The Battle between Bacteria and Bacteriophages: A Conundrum to Their Immune System
Abstract
1. Introduction
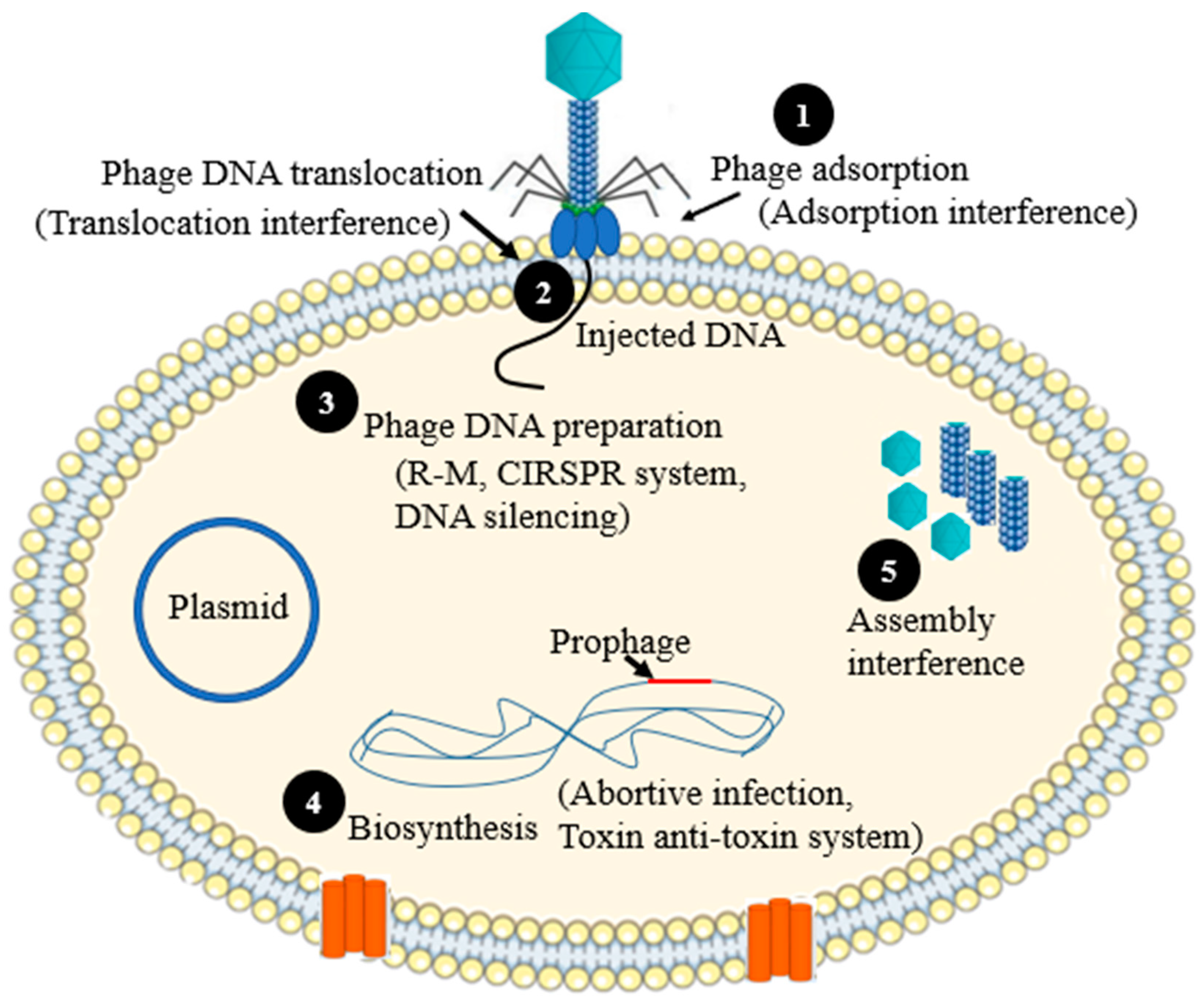
2. Mechanisms of Phage Resistances
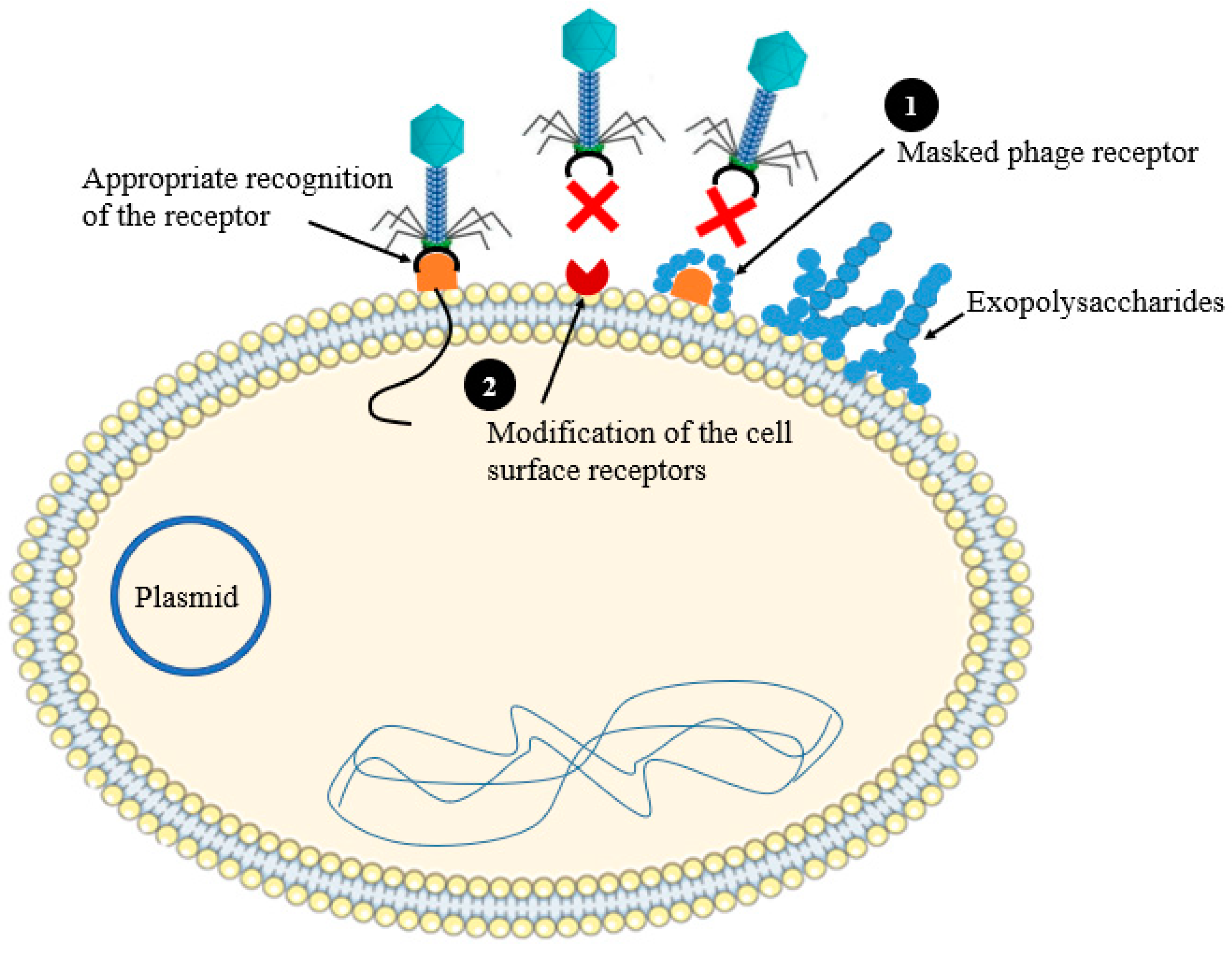
2.1. Preventing Phage Adsorption
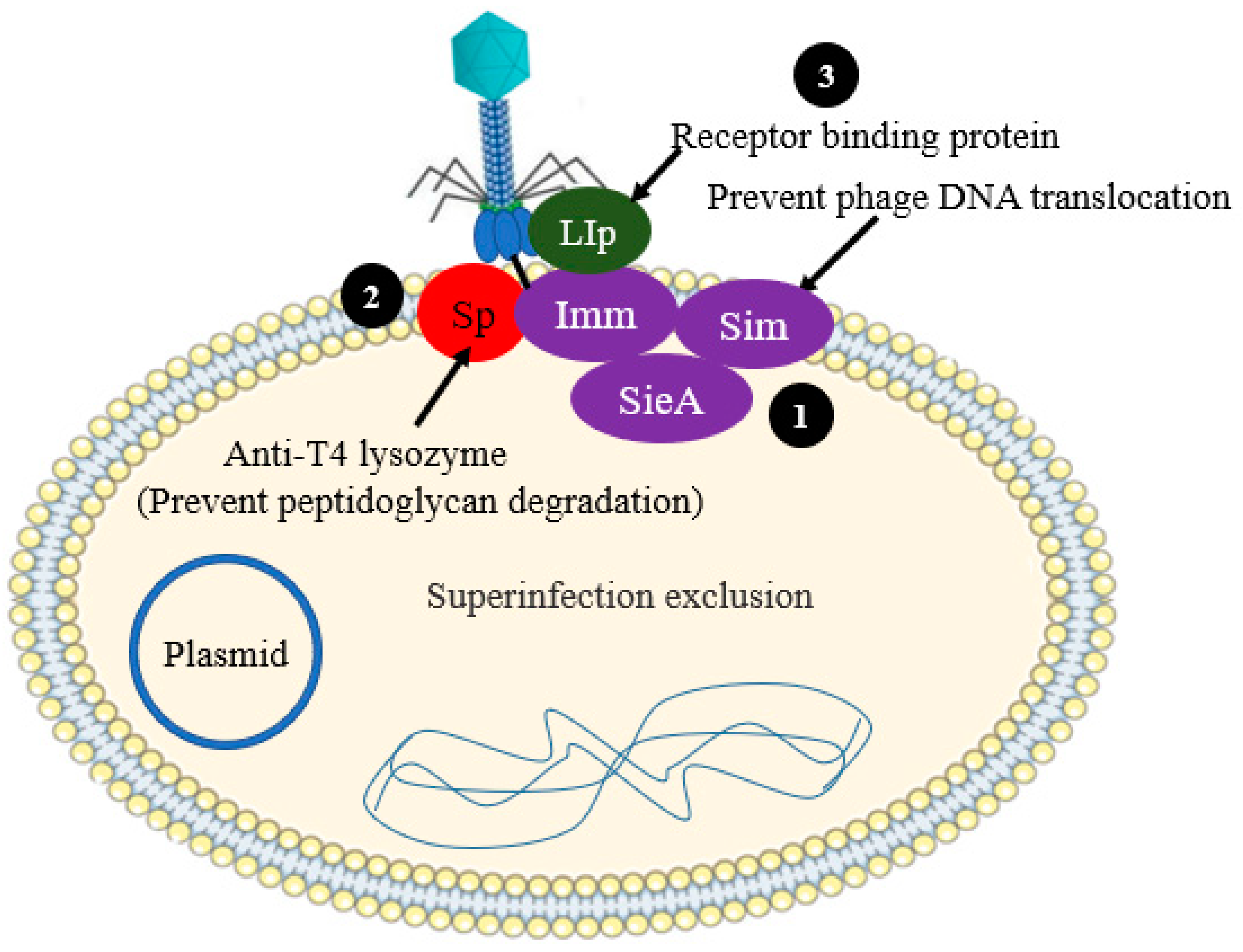
2.2. Preventing Phage DNA Entry
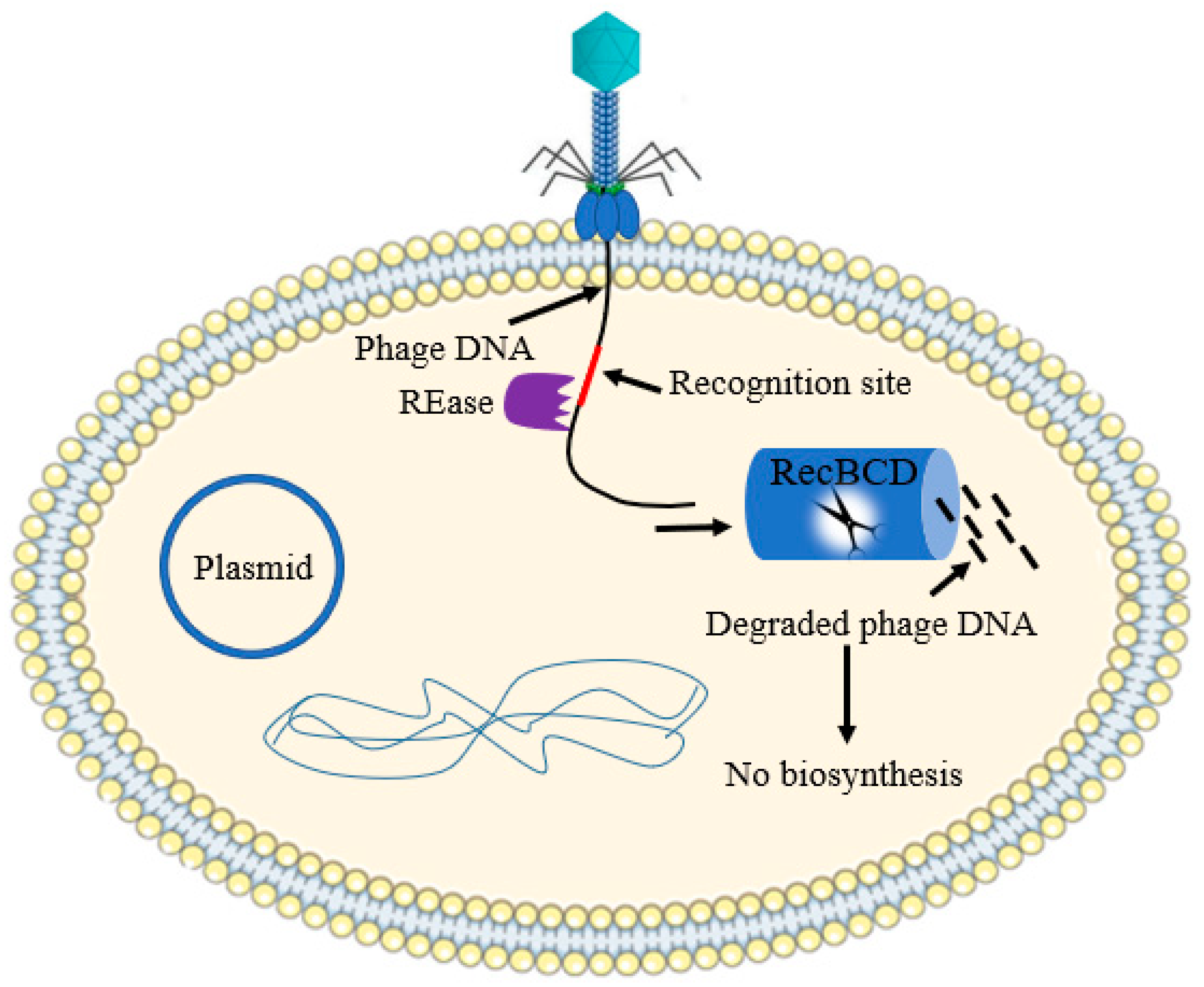
2.3. Nucleic Acid Interference
2.4. Assembly Interference
2.5. CRISPR-Cas Systems
2.6. Abortive Infection
2.7. Toxin-Antitoxin Systems
2.8. Bacterial Retrons
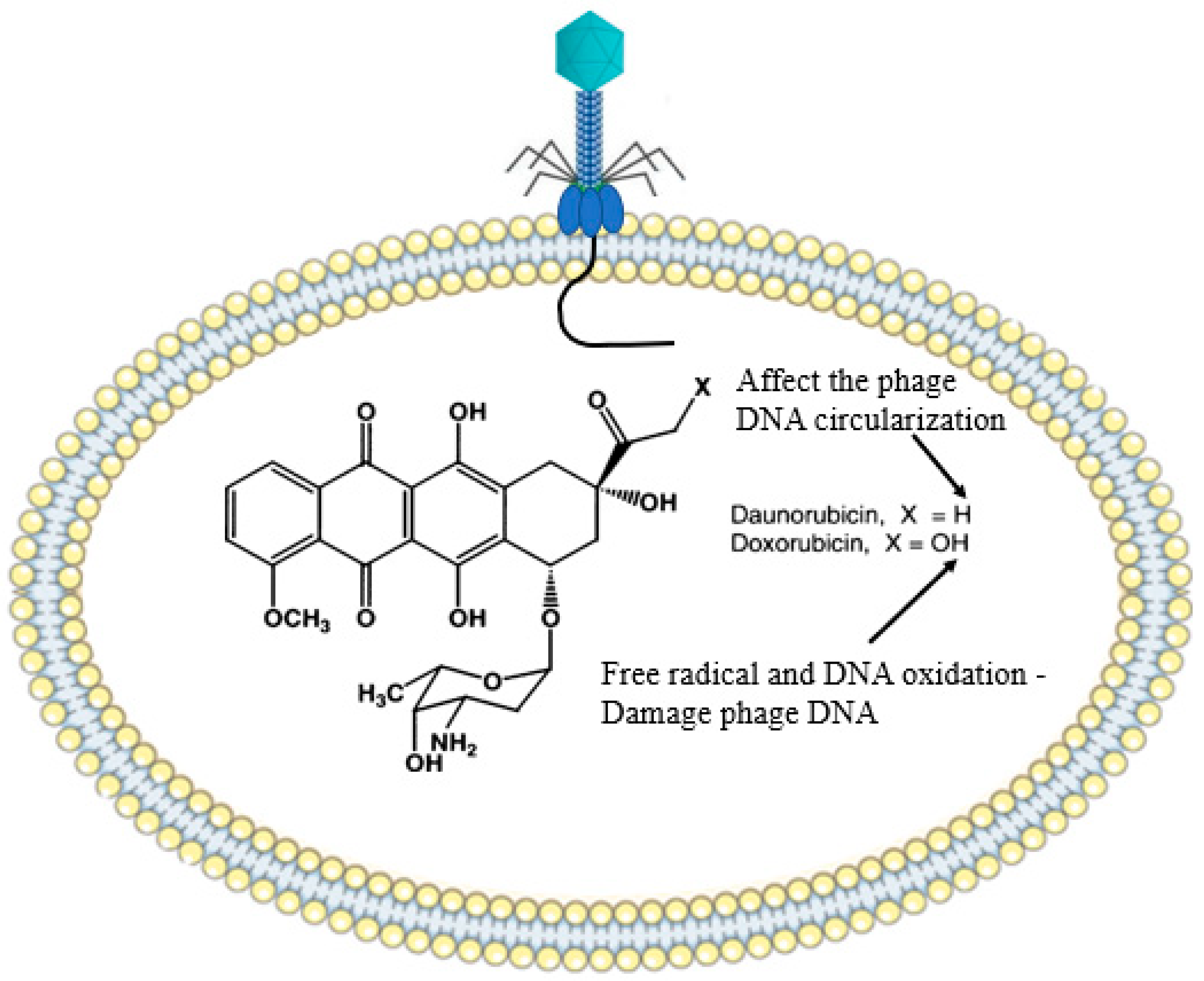
2.9. Bacterial Secondary Metabolites (Chemical Agents)
3. Phage Counteracting Mechanisms
3.1. Access to Host Receptors
3.2. Anti-Restriction—Modification Systems
3.2.1. Active Evasion Mechanisms
3.2.2. Passive Mechanisms of Phage Evasion
3.3. Escaping Abortive-Infection Mechanisms
3.4. Evading CRISPR–Cas Systems
3.4.1. Evasion by Mutation
3.4.2. Evasion by Anti-CRISPR Genes
4. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Correction Statement
References
- De Kraker, M.E.; Stewardson, A.J.; Harbarth, S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A. Marine viruses—Major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Hatfull, G.F.; Hendrix, R.W. Bacteriophages and their genomes. Curr. Opin. Virol. 2011, 1, 298–303. [Google Scholar] [CrossRef]
- Górski, A.; Międzybrodzki, R.; Węgrzyn, G.; Jończyk-Matysiak, E.; Borysowski, J.; Weber-Dąbrowska, B. Phage therapy: Current status and perspectives. Med. Res. Rev. 2020, 40, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.P.; Costa, A.R.; Pinto, G.; Meneses, L.; Azeredo, J. Current challenges and future opportunities of phage therapy. FEMS Microbiol. Rev. 2020, 44, 684–700. [Google Scholar] [CrossRef]
- Seed, K.D. Battling phages: How bacteria defend against viral attack. PLoS Pathog. 2015, 11, e1004847. [Google Scholar] [CrossRef] [PubMed]
- Bondy-Denomy, J.; Qian, J.; Westra, E.R.; Buckling, A.; Guttman, D.S.; Davidson, A.R.; Maxwell, K.L. Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 2016, 10, 2854–2866. [Google Scholar] [CrossRef]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef]
