Comparative Assessment of Docking Programs for Docking and Virtual Screening of Ribosomal Oxazolidinone Antibacterial Agents
Abstract
1. Introduction
2. Results and Discussion
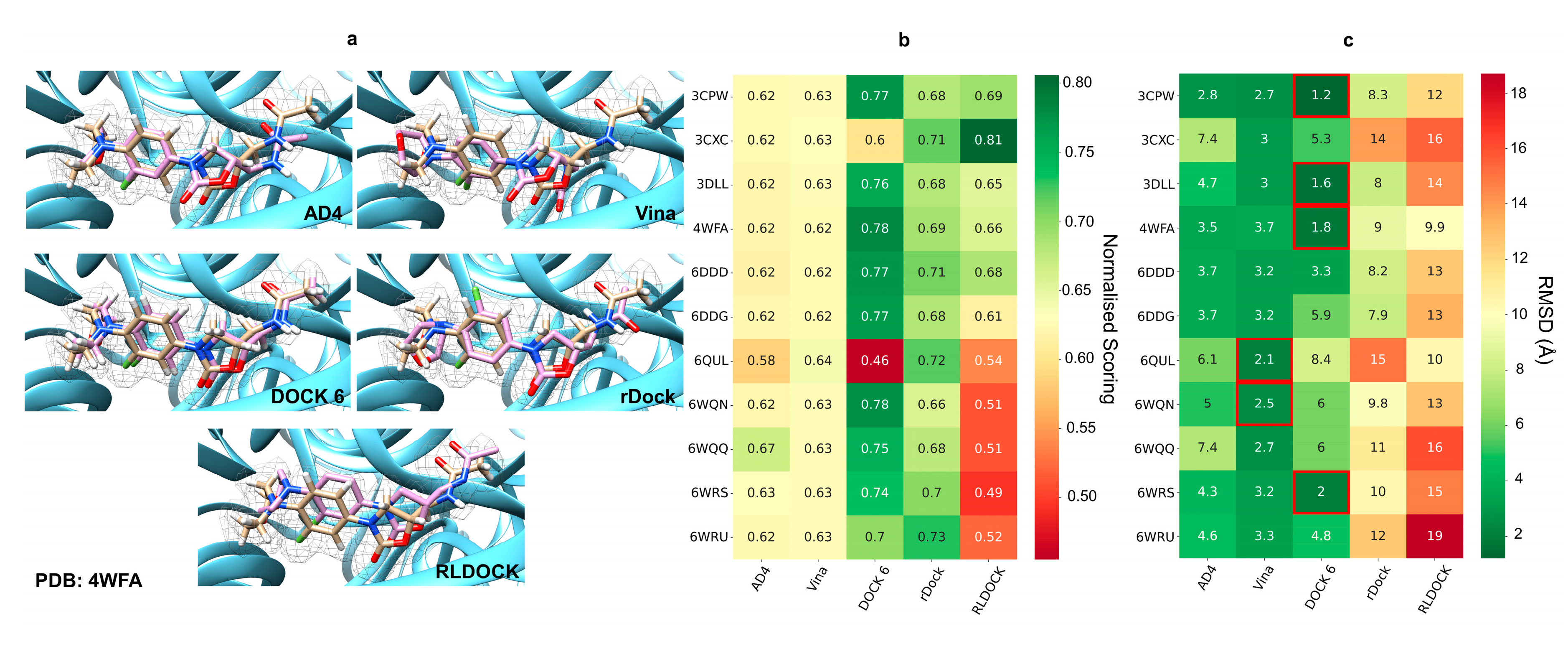
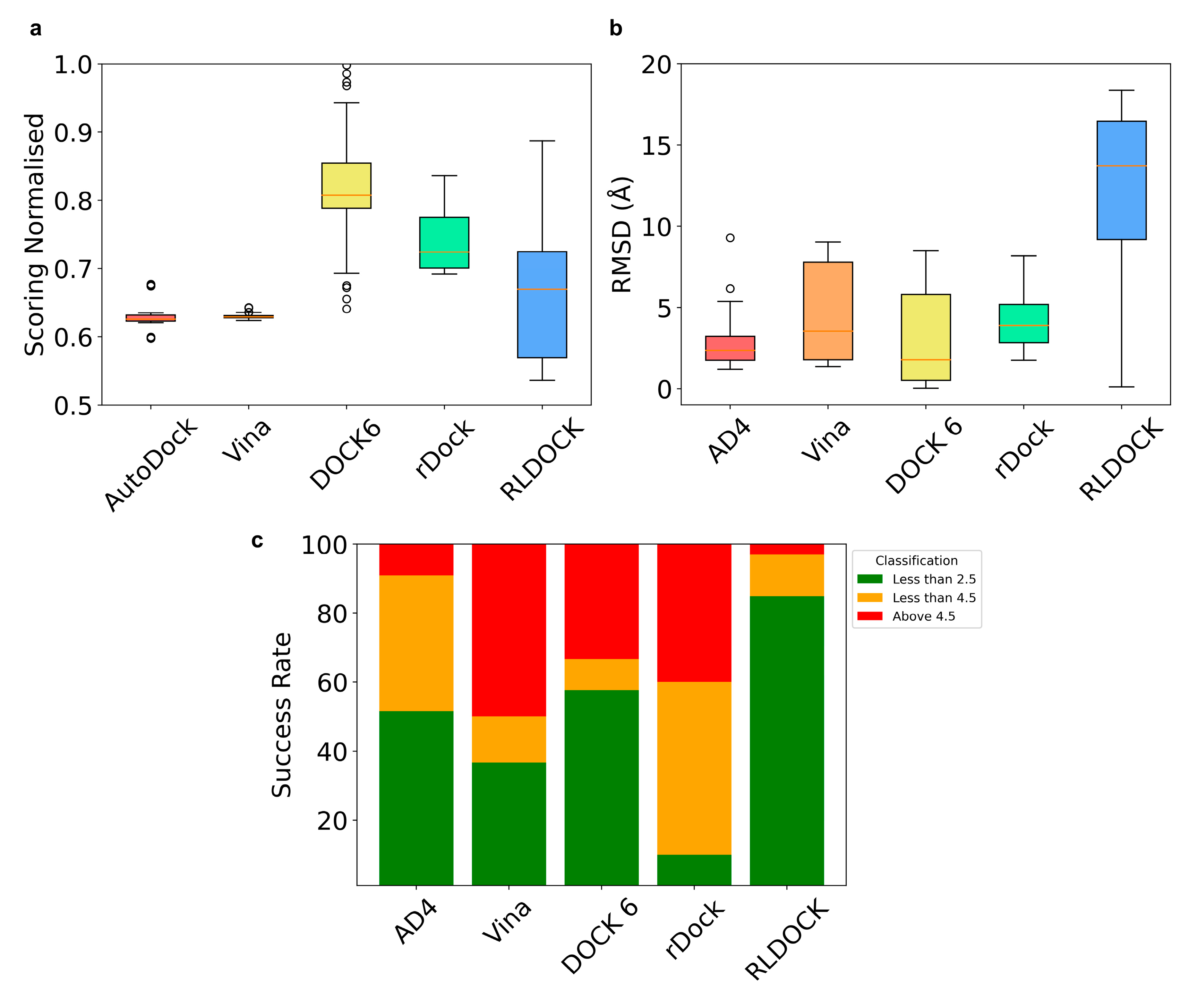
2.1. Pose Prediction Using Five Commonly Used RNA Docking Programs
2.2. Selection of Ribosomal Structure for Virtual Screening
2.3. Virtual Screening (VS) of the Oxazolidinone Dataset
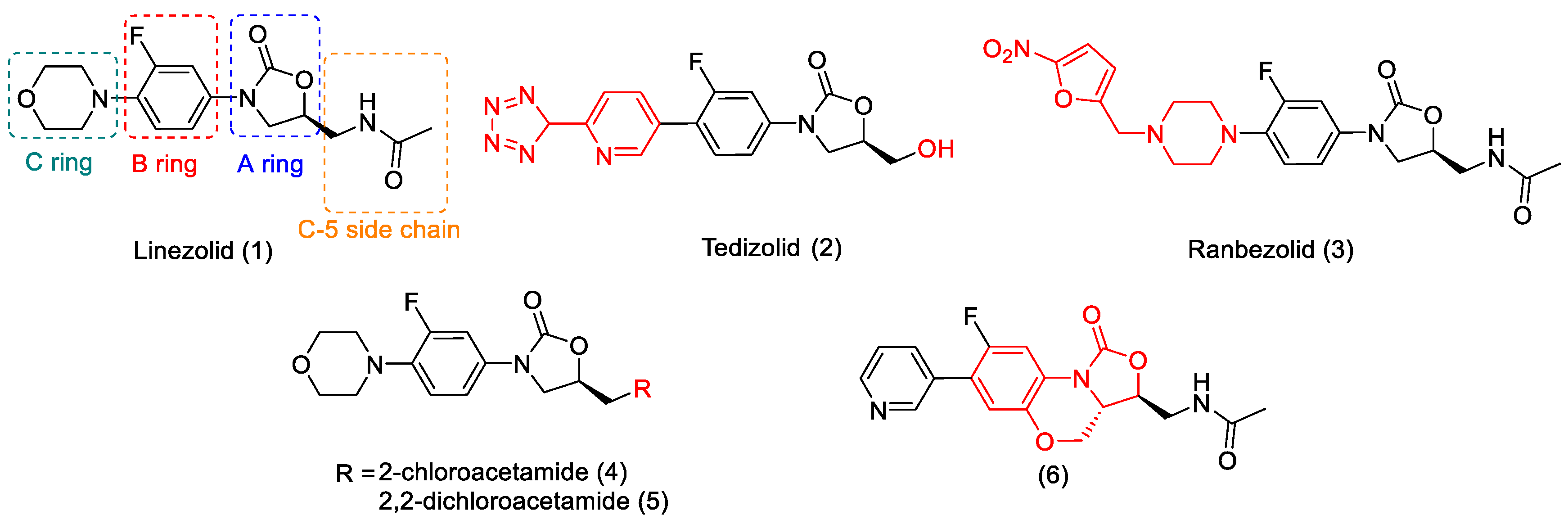
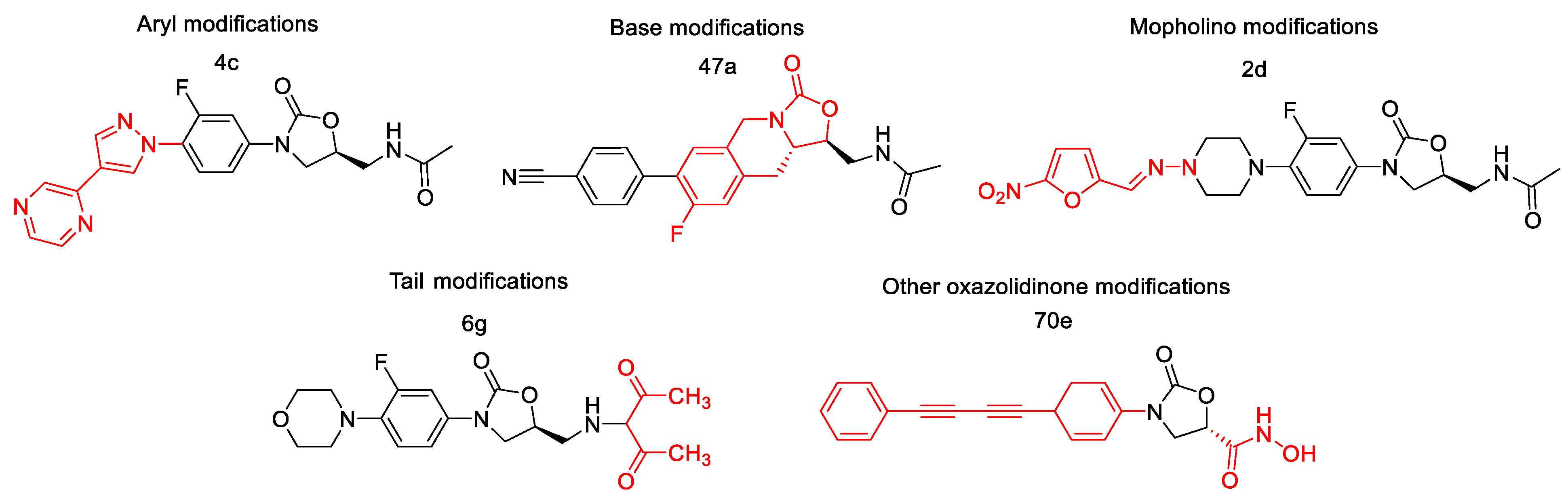
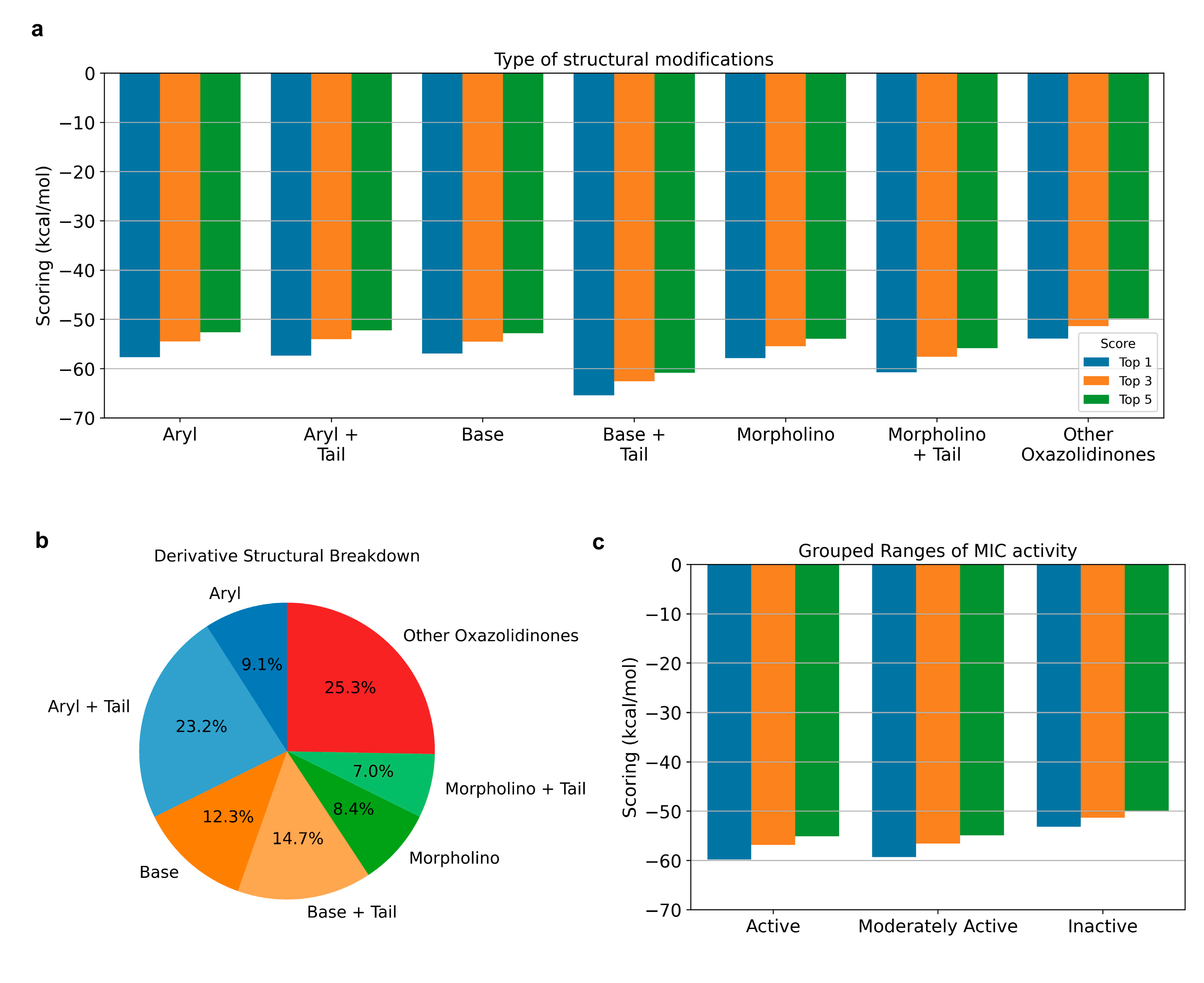
2.3.1. Structural Modification and MIC Activity
2.3.2. Additional Structural Analysis
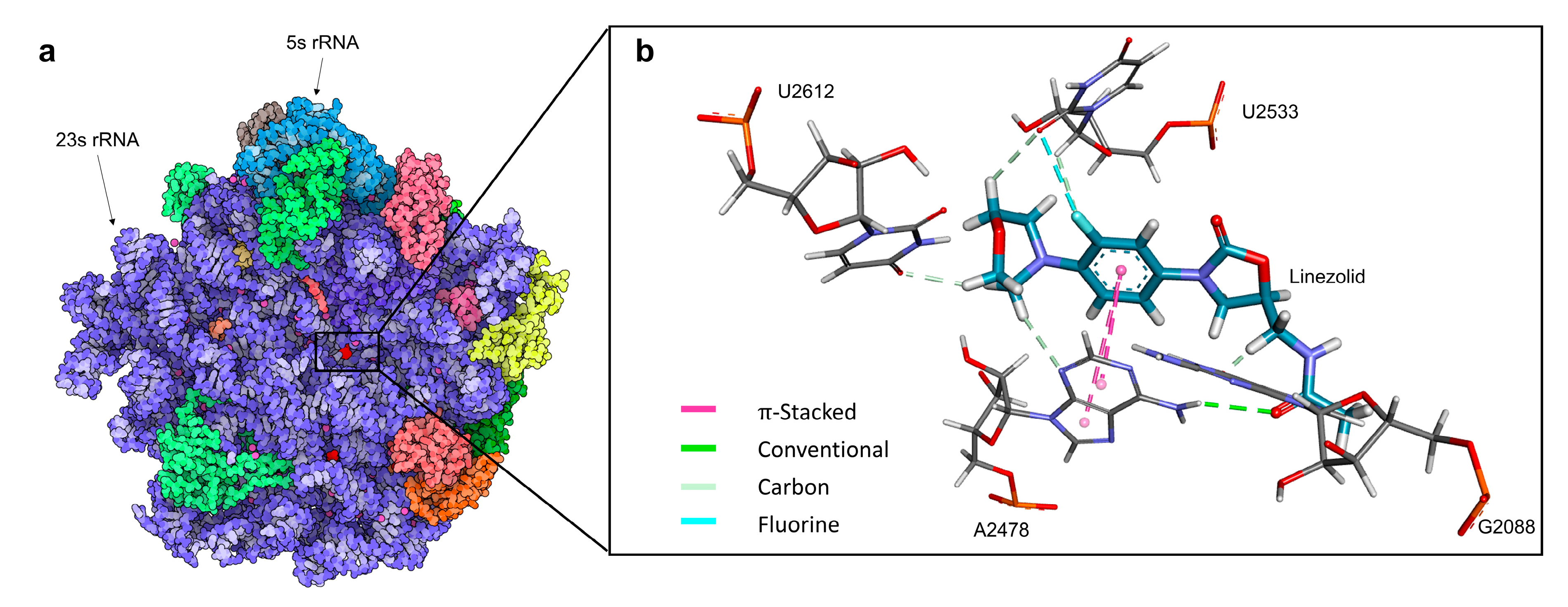
2.3.3. Top-Performing Derivatives and Their Interactions
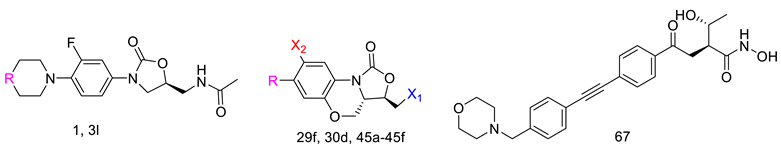 | |||||
|---|---|---|---|---|---|
| Derivative | R Group | X1 Group | X2 Group | Docking Score (kcal/mol) | S. aureus MIC (µg/mL) |
| 1 [45] | 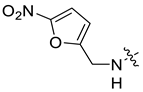 | - | - | −94.578 | 1 |
| 3l [46] |  | - | - | −90.462 | 2 |
| 29f [10] |  |  | H | −96.613 | 0.25 |
| 30d [47] | 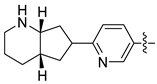 | 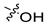 | H | −101.091 | - |
| 45a [48] | 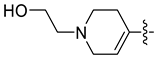 | 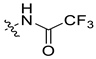 | F | −85.279 | - |
| 45b [48] | 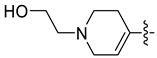 | 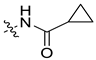 | F | −89.955 | 1.916 |
| 45c [48] |  |  | F | −93.988 | 0.473 |
| 45d [48] | 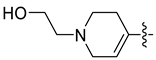 |  | F | −91.024 | 1.104 |
| 45e [48] |  | 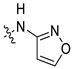 | F | −101.503 | 2.223 |
| 45f [48] | 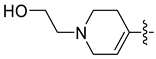 | 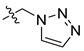 | F | −99.681 | 0.98 |
| 67 [49] | - | - | - | −92.438 | - |
2.3.4. Scoring Functions
2.3.5. Random Forest to Systematically Classify Bias in Scoring Functions
2.3.6. Tuning the Scoring Function/Re-scoring Function
2.4. Limitations of Study
3. Materials and Methods
3.1. System Selection
3.2. Pocket Location, RNA–Ligand Preparation, and Docking Protocols for Native Ligands
3.3. Molecular Docking
3.3.1. Ligand Docking with AutoDock Vina (Version 1.2.0)
3.3.2. Ligand Docking with AutoDock4 (Version 4.2.6)
3.3.3. Ligand Docking with DOCK (Version 6.9)
3.3.4. Ligand Docking with rDock
3.3.5. Ligand Docking with RLDOCK
3.3.6. Normalisation and Re-scoring of Redocking
3.4. Virtual Screening (VS) with Oxazolidinone Derivatives
3.5. Re-scoring of Docking Results with AnnapuRNA
3.6. Statistical Analysis and Classifier
