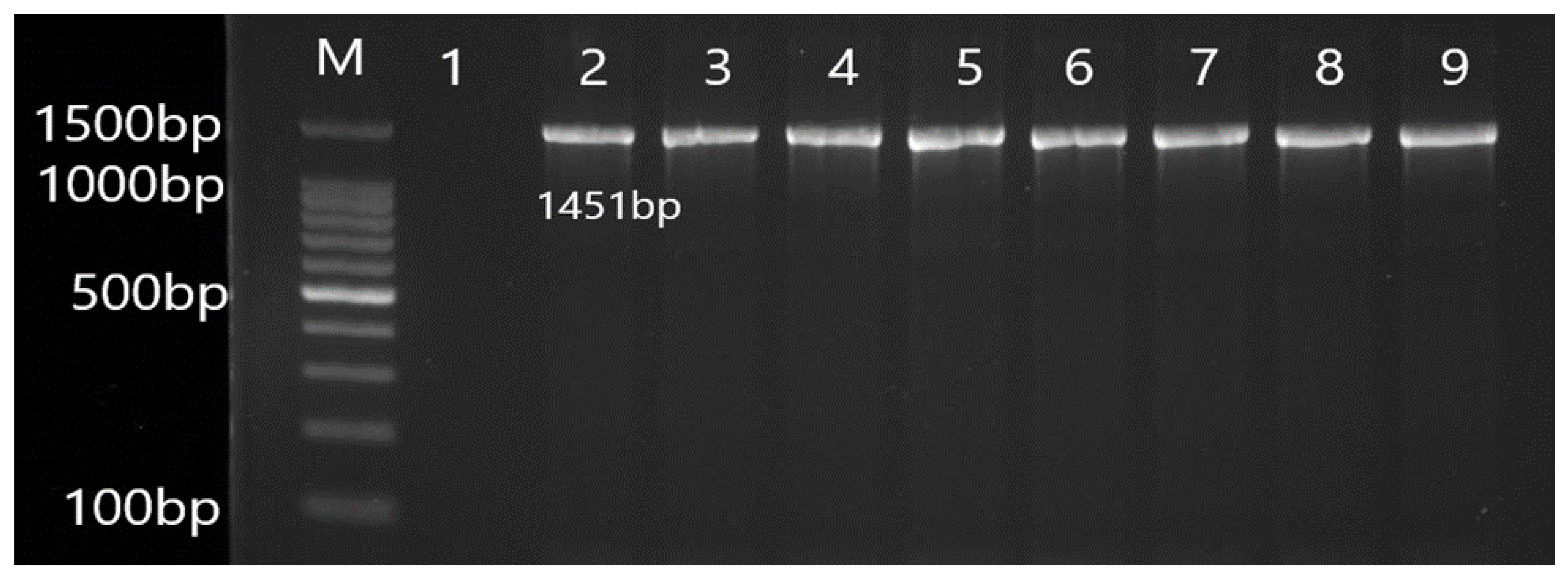


| Gene | Primer | Product Size (bp) | Annealing Temperature (°C) |
|---|---|---|---|
| OprL | F ATGGAAATGCTGAAATTCGGC R CTTCTTCAGCTCGACGCGACG | 504 | 55 |
| MexA | F CTATGCAACGAACGCCAGC R AGCCCTTGCTGTCGGTTTTC | 1152 | 56 |
| MexB | F TAGGCCCATTTTCGCGTGG R CGGTACCCAGAAGATCGCC | 3043 | 56 |
| OprM | F CGGTCCTTCCTTTCCCTGG R CAAGCCTGGGGATCTTCCTT | 1451 | 55 |
| MexR | F CAAGCGGTTGCGCGG R CCCCGTGAATCCCGACCTG | 425 | 56 |
| MexC | F TTACTGTTGCGGCGCAGG R CGTGCAATAGGAAGGATCGG | 1152 | 55 |
| MexD | F CAGCAGCCAGACGAAACAGA R TTCTTCATCAAGCGGCCGAA | 3066 | 56 |
| OprJ | F CTGCCGCCTCGATGTACC R GTATCGGCGCTGCTGATCG | 1412 | 55 |
| NfxB | F GACCCTGATTTCCCATGACG R GGAACATCTGCTCCAGGGTAT | 530 | 56 |
| S. No | Antibiotics (µg) | Family (Symbol) |
|---|---|---|
| 1 | Amikacin (20) | Aminoglycoside (AK) |
| 2 | Gentamicin (10) | Aminoglycoside (CN) |
| 3 | Azithromycin (30) | Macrolide (AZM) |
| 4 | Tigecycline (15) | Tetracycline (TGC) |
| 5 | Chloramphenicol (30) | Chloramphenicol (C) |
| 6 | Ciprofloxacin (5) | Fluoroquinolone (CIP) |
| 7 | Levofloxacin (5) | Fluoroquinolone (LEV) |
| 8 | Moxifloxacin (5) | Fluoroquinolone (MXF) |
| 9 | Amoxicillin (25) | β-lactam (penicillin) (AML) |
| 10 | Amoxicillin-clavulanic acid (30) | β-lactam (penicillin) (AMC) |
| 11 | Piperacillin-tazobactam (110) | β-lactam (penicillin) (TZP) |
| 12 | Aztreonam (30) | β-lactam (monobactams) (ATM) |
| 13 | Cefotaxime (30) | β-lactam (cephalosporin) (CTX) |
| 14 | Cefepime (30) | β-lactam (cephalosporin) (FEP) |
| 15 | Ceftazidime (30) | β-lactam (cephalosporin) (CAZ) |
| 16 | Cefoperazone (75) | β-lactam (cephalosporin) (CFP) |
| 17 | Cefoperazone-sulbactam (105) | β-lactam (cephalosporin) (SCF) |
| 18 | Ceftriaxone (30) | β-lactam (cephalosporin) (CRO) |
| 19 | Cefixime (5) | β-lactam (cephalosporin) (CFM) |
| 20 | Meropenem (10) | β-lactam (carbapenem) (MEM) |
| 21 | Imipenem (10) | β-lactam (carbapenem) (IMP) |
| 22 | Fosfomycin (50) | Fosfomycin (FOS) |
| 23 | Colistin (10) | Polymyxin (CT) |
| 24 | Polymyxin B (300) | Polymyxin (PB) |
| 25 | Trimethoprim-sulfamethoxazole (25) | Sulfonamide (SXT) |