- Bono, L.M.; Gensel, C.L.; Pfennig, D.W.; Burch, C.L. Competition and the origins of novelty: Experimental evolution of niche-width expansion in a virus. Biol. Lett. 2013, 9, 20120616. [Google Scholar] [CrossRef]
- Simmons, E.L.; Drescher, K.; Nadell, C.D.; Bucci, V. Phage mobility is a core determinant of phage–bacteria coexistence in biofilms. ISME J. 2018, 12, 531–543. [Google Scholar] [CrossRef]
- Egido, J.E.; Costa, A.R.; Aparicio-Maldonado, C.; Haas, P.; Brouns, S.J. Mechanisms and clinical importance of bacteriophage resistance. FEMS Microbiol. Rev. 2022, 46, fuab048. [Google Scholar] [CrossRef] [PubMed]
- Scholl, D.; Adhya, S.; Merril, C. Escherichia coli K1’s capsule is a barrier to bacteriophage T7. Appl. Environ. Microbiol. 2005, 71, 4872–4874. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Robles, T.; Dillard, R.S.; Cairns, L.S.; Silva-Valenzuela, C.A.; Housman, M.; Ali, A.; Wright, E.R.; Camilli, A. Vibrio cholerae outer membrane vesicles inhibit bacteriophage infection. J. Bacteriol. 2018, 200, e00792-17. [Google Scholar] [CrossRef]
- Gencay, Y.E.; Sørensen, M.C.; Wenzel, C.Q.; Szymanski, C.M.; Brøndsted, L. Phase variable expression of a single phage receptor in Campylobacter jejuni NCTC12662 influences sensitivity toward several diverse CPS-dependent phages. Front. Microbiol. 2018, 9, 82. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Arambula, D.; Ghosh, P.; Miller, J.F. Diversity-generating retroelements in phage and bacterial genomes. Mob. DNA III 2015, 2, 1237–1252. [Google Scholar] [CrossRef]
- Shi, K.; Oakland, J.T.; Kurniawan, F.; Moeller, N.H.; Banerjee, S.; Aihara, H. Structural basis of superinfection exclusion by bacteriophage T4 Spackle. Commun. Biol. 2020, 3, 691. [Google Scholar] [CrossRef]
- Lu, M.J.; Stierhof, Y.D.; Henning, U. Location and unusual membrane topology of the immunity protein of the Escherichia coli phage T4. J. Virol. 1993, 67, 4905–4913. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Henning, U. Superinfection exclusion by T-even-type coliphages. Trends Microbiol. 1994, 2, 137–139. [Google Scholar] [CrossRef]