3.7. Principal Component Analysis (PCA) and Re-scoring Function
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD4 | AutoDock 4 |
| ATCC | American Type Culture Collection |
| D. radiodurans | Deinococcus radiodurans |
| DMS | Dot molecular surfaces |
| E. coli | Escherichia coli |
| E. faecalis | Enterococcus faecalis |
| H. marismortui | Haloarcula marismortui |
| HBA | Number of H bond acceptors |
| HBD | Number of H bond acceptors |
| HPC | High-performance computing |
| K. pneumoniae | Klebsiella pneumoniae |
| kNN | k-nearest neighbors |
| MD | Molecular dynamics |
| MIC | Minimum inhibitory concentration |
| ML | Machine learning |
| MolWt | Molecular weight |
| mRNA | Messenger ribonucleic acid |
| MRSA | Methicillin-resistant Staphylococcus aureus |
| NA | Nucleic acid |
| NCTC | National Collection of Type Cultures |
| NumRings | Number of rings |
| P. aeruginosa | Pseudomonas aeruginosa |
| PCA | Principle component analysis |
| PDB | Protein Data Bank |
| pMIC | Log of the minimum inhibitory concentration |
| POAP | Parallelized Open Babel & Autodock Suite Pipeline |
| PTC | Peptidyl transferase centre |
| QSAR | Quantitative structure-activity relationship |
| RCSB | Research Collaboratory for Structural Bioinformatics |
| RF | Random forest |
| RMSD | Root-mean-square deviation |
| RNA | Ribonucleic acid |
| S. aureus | Staphylococcus aureus |
| S. capitis | Staphylococcus capitis |
| S. epidermidis | Staphylococcus epidermidis |
| S. pyogenes | Streptococcus pyogenes |
| SAR | Structure–activity relationships |
| SF | Scoring function |
| SMILE | Simplified molecular-input line-entry system |
| TPSA | Topological polar surface area |
| tRNA | Transfer ribonucleic acid |
| VDW | Van der Waals |
| Vina | AutoDock Vina |
| VRE | Vancomycin-resistant Enterococci |
| VS | Virtual screening |
References
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How Antibiotics Kill Bacteria: From Targets to Networks. Nat. Rev. Microbiol. 2010, 8, 423–435. [Google Scholar] [CrossRef]
- Stefani, S.; Bongiorno, D.; Mongelli, G.; Campanile, F. Linezolid Resistance in Staphylococci. Pharmaceuticals 2010, 3, 1988–2006. [Google Scholar] [CrossRef] [PubMed]
- van Bambeke, F.; Mingeot-Leclercq, M.-P.; Glupczynski, Y.; Tulkens, P.M. 137-Mechanisms of Action. In Infectious Diseases, 4th ed.; Cohen, J., Powderly, W.G., Opal, S.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 1162–1180.e1. ISBN 978-0-7020-6285-8. [Google Scholar]
- Tomasello, G.; Armenia, I.; Molla, G. The Protein Imager: A Full-Featured Online Molecular Viewer Interface with Server-Side HQ-Rendering Capabilities. Bioinformatics 2020, 36, 2909–2911. [Google Scholar] [CrossRef]