| 26 | Nitrofurantoin (300) | Nitrofurantoin (F) |
| Source | Number (Percentage) |
|---|---|
| Urine catheter | 1 (0.5) |
| Stone analysis | 1 (0.5) |
| Urine | 28 (14) |
| Pus | 57 (28.5) |
| Wound swab | 94 (47) |
| Blood | 7 (3.5) |
| Sputum | 9 (4.5) |
| CSF | 1 (0.5) |
| Ear swab | 2 (1.0) |
| Total | 200 |
| Parameter | Frequency | Percentage | |
|---|---|---|---|
| Gender | Male | 108 | 54.0 |
| Female | 92 | 46.0 | |
| Age Group (Years) | 1–10 | 12 | 6 |
| 11–20 | 30 | 15 | |
| 21–30 | 43 | 21.5 | |
| 31–40 | 37 | 18.5 | |
| 41–50 | 23 | 11.5 | |
| 51–60 | 25 | 12.5 | |
| 61–70 | 21 | 10.5 | |
| 71–80 | 8 | 4 | |
| 81–90 | 1 | 0.5 | |
| Antibiotics | Resistant (n) | Percentage (%) | Intermediate (n) | Percentage (%) | Susceptible (n) | Percentage (%) |
|---|---|---|---|---|---|---|
| AK | 40 | 20 | 4 | 2 | 156 | 78 |
| CN | 88 | 44 | 10 | 5 | 102 | 51 |
| CIP | 79 | 39.5 | 9 | 4.5 | 112 | 58 |
| LEV | 71 | 35.5 | 23 | 11.5 | 106 | 53 |
| MXF | 80 | 40 | 11 | 5.5 | 109 | 54.5 |
| AML | 6 | 3 | - | - | 1 | 0.5 |
| AMC | 178 | 89 | 1 | 0.5 | 21 | 10.5 |
| TZP | 49 | 24.5 | 5 | 2.5 | 146 | 73 |
| ATM | 71 | 35.5 | 16 | 8.0 | 113 | 56.5 |
| CTX | 128 | 64 | 5 | 2.5 | 67 | 33.5 |
| FEP | 72 | 36 | 7 | 3.5 | 121 | 60.5 |
| CAZ | 73 | 36.5 | 11 | 5.5 | 116 | 58 |
| CEP | 72 | 36 | 15 | 7.5 | 113 | 56.5 |
| SCF | 49 | 24.5 | 10 | 5.0 | 141 | 70.5 |
| CRO | 96 | 48 | 11 | 5.5 | 93 | 46.5 |
| CFM | 158 | 79 | 7 | 3.5 | 35 | 17.5 |
| MEM | 63 | 31.5 | 8 | 4.0 | 129 | 64.5 |
| IMP | 63 | 31.5 | 11 | 5.5 | 126 | 63 |
| AZM | - | - | - | - | 7 | 3.5 |
| TGC | 100 | 50 | 12 | 6 | 88 | 44 |
| CT | 62 | 31 | 17 | 8.5 | 121 | 60.5 |
| PB | 63 | 31.5 | 21 | 10.5 | 116 | 58 |
| FOS | 6 | 3 | 2 | 1 | 22 | 11 |
| C | 2 | 1 | - | - | 5 | 2.5 |
| SXT | 125 | 62.5 | 5 | 2.5 | 70 | 35 |
| F | 15 | 7.5 | - | - | 15 | 7.5 |
| Positive Isolates of Efflux Pump Genes | Genes | Positive Result |
|---|---|---|
| AMC-resistant isolates | MexA | 178 (89%) |
| MexB | 178 (89%) | |
| OprM | 178 (89%) | |
| MexR | 178 (89%) | |
| MexC | 178 (89%) | |
| MexD | 178 (89%) | |
| OprJ | 178 (89%) | |
| NfxB | 178 (89%) |