- Moak, M.; Molineux, I.J. Role of the Gp16 lytic transglycosylase motif in bacteriophage T7 virions at the initiation of infection. Mol. Microbiol. 2000, 37, 345–355. [Google Scholar] [CrossRef]
- Vasu, K.; Nagaraja, V. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol. Mol. Biol. Rev. 2013, 77, 53–72. [Google Scholar] [CrossRef]
- Stein, D.C.; Gunn, J.S.; Radlinska, M.; Piekarowicz, A. Restriction and modification systems of Neisseria gonorrhoeae. Gene 1995, 157, 19–22. [Google Scholar] [CrossRef] [PubMed]
- Tock, M.R.; Dryden, D.T. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 2005, 8, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Lepikhov, K.; Tchernov, A.; Zheleznaja, L.; Matvienko, N.; Walter, J.; Trautner, T.A. Characterization of the type IV restriction modification system Bsp LU11III from Bacillus sp. LU11. Nucleic Acids Res. 2001, 29, 4691–4698. [Google Scholar] [CrossRef] [PubMed]
- Wilson, G.G.; Murray, N.E. Restriction and modification systems. Annu. Rev. Genet. 1991, 25, 585–627. [Google Scholar] [CrossRef] [PubMed]
- McGrath, S.; Seegers, J.F.; Fitzgerald, G.F.; van Sinderen, D. Molecular characterization of a phage-encoded resistance system in Lactococcus lactis. Appl. Environ. Microbiol. 1999, 65, 1891–1899. [Google Scholar] [CrossRef]
- Bickle, T.A.; Krüger, D.H. Biology of DNA restriction. Microbiol. Rev. 1993, 57, 434–450. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rostøl, J.T.; Marraffini, L. (Ph) ighting phages: How bacteria resist their parasites. Cell Host Microbe 2019, 25, 184–194. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Haft, D.H. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Marraffini, L.A.; Sontheimer, E.J. CRISPR interference limits horizontal gene transfer in Staphylococci by targeting DNA. Science 2008, 322, 1843–1845. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification, and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Lopatina, A.; Tal, N.; Sorek, R. Abortive infection: Bacterial suicide as an antiviral immune strategy. Annu. Rev. Virol. 2020, 7, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Snyder, L. Phage-exclusion enzymes: A bonanza of biochemical and cell biology reagents? Mol. Microbiol. 1995, 15, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, C.K.; Molineux, I.J. Expression of gene 1.2 and gene 10 of bacteriophage T7 is lethal to F plasmid-containing Escherichia coli. J. Bacteriol. 1991, 173, 1536–1543. [Google Scholar] [CrossRef]
- Bingham, R.; Ekunwe, S.I.; Falk, S.; Snyder, L.; Kleanthous, C. The major head protein of bacteriophage T4 binds specifically to elongation factor Tu. J. Biol. Chem. 2000, 275, 23219–23226. [Google Scholar] [CrossRef]
- Depardieu, F.; Didier, J.; Bernheim, A.; Sherlock, A.; Molina, H.; Duclos, B.; Bikard, D. A eukaryotic-like serine/threonine kinase protects staphylococci against phages. Cell Host Microbe 2016, 20, 471–481. [Google Scholar] [CrossRef]
- Durmaz, E.; Klaenhammer, T.R. Abortive phage resistance mechanism AbiZ speeds the lysis clock to cause premature lysis of phage-infected Lactococcus lactis. J. Bacteriol. 2007, 189, 1417–1425. [Google Scholar] [CrossRef]
- Kazlauskiene, M.; Kostiuk, G.; Venclovas, Č.; Tamulaitis, G.; Siksnys, V. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 2017, 357, 605–609. [Google Scholar] [CrossRef]
- Niewoehner, O.; Garcia-Doval, C.; Rostøl, J.T.; Berk, C.; Schwede, F.; Bigler, L.; Hall, J.; Marraffini, L.A.; Jinek, M. Type III CRISPR–Cas systems produce cyclic oligoadenylate second messengers. Nature 2017, 548, 543–548. [Google Scholar] [CrossRef]
- Koga, M.; Otsuka, Y.; Lemire, S.; Yonesaki, T. Escherichia coli rnlA and rnlB compose a novel toxin–antitoxin system. Genetics 2011, 187, 123–130. [Google Scholar] [CrossRef]
- Dai, G.; Su, P.; Allison, G.E.; Geller, B.L.; Zhu, P.; Kim, W.S.; Dunn, N.W. Molecular characterization of a new abortive infection system (AbiU) from Lactococcus lactis LL51-1. Appl. Environ. Microbiol. 2001, 67, 5225–5232. [Google Scholar] [CrossRef] [PubMed]