- Barbachyn, M.R.; Ford, C.W. Oxazolidinone Structure–Activity Relationships Leading to Linezolid. Angew. Chem. Int. Ed. 2003, 42, 2010–2023. [Google Scholar] [CrossRef]
- Zhao, Q.; Xin, L.; Liu, Y.; Liang, C.; Li, J.; Jian, Y.; Li, H.; Shi, Z.; Liu, H.; Cao, W. Current Landscape and Future Perspective of Oxazolidinone Scaffolds Containing Antibacterial Drugs. J. Med. Chem. 2021, 64, 10557–10580. [Google Scholar] [CrossRef]
- Long, K.S.; Vester, B. Resistance to Linezolid Caused by Modifications at Its Binding Site on the Ribosome. Antimicrob Agents Chemother 2012, 56, 603–612. [Google Scholar] [CrossRef] [PubMed]
- BIOVIA. Dassault Systèmes, Discovery Studio Visualiser, v21.1.0.20298; Dassault Systèmes: San Diego, CA, USA, 2022. [Google Scholar]
- Brickner, S.J.; Hutchinson, D.K.; Barbachyn, M.R.; Manninen, P.R.; Ulanowicz, D.A.; Garmon, S.A.; Grega, K.C.; Hendges, S.K.; Toops, D.S.; Ford, C.W.; et al. Synthesis and Antibacterial Activity of U-100592 and U-100766, Two Oxazolidinone Antibacterial Agents for the Potential Treatment of Multidrug-Resistant Gram-Positive Bacterial Infections. J. Med. Chem. 1996, 39, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Xin, Q.; Fan, H.; Guo, B.; He, H.; Gao, S.; Wang, H.; Huang, Y.; Yang, Y. Design, Synthesis, and Structure-Activity Relationship Studies of Highly Potent Novel Benzoxazinyl-Oxazolidinone Antibacterial Agents. J. Med. Chem. 2011, 54, 7493–7502. [Google Scholar] [CrossRef]
- Morris, G.M.; Lim-Wilby, M. Molecular Docking. Methods Mol. Biol. 2008, 443, 365–382. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization and Multithreading. J Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and Validation of a Genetic Algorithm for Flexible Docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Lang, P.T.; Brozell, S.R.; Mukherjee, S.; Pettersen, E.F.; Meng, E.C.; Thomas, V.; Rizzo, R.C.; Case, D.A.; James, T.L.; Kuntz, I.D. DOCK 6: Combining Techniques to Model RNA-Small Molecule Complexes. RNA 2009, 15, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Corbeil, C.R.; Englebienne, P.; Moitessier, N. Docking Ligands into Flexible and Solvated Macromolecules. 1. Development and Validation of FITTED 1.0. J. Chem. Inf. Model. 2007, 47, 435–449. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Zhang, K.; Wu, Q.; Huang, S.-Y. NLDock: A Fast Nucleic Acid–Ligand Docking Algorithm for Modeling RNA/DNA–Ligand Complexes. J. Chem. Inf. Model. 2021, 61, 4771–4782. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Carmona, S.; Alvarez-Garcia, D.; Foloppe, N.; Garmendia-Doval, A.B.; Juhos, S.; Schmidtke, P.; Barril, X.; Hubbard, R.E.; Morley, S.D. RDock: A Fast, Versatile and Open Source Program for Docking Ligands to Proteins and Nucleic Acids. PLOS Comput. Biol. 2014, 10, e1003571. [Google Scholar] [CrossRef]