| Codon Position | Reference Amino Acid | Altered Amino Acid | Amino Acid Position |
|---|---|---|---|
| 389 | GGT (Glycine) | AGT (Serine) | 368 |
| Wild Type | New | I-Mutant Prediction Effect | DDG Value | Reliability Index (RI) | Temperature | pH |
|---|---|---|---|---|---|---|
| G (Glycine) | S (Serine) | Decrease | −1 | 8 | 25 | 7 |
| Codon Position | Reference Amino Acid Position | Altered Amino Acid Position | Amino Acid Position |
|---|---|---|---|
| Synonymous mutation of mexB gene | |||
| 148 | TCC-TCG | Serine | 129 |
| 154 | AGC-AGT | Serine | 130 |
| 184 | GTC-GTG | Valine | 142 |
| 256 | CCT-CCG | Proline | 166 |
| 259 | CTC-CTA | Leucine | 167 |
| 302 | AAA-AAG | Lysine | 290 |
| 308 | GTA-GTC | Valine | 291 |
| 635 | CAA-CAG | Glutamine | 673 |
| Non-synonymous mutation of the mexB gene | |||
| 126 | Asparagine (AAC) | Aspartate (GAC) | 123 |
| 129 | Tyrosine (TAT) | Asparagine (AAT) | 124 |
| 136 | Leucine (CTC) | Arginine (CGC) | 126 |
| 138 | Phenylalanine (TTC) | Tyrosine (TAC) | 127 |
| 140 | Phenylalanine (TTC) | Isoleucine (ATC) | 128 |
| 151 | Aspartate (GAC) | Glutamate (GAG) | 131 |
| 165 | Alanine (GCC) | Glycine (GGC) | 136 |
| 167 | Cysteine (TGC) | Serine (AGC) | 137 |
| 170 | Proline (CCG) | Methionine (ATG) | 138 |
| 191 | Glutamine (CAA) | Glutamate (GAA) | 145 |
| 197 | Leucine (CTC) | Glycine (GGC) | 147 |
| 200 | Proline (CCC) | Threonine (ACC) | 148 |
| 203 | Asparagine (AAC) | Aspartate (GAC) | 149 |
| 215 | Proline (CCC) | Alanine (GCC) | 143 |
| 219 | Leucine (CTG) | Glutamine (CAG) | 154 |
| 228 | Alanine (GCC) | Valine (GTG) | 157 |
| 231 | Leucine (CTC) | Glutamine (CAG) | 158 |
| 244 | Histidine (CAC) | Glutamine (CAA) | 162 |
| 269 | Glutamine (CAA) | Glutamate (GAA) | 171 |
| 283 | Histidine (CAT) | Glutamine (CAG) | 175 |
| 292 | Histidine (CAC) | Arginine (CGG) | 287 |
| 303 | Serine (TCG) | Alanine (GCG) | 291 |
| 321 | Leucine (CTG) | Methionine (ATG) | 296 |
| 324 | Leucine (CTG) | Valine (GTG) | 298 |
| 327 | Leucine (CTG) | Valine (GTG) | 299 |
| 330 | Arginine (CGT) | Glycine (GGT) | 300 |
| 340 | Proline (CCT) | Valine (GTT) | 302 |