- Harms, A.; Brodersen, D.E.; Mitarai, N.; Gerdes, K. Toxins, targets, and triggers: An overview of toxin-antitoxin biology. Mol. Cell 2018, 70, 768–784. [Google Scholar] [CrossRef] [PubMed]
- Alawneh, A.M.; Qi, D.; Yonesaki, T.; Otsuka, Y. An ADP-ribosyltransferase A lt of bacteriophage T 4 negatively regulates the Escherichia coli MazF toxin of a toxin–antitoxin module. Mol. Microbiol. 2016, 99, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Blower, T.R.; Pei, X.Y.; Short, F.L.; Fineran, P.C.; Humphreys, D.P.; Luisi, B.F.; Salmond, G.P. A processed noncoding RNA regulates an altruistic bacterial antiviral system. Nat. Struct. Mol. Biol. 2011, 18, 185–190. [Google Scholar] [CrossRef]
- Tormo-Más, M.Á.; Mir, I.; Shrestha, A.; Tallent, S.M.; Campoy, S.; Lasa, Í.; Barbé, J.; Novick, R.P.; Christie, G.E.; Penadés, J.R. Moonlighting bacteriophage proteins derepress staphylococcal pathogenicity islands. Nature 2010, 465, 779–782. [Google Scholar] [CrossRef]
- Ram, G.; Chen, J.; Kumar, K.; Ross, H.F.; Ubeda, C.; Damle, P.K.; Lane, K.D.; Penadés, J.R.; Christie, G.E.; Novick, R.P. Staphylococcal pathogenicity island interference with helper phage reproduction is a paradigm of molecular parasitism. Proc. Natl. Acad. Sci. USA 2012, 109, 16300–16305. [Google Scholar] [CrossRef]
- Wang, C.; Villion, M.; Semper, C.; Coros, C.; Moineau, S.; Zimmerly, S. A reverse transcriptase-related protein mediates phage resistance and polymerizes untemplated DNA in vitro. Nucleic Acids Res. 2011, 39, 7620–7629. [Google Scholar] [CrossRef]
- Kaufmann, G. Anticodon nucleases. Trends Biochem. Sci. 2000, 25, 70–74. [Google Scholar] [CrossRef]
- Chinenova, T.A.; Mkrtumian, N.M.; Lomovskaia, N.D. Genetic characteristics of a new phage resistance trait in Streptomyces coelicolor A3 (2). Genetika 1982, 18, 1945–1952. [Google Scholar]
- Harvey, H.; Bondy-Denomy, J.; Marquis, H.; Sztanko, K.M.; Davidson, A.R.; Burrows, L.L. Pseudomonas aeruginosa defends against phages through type IV pilus glycosylation. Nat. Microbiol. 2018, 3, 47–52. [Google Scholar] [CrossRef]