- Sun, L.-Z.; Jiang, Y.; Zhou, Y.; Chen, S.-J. RLDOCK: A New Method for Predicting RNA–Ligand Interactions. J. Chem. Theory Comput. 2020, 16, 7173–7183. [Google Scholar] [CrossRef]
- Feng, Y.; Huang, S.-Y. ITScore-NL: An Iterative Knowledge-Based Scoring Function for Nucleic Acid–Ligand Interactions. J. Chem. Inf. Model. 2020, 60, 6698–6708. [Google Scholar] [CrossRef] [PubMed]
- Philips, A.; Milanowska, K.; Lach, G.; Bujnicki, J.M. LigandRNA: Computational Predictor of RNA-Ligand Interactions. RNA 2013, 19, 1605–1616. [Google Scholar] [CrossRef]
- Yan, Z.; Wang, J. SPA-LN: A Scoring Function of Ligand–Nucleic Acid Interactions via Optimizing Both Specificity and Affinity. Nucleic. Acids Res. 2017, 45, e110. [Google Scholar] [CrossRef]
- Pfeffer, P.; Gohlke, H. DrugScoreRNA--Knowledge-Based Scoring Function to Predict RNA-Ligand Interactions. J. Chem. Inf. Model. 2007, 47, 1868–1876. [Google Scholar] [CrossRef]
- Stefaniak, F.; Bujnicki, J.M. AnnapuRNA: A Scoring Function for Predicting RNA-Small Molecule Binding Poses. PLOS Comput. Biol. 2021, 17, e1008309. [Google Scholar] [CrossRef]
- Chhabra, S.; Xie, J.; Frank, A.T. RNAPosers: Machine Learning Classifiers for Ribonucleic Acid–Ligand Poses. J. Phys. Chem. B 2020, 124, 4436–4445. [Google Scholar] [CrossRef]
- Gandhi, N. 3D QSAR Analysis of Oxazolidinone Antibacterials: Can We Predict? Arkivoc 2007, 2006, 109–121. [Google Scholar] [CrossRef]
- Lohray, B.B.; Gandhi, N.; Srivastava, B.K.; Lohray, V.B. 3D QSAR Studies of N-4-Arylacryloylpiperazin-1-Yl-Phenyl-Oxazolidinones: A Novel Class of Antibacterial Agents. Bioorganic Med. Chem. Lett. 2006, 16, 3817–3823. [Google Scholar] [CrossRef]
- Pae, A.N.; Kim, S.Y.; Kim, H.Y.; Joo, H.J.; Cho, Y.S.; Choi, K.I.; Choi, J.H.; Koh, H.Y. 3D QSAR Studies on New Oxazolidinone Antibacterial Agents by Comparative Molecular Field Analysis. Bioorganic Med. Chem. Lett. 1999, 9, 2685–2690. [Google Scholar] [CrossRef]
- Deshmukh, M.S.; Jain, N. Correction to “Design, Synthesis, and Antibacterial Evaluation of Oxazolidinones with Fused Heterocyclic C-Ring Substructure. ” ACS Med. Chem. Lett. 2018, 9, 512. [Google Scholar] [CrossRef]