| 365 | Asparagine (AAC) | Lysine (AAG) | 311 |
| 378 | Histidine (CAC) | Asparagine (AAC) | 316 |
| 388 | Alanine (GCT) | Valine (GTT) | 319 |
| 424 | Alanine (GCC) | Glycine (GGC) | 331 |
| 429 | Cysteine (TGC) | Glycine (GGT) | 333 |
| 439 | Proline (CCG) | Glutamine (CAG) | 336 |
| 441 | Leucine (CTG) | Valine (GTG) | 337 |
| 456 | Histidine (CAC) | Tyrosine (TAC) | 342 |
| 488 | Asparagine (AAT) | Lysine (AAG) | 472 |
| 536 | Histidine (CAT) | Glutamine (CAG) | 488 |
| 590 | Asparagine (AAC) | Lysine (AAG) | 506 |
| 599 | Histidine (CAT) | Tyrosine (CAG) | 509 |
| 732 | Histidine (CAT) | Tyrosine (CAG) | 673 |
| Wild Type | New Type | I-Mutant Prediction Effect | DDG Value | Reliability Index (RI) | Temperature | pH |
|---|---|---|---|---|---|---|
| N | D | Decrease | −0.95 | 7 | 25 | 7 |
| Y | N | Increase | −0.24 | 0 | 25 | 7 |
| L | R | Decrease | −0.95 | 7 | 25 | 7 |
| F | Y | Decrease | −0.85 | 7 | 25 | 7 |
| F | I | Decrease | −1.99 | 9 | 25 | 7 |
| D | E | Decrease | −0.59 | 7 | 25 | 7 |
| A | G | Decrease | −1.03 | 7 | 25 | 7 |
| C | S | Decrease | −0.53 | 1 | 25 | 7 |
| P | M | Decrease | −0.96 | 1 | 25 | 7 |
| Q | E | Decrease | −0.29 | 4 | 25 | 7 |
| L | G | Increase | 0.22 | 2 | 25 | 7 |
| P | T | Decrease | −0.02 | 1 | 25 | 7 |
| N | D | Increase | 0.11 | 5 | 25 | 7 |
| P | A | Decrease | −1.02 | 4 | 25 | 7 |
| L | Q | Decrease | 0.14 | 1 | 25 | 7 |
| A | V | Decrease | −0.93 | 6 | 25 | 7 |
| L | Q | Decrease | 0.00 | 3 | 25 | 7 |
| H | Q | Decrease | −0.61 | 7 | 25 | 7 |
| Q | E | Decrease | −0.11 | 1 | 25 | 7 |
| H | Q | Decrease | −0.61 | 7 | 25 | 7 |
| H | R | Decrease | −1.37 | 9 | 25 | 7 |
| S | A | Decrease | −0.90 | 8 | 25 | 7 |
| L | M | Decrease | −0.80 | 5 | 25 | 7 |
| L | V | Decrease | −1.30 | 6 | 25 | 7 |
| L | V | Decrease | −1.32 | 6 | 25 | 7 |
| R | G | Decrease | −0.48 | 1 | 25 | 7 |
| P | V | Decrease | −1.57 | 4 | 25 | 7 |
| N | K | Increase | −0.48 | 3 | 25 | 7 |
| H | N | Decrease | −0.66 | 9 | 25 | 7 |
| A | V | Decrease | −1.37 | 7 | 25 | 7 |
| A | G | Increase | −0.51 | 1 | 25 | 7 |
| C | G | Decrease | −0.76 | 0 | 25 | 7 |
| P | Q | Decrease | −0.41 | 6 | 25 | 7 |