- McGrath, S.; Fitzgerald, G.F.; Sinderen, D.V. Identification and characterization of phage-resistance genes in temperate lactococcal bacteriophages. Mol. Microbiol. 2002, 43, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Mahony, J.; McGrath, S.; Fitzgerald, G.F.; van Sinderen, D. Identification and characterization of lactococcal-prophage-carried superinfection exclusion genes. Appl. Environ. Microbiol. 2008, 74, 6206–6215. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Göhler, A.; Heller, K.J.; Neve, H. The ltp gene of temperate Streptococcus thermophilus phage TP-J34 confers superinfection exclusion to Streptococcus thermophilus and Lactococcus lactis. Virology 2006, 350, 146–157. [Google Scholar] [CrossRef]
- Vovis, G.F.; Lacks, S. Complementary action of restriction enzymes endo R· DpnI and endo R· DpnII on bacteriophage f1 DNA. J. Mol. Biol. 1977, 115, 525–538. [Google Scholar] [CrossRef] [PubMed]
- Hegge, J.W.; Swarts, D.C.; van der Oost, J. Prokaryotic Argonaute proteins: Novel genome-editing tools? Nat. Rev. Microbiol. 2018, 16, 5–11. [Google Scholar] [CrossRef]
- Simon, A.J.; Ellington, A.D.; Finkelstein, I.J. Retrons and their applications in genome engineering. Nucleic Acids Res. 2019, 47, 11007–11019. [Google Scholar] [CrossRef]
- Lampson, B.; Inouye, M.; Inouye, S. The msDNAs of bacteria. Prog. Nucleic Acid Res. Mol. Biol. 2001, 67, 65–91. [Google Scholar]
- Millman, A.; Bernheim, A.; Stokar-Avihail, A.; Fedorenko, T.; Voichek, M.; Leavitt, A.; Oppenheimer-Shaanan, Y.; Sorek, R. Bacterial retrons function in anti-phage defense. Cell 2020, 183, 1551–1561.e12. [Google Scholar] [CrossRef]
- Kronheim, S.; Daniel-Ivad, M.; Duan, Z.; Hwang, S.; Wong, A.I.; Mantel, I.; Nodwell, J.R.; Maxwell, K.L. A chemical defence against phage infection. Nature 2018, 564, 283–286. [Google Scholar] [CrossRef]
- York, A. Chemical warfare against phages. Nat. Rev. Microbiol. 2019, 17, 64. [Google Scholar] [CrossRef]