- Kalia, V.; Miglani, R.; Purnapatre, K.P.; Mathur, T.; Singhal, S.; Khan, S.; Voleti, S.R.; Upadhyay, D.J.; Saini, K.S.; Rattan, A.; et al. Mode of Action of Ranbezolid against Staphylococci and Structural Modeling Studies of Its Interaction with Ribosomes. Antimicrobx. Agents Chemother. 2009, 53, 1427–1433. [Google Scholar] [CrossRef]
- Fortuna, C.G.; Berardozzi, R.; Bonaccorso, C.; Caltabiano, G.; Di Bari, L.; Goracci, L.; Guarcello, A.; Pace, A.; Palumbo Piccionello, A.; Pescitelli, G.; et al. New Potent Antibacterials against Gram-Positive Multiresistant Pathogens: Effects of Side Chain Modification and Chirality in Linezolid-like 1,2,4-Oxadiazoles. Bioorg. Med. Chem. 2014, 22, 6814–6825. [Google Scholar] [CrossRef]
- Orac, C.M.; Zhou, S.; Means, J.A.; Boehm, D.; Bergmeier, S.C.; Hines, J.V. Synthesis and Stereospecificity of 4,5-Disubstituted Oxazolidinone Ligands Binding to T-Box Riboswitch RNA. J. Med. Chem. 2011, 54, 6786–6795. [Google Scholar] [CrossRef]
- Locke, J.B.; Finn, J.; Hilgers, M.; Morales, G.; Rahawi, S.; Kedar, G.C.; Picazo, J.J.; Im, W.; Shaw, K.J.; Stein, J.L. Structure-Activity Relationships of Diverse Oxazolidinones for Linezolid-Resistant Staphylococcus Aureus Strains Possessing the Cfr Methyltransferase Gene or Ribosomal Mutations. Antimicrob. Agents Chemother. 2010, 54, 5337–5343. [Google Scholar] [CrossRef]
- Jin, B.; Wang, T.; Chen, J.; Liu, X.; Zhang, Y.; Zhang, X.; Sheng, Z.; Yang, H.-L. Synthesis and Biological Evaluation of 3-(Pyridine-3-Yl)-2-Oxazolidinone Derivatives as Antibacterial Agents. Front. Chem. 2022, 10, 949813. [Google Scholar] [CrossRef] [PubMed]
- Dickerhoff, J.; Warnecke, K.R.; Wang, K.; Deng, N.; Yang, D. Evaluating Molecular Docking Software for Small Molecule Binding to G-Quadruplex DNA. Int. J. Mol. Sci. 2021, 22, 10801. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Franceschi, F.; Duffy, E.M. Structure-Based Drug Design Meets the Ribosome. Biochem. Pharmacol. 2006, 71, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Eyal, Z.; Matzov, D.; Krupkin, M.; Wekselman, I.; Paukner, S.; Zimmerman, E.; Rozenberg, H.; Bashan, A.; Yonath, A. Structural Insights into Species-Specific Features of the Ribosome from the Pathogen Staphylococcus Aureus. Proc. Natl. Acad. Sci. USA 2015, 112, E5805–E5814. [Google Scholar] [CrossRef]
- Bulkley, D.; Innis, C.A.; Blaha, G.; Steitz, T.A. Revisiting the Structures of Several Antibiotics Bound to the Bacterial Ribosome. Proc. Natl. Acad. Sci. USA 2010, 107, 17158–17163. [Google Scholar] [CrossRef]