| L | V | Decrease | −0.74 | 4 | 25 | 7 |
| H | Y | Decrease | 0.04 | 1 | 25 | 7 |
| N | K | Increase | 0.04 | 4 | 25 | 7 |
| H | Q | Decrease | −0.53 | 6 | 25 | 7 |
| N | K | Decrease | −0.55 | 2 | 25 | 7 |
| H | Q | Decrease | −0.97 | 8 | 25 | 7 |
| H | Q | Decrease | −0.91 | 6 | 25 | 7 |
| Codon Position | Reference Amino Acid Position | Altered Amino Acid Position | Amino Acid Position |
|---|---|---|---|
| Non-synonymous mutation of the OprM gene | |||
| 11 | Glutamine (CAA) | Arginine (CGC) | 7 |
| 50 | Valine (GTG) | Alanine (GCG) | 20 |
| Synonymous mutation of the OprM gene | |||
| 43 | ACT-ACC | T | 17 |
| Wild Type | New Type | I-Mutant Prediction Effect | DDG Value | Reliability Index (RI) | Temperature | PH |
|---|---|---|---|---|---|---|
| Q (Glutamine) | R (Arginine) | Increase | −0.11 | 1 | 25 | 7 |
| V (Valine) | A (Alanine) | Decrease | −1.66 | 8 | 25 | 7 |
| Complex | Docking Score |
|---|---|
| max-A wild_meropenem | −6.1 |
| max-A mutant (E178K) meropenem | −6.5 |
| max-B wild_meropenem | −5.7 |
| max-B mutant_meropenem | −8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quddus, S.; Liaqat, Z.; Azam, S.; Haq, M.U.; Ahmad, S.; Alharbi, M.; Khan, I. Identification of Efflux Pump Mutations in Pseudomonas aeruginosa from Clinical Samples. Antibiotics 2023, 12, 486. https://doi.org/10.3390/antibiotics12030486
Quddus S, Liaqat Z, Azam S, Haq MU, Ahmad S, Alharbi M, Khan I. Identification of Efflux Pump Mutations in Pseudomonas aeruginosa from Clinical Samples. Antibiotics. 2023; 12(3):486. https://doi.org/10.3390/antibiotics12030486
Chicago/Turabian StyleQuddus, Sonia, Zainab Liaqat, Sadiq Azam, Mahboob Ul Haq, Sajjad Ahmad, Metab Alharbi, and Ibrar Khan. 2023. "Identification of Efflux Pump Mutations in Pseudomonas aeruginosa from Clinical Samples" Antibiotics 12, no. 3: 486. https://doi.org/10.3390/antibiotics12030486
APA StyleQuddus, S., Liaqat, Z., Azam, S., Haq, M. U., Ahmad, S., Alharbi, M., & Khan, I. (2023). Identification of Efflux Pump Mutations in Pseudomonas aeruginosa from Clinical Samples. Antibiotics, 12(3), 486. https://doi.org/10.3390/antibiotics12030486