- Davies, J. Specialized microbial metabolites: Functions and origins. J. Antibiot. 2013, 66, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Rothenberg, E. Interaction of bacteriophage λ with its E. coli receptor, LamB. Viruses 2012, 4, 3162–3178. [Google Scholar] [CrossRef] [PubMed]
- Laanto, E.; Mäkelä, K.; Hoikkala, V.; Ravantti, J.J.; Sundberg, L. Adapting a phage to combat phage resistance. Antibiotics 2020, 9, 291. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.J.; Belfort, M.; Bestor, T.; Bhagwat, A.S.; Bickle, T.A.; Bitinaite, J.; Blumenthal, R.M.; Degtyarev, S.K.; Dryden, D.T.; Dybvig, K. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003, 31, 1805–1812. [Google Scholar] [CrossRef] [PubMed]
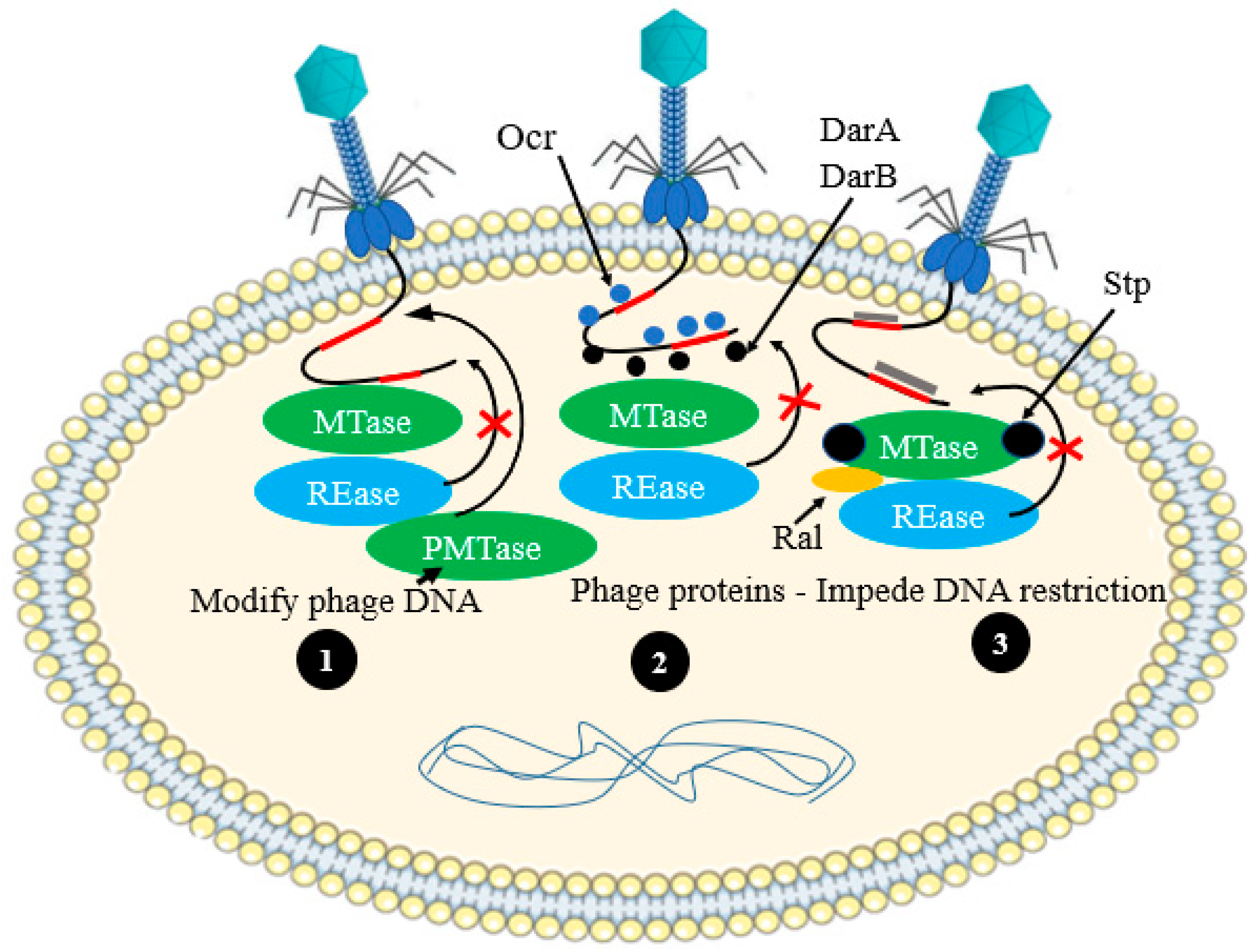
- Iida, S.; Streiff, M.B.; Bickle, T.A.; Arber, W. Two DNA antirestriction systems of bacteriophage P1, darA, and darB: Characterization of darA−phages. Virology 1987, 157, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Atanasiu, C.; Su, T.; Sturrock, S.S.; Dryden, D. Interaction of the ocr gene 0.3 protein of bacteriophage T7 with Eco KI restriction/modification enzyme. Nucleic Acids Res. 2002, 30, 3936–3944. [Google Scholar] [CrossRef]
- Walkinshaw, M.D.; Taylor, P.; Sturrock, S.S.; Atanasiu, C.; Berge, T.; Henderson, R.M.; Edwardson, J.M.; Dryden, D. Structure of Ocr from bacteriophage T7, a protein that mimics B-form DNA. Mol. Cell 2002, 9, 187–194. [Google Scholar] [CrossRef]
- Lacks, S.; Greenberg, B. A deoxyribonuclease of Diplococcus pneumoniae specific for methylated DNA. J. Biol. Chem. 1975, 250, 4060–4066. [Google Scholar] [CrossRef]
- Sberro, H.; Leavitt, A.; Kiro, R.; Koh, E.; Peleg, Y.; Qimron, U.; Sorek, R. Discovery of functional toxin/antitoxin systems in bacteria by shotgun cloning. Mol. Cell 2013, 50, 136–148. [Google Scholar] [CrossRef]
- Samson, J.E.; Bélanger, M.; Moineau, S. Effect of the abortive infection mechanism and type III toxin/antitoxin system AbiQ on the lytic cycle of Lactococcus lactis phages. J. Bacteriol. 2013, 195, 3947–3956. [Google Scholar] [CrossRef]