- Wilson, D.N.; Schluenzen, F.; Harms, J.M.; Starosta, A.L.; Connell, S.R.; Fucini, P. The Oxazolidinone Antibiotics Perturb the Ribosomal Peptidyl-Transferase Center and Effect TRNA Positioning. Proc. Natl. Acad. Sci. USA 2008, 105, 13339–13344. [Google Scholar] [CrossRef]
- Ippolito, J.A.; Kanyo, Z.F.; Wang, D.; Franceschi, F.J.; Moore, P.B.; Steitz, T.A.; Duffy, E.M. Crystal Structure of the Oxazolidinone Antibiotic Linezolid Bound to the 50S Ribosomal Subunit. J. Med. Chem. 2008, 51, 3353–3356. [Google Scholar] [CrossRef]
- Leach, K.L.; Swaney, S.M.; Colca, J.R.; McDonald, W.G.; Blinn, J.R.; Thomasco, L.M.; Gadwood, R.C.; Shinabarger, D.; Xiong, L.; Mankin, A.S. The Site of Action of Oxazolidinone Antibiotics in Living Bacteria and in Human Mitochondria. Mol. Cell 2007, 26, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Bashan, A.; Agmon, I.; Zarivach, R.; Schluenzen, F.; Harms, J.; Berisio, R.; Bartels, H.; Franceschi, F.; Auerbach, T.; Hansen, H.A.S.; et al. Structural Basis of the Ribosomal Machinery for Peptide Bond Formation, Translocation, and Nascent Chain Progression. Mol. Cell 2003, 11, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Hoellman, D.B.; Lin, G.; Ednie, L.M.; Rattan, A.; Jacobs, M.R.; Appelbaum, P.C. Antipneumococcal and Antistaphylococcal Activities of Ranbezolid (RBX 7644), a New Oxazolidinone, Compared to Those of Other Agents. Antimicrob. Agents Chemother. 2003, 47, 1148–1150. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Dong, Y.; Ye, T.; Jiang, J.; Ding, L.; Qin, M.; Ding, X.; Zhao, Y. Synthesis and Antibacterial Evaluation of Novel Oxazolidinone Derivatives Containing a Piperidinyl Moiety. Bioorg. Med. Chem. Lett. 2019, 29, 126746. [Google Scholar] [CrossRef]
- Guo, B.; Fan, H.; Xin, Q.; Chu, W.; Wang, H.; Huang, Y.; Chen, X.; Yang, Y. Solubility-Driven Optimization of (Pyridin-3-Yl) Benzoxazinyl-Oxazolidinones Leading to a Promising Antibacterial Agent. J. Med. Chem. 2013, 56, 2642–2650. [Google Scholar] [CrossRef]
- Zhao, H.; Lu, Y.; Sheng, L.; Yuan, Z.; Wang, B.; Wang, W.; Li, Y.; Ma, C.; Wang, X.; Zhang, D.; et al. Discovery of Fluorine-Containing Benzoxazinyl-Oxazolidinones for the Treatment of Multidrug Resistant Tuberculosis. ACS Med. Chem. Lett. 2017, 8, 533–537. [Google Scholar] [CrossRef]
- Barb, A.W.; Jiang, L.; Raetz, C.R.H.; Zhou, P. Structure of the Deacetylase LpxC Bound to the Antibiotic CHIR-090: Time-Dependent Inhibition and Specificity in Ligand Binding. Proc. Natl. Acad. Sci. USA 2007, 104, 18433–18438. [Google Scholar] [CrossRef]
- Feng, Y.; Yan, Y.; He, J.; Tao, H.; Wu, Q.; Huang, S.-Y. Docking and Scoring for Nucleic Acid-Ligand Interactions: Principles and Current Status. Drug Discov. Today 2022, 27, 838–847. [Google Scholar] [CrossRef]
- Morgan, H.L. The Generation of a Unique Machine Description for Chemical Structures-A Technique Developed at Chemical Abstracts Service. J. Chem. Doc. 1965, 5, 107–113. [Google Scholar] [CrossRef]
- Boittier, E.D.; Tang, Y.Y.; Buckley, M.E.; Schuurs, Z.P.; Richard, D.J.; Gandhi, N.S. Assessing Molecular Docking Tools to Guide Targeted Drug Discovery of CD38 Inhibitors. Int. J. Mol. Sci. 2020, 21, 5183. [Google Scholar] [CrossRef]
- Patel, L.; Shukla, T.; Huang, X.; Ussery, D.W.; Wang, S. Machine Learning Methods in Drug Discovery. Molecules 2020, 25, 5277. [Google Scholar] [CrossRef]
- Zhou, Y.; Jiang, Y.; Chen, S.-J. RNA–Ligand Molecular Docking: Advances and Challenges. WIREs Comput. Mol. Sci. 2022, 12, e1571. [Google Scholar] [CrossRef]
- Luo, J.; Wei, W.; Waldispühl, J.; Moitessier, N. Challenges and Current Status of Computational Methods for Docking Small Molecules to Nucleic Acids. Eur. J. Med. Chem. 2019, 168, 414–425. [Google Scholar] [CrossRef] [PubMed]
- Tessaro, F.; Scapozza, L. How ‘Protein-Docking’ Translates into the New Emerging Field of Docking Small Molecules to Nucleic Acids? Molecules 2020, 25, 2749. [Google Scholar] [CrossRef]
- Park, S.-J.; Kim, Y.-G.; Park, H.-J. Identification of RNA Pseudoknot-Binding Ligand That Inhibits the −1 Ribosomal Frameshifting of SARS-Coronavirus by Structure-Based Virtual Screening. J. Am. Chem. Soc. 2011, 133, 10094–10100. [Google Scholar] [CrossRef] [PubMed]
- Moitessier, N.; Westhof, E.; Hanessian, S. Docking of Aminoglycosides to Hydrated and Flexible RNA. J. Med. Chem. 2006, 49, 1023–1033. [Google Scholar] [CrossRef] [PubMed]
- Belousoff, M.J.; Venugopal, H.; Wright, A.; Seoner, S.; Stuart, I.; Stubenrauch, C.; Bamert, R.S.; Lupton, D.W.; Lithgow, T. CryoEM-Guided Development of Antibiotics for Drug-Resistant Bacteria. ChemMedChem 2019, 14, 527–531. [Google Scholar] [CrossRef] [PubMed]
- Kallert, E.; Fischer, T.R.; Schneider, S.; Grimm, M.; Helm, M.; Kersten, C. Protein-Based Virtual Screening Tools Applied for RNA–Ligand Docking Identify New Binders of the PreQ1-Riboswitch. J. Chem. Inf. Model. 2022, 62, 4134–4148. [Google Scholar] [CrossRef]
- Chen, L.; Calin, G.A.; Zhang, S. Novel Insights of Structure-Based Modeling for RNA-Targeted Drug Discovery. J. Chem. Inf. Model. 2012, 52, 2741–2753. [Google Scholar] [CrossRef]
- Context-Specific Inhibition of Translation by Ribosomal Antibiotics Targeting the Peptidyl Transferase Center. Available online: https://www.pnas.org/doi/10.1073/pnas.1613055113 (accessed on 6 January 2023).
- Bernhardt, H.S.; Tate, W.P. Primordial Soup or Vinaigrette: Did the RNA World Evolve at Acidic PH? Biol. Direct. 2012, 7, 4. [Google Scholar] [CrossRef]
- Makarov, G.I.; Reshetnikova, R.V. Investigation of Radezolid Interaction with Non-Canonical Chloramphenicol Binding Site by Molecular Dynamics Simulations. J. Mol. Graph. Model. 2021, 105, 107902. [Google Scholar] [CrossRef] [PubMed]
- Makarov, G.I.; Makarova, T.M. A Noncanonical Binding Site of Linezolid Revealed via Molecular Dynamics Simulations. J. Comput. Aided. Mol. Des. 2020, 34, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Saini, J.S.; Homeyer, N.; Fulle, S.; Gohlke, H. Determinants of the Species Selectivity of Oxazolidinone Antibiotics Targeting the Large Ribosomal Subunit. Biol. Chem. 2013, 394, 1529–1541. [Google Scholar] [CrossRef] [PubMed]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef]
- Srinivasan, J.; Cheatham, T.E.; Cieplak, P.; Kollman, P.A.; Case, D.A. Continuum Solvent Studies of the Stability of DNA, RNA, and Phosphoramidate−DNA Helices. J. Am. Chem. Soc. 1998, 120, 9401–9409. [Google Scholar] [CrossRef]
- Tuszynska, I.; Magnus, M.; Jonak, K.; Dawson, W.; Bujnicki, J.M. NPDock: A Web Server for Protein–Nucleic Acid Docking. Nucleic. Acids Res. 2015, 43, W425–W430. [Google Scholar] [CrossRef]
- He, J.; Wang, J.; Tao, H.; Xiao, Y.; Huang, S.-Y. HNADOCK: A Nucleic Acid Docking Server for Modeling RNA/DNA–RNA/DNA 3D Complex Structures. Nucleic. Acids Res. 2019, 47, W35–W42. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.-Y. The HDOCK Server for Integrated Protein–Protein Docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Scaiola, A.; Leibundgut, M.; Boehringer, D.; Caspers, P.; Bur, D.; Locher, H.H.; Rueedi, G.; Ritz, D. Structural Basis of Translation Inhibition by Cadazolid, a Novel Quinoxolidinone Antibiotic. Sci. Rep. 2019, 9, 5634. [Google Scholar] [CrossRef]
- Zhou, J.; Bhattacharjee, A.; Chen, S.; Chen, Y.; Duffy, E.; Farmer, J.; Goldberg, J.; Hanselmann, R.; Ippolito, J.A.; Lou, R.; et al. Design at the Atomic Level: Design of Biaryloxazolidinones as Potent Orally Active Antibiotics. Bioorg. Med. Chem. Lett. 2008, 18, 6175–6178. [Google Scholar] [CrossRef]
- Wright, A.; Deane-Alder, K.; Marschall, E.; Bamert, R.; Venugopal, H.; Lithgow, T.; Lupton, D.W.; Belousoff, M.J. Characterization of the Core Ribosomal Binding Region for the Oxazolidone Family of Antibiotics Using Cryo-EM. ACS Pharmacol. Transl. Sci. 2020, 3, 425–432. [Google Scholar] [CrossRef]
- Samdani, A.; Vetrivel, U. POAP: A GNU Parallel Based Multithreaded Pipeline of Open Babel and AutoDock Suite for Boosted High Throughput Virtual Screening. Comput. Biol. Chem. 2018, 74, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Taminau, J.; Thijs, G.; De Winter, H. Pharao: Pharmacophore Alignment and Optimization. J. Mol. Graph. Model. 2008, 27, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Cresset Group. FlareTM, v3.; Cresset Group: Peterborough, UK, 2022. [Google Scholar]
- Rahuman, M.H.; Muthu, S.; Raajaraman, B.R.; Raja, M.; Umamahesvari, H. Investigations on 2-(4-Cyanophenylamino) Acetic Acid by FT-IR,FT-Raman, NMR and UV-Vis Spectroscopy, DFT (NBO, HOMO-LUMO, MEP and Fukui Function) and Molecular Docking Studies. Heliyon 2020, 6, e04976. [Google Scholar] [CrossRef] [PubMed]
- Chami Khazraji, A.; Robert, S. Self-Assembly and Intermolecular Forces When Cellulose and Water Interact Using Molecular Modeling. J. Nanomater. 2013, 2013, e745979. [Google Scholar] [CrossRef]
- Peach, M.L.; Nicklaus, M.C. Combining Docking with Pharmacophore Filtering for Improved Virtual Screening. J. Cheminform. 2009, 1, 6. [Google Scholar] [CrossRef] [PubMed]


| Program | Target | Scoring Function | Search Algorithm |
|---|---|---|---|
| AutoDock 4 | Protein | Physics-based + empirical | Lamarckian genetic algorithm |
| AutoDock Vina | Protein | Physics-based + empirical | Monte Carlo and quasi-Newton |
| DOCK 6 | RNA | Physics-based + force field | Incremental construction |
| rDOCK | Protein/RNA | Physics-based + empirical | Genetic algorithm, Monte Carlo and simplex minimization |
| RLDOCK | RNA | Physics-based + empirical | Multiconformer docking |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buckley, M.E.; Ndukwe, A.R.N.; Nair, P.C.; Rana, S.; Fairfull-Smith, K.E.; Gandhi, N.S. Comparative Assessment of Docking Programs for Docking and Virtual Screening of Ribosomal Oxazolidinone Antibacterial Agents. Antibiotics 2023, 12, 463. https://doi.org/10.3390/antibiotics12030463
Buckley ME, Ndukwe ARN, Nair PC, Rana S, Fairfull-Smith KE, Gandhi NS. Comparative Assessment of Docking Programs for Docking and Virtual Screening of Ribosomal Oxazolidinone Antibacterial Agents. Antibiotics. 2023; 12(3):463. https://doi.org/10.3390/antibiotics12030463
Chicago/Turabian StyleBuckley, McKenna E., Audrey R. N. Ndukwe, Pramod C. Nair, Santu Rana, Kathryn E. Fairfull-Smith, and Neha S. Gandhi. 2023. "Comparative Assessment of Docking Programs for Docking and Virtual Screening of Ribosomal Oxazolidinone Antibacterial Agents" Antibiotics 12, no. 3: 463. https://doi.org/10.3390/antibiotics12030463
APA StyleBuckley, M. E., Ndukwe, A. R. N., Nair, P. C., Rana, S., Fairfull-Smith, K. E., & Gandhi, N. S. (2023). Comparative Assessment of Docking Programs for Docking and Virtual Screening of Ribosomal Oxazolidinone Antibacterial Agents. Antibiotics, 12(3), 463. https://doi.org/10.3390/antibiotics12030463