- Otsuka, Y.; Yonesaki, T. Dmd of bacteriophage T4 functions as an antitoxin against Escherichia coli LsoA and RnlA toxins. Mol. Microbiol. 2012, 83, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Short, F.L.; Akusobi, C.; Broadhurst, W.R.; Salmond, G.P. The bacterial Type III toxin-antitoxin system, ToxIN, is a dynamic protein-RNA complex with stability-dependent antiviral abortive infection activity. Sci. Rep. 2018, 8, 1013. [Google Scholar] [CrossRef]
- Deveau, H.; Barrangou, R.; Garneau, J.E.; Labonté, J.; Fremaux, C.; Boyaval, P.; Romero, D.A.; Horvath, P.; Moineau, S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008, 190, 1390–1400. [Google Scholar] [CrossRef] [PubMed]
- Sapranauskas, R.; Gasiunas, G.; Fremaux, C.; Barrangou, R.; Horvath, P.; Siksnys, V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011, 39, 9275–9282. [Google Scholar] [CrossRef] [PubMed]
- Bondy-Denomy, J.; Pawluk, A.; Maxwell, K.L.; Davidson, A.R. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 2013, 493, 429–432. [Google Scholar] [CrossRef]
- Pawluk, A.; Davidson, A.R.; Maxwell, K.L. Anti-CRISPR: Discovery, mechanism and function. Nat. Rev. Microbiol. 2018, 16, 12–17. [Google Scholar] [CrossRef]
- Krüger, D.H.; Barcak, G.J.; Reuter, M.; Smith, H.O. Eco RII can be activated to cleave refractory DNA recognition sites. Nucleic Acids Res. 1988, 16, 3997–4008. [Google Scholar] [CrossRef]
- Stummeyer, K.; Schwarzer, D.; Claus, H.; Vogel, U.; Gerardy-Schahn, R.; Mühlenhoff, M. Evolution of bacteriophages infecting encapsulated bacteria: Lessons from Escherichia coli K1-specific phages. Mol. Microbiol. 2006, 60, 1123–1135. [Google Scholar] [CrossRef]
- Meyer, J.R.; Dobias, D.T.; Weitz, J.S.; Barrick, J.E.; Quick, R.T.; Lenski, R.E. Repeatability and contingency in the evolution of a key innovation in phage lambda. Science 2012, 335, 428–432. [Google Scholar] [CrossRef]
- Ravin, V.; Räisänen, L.; Alatossava, T. A conserved C-terminal region in Gp71 of the small isometric-head phage LL-H and ORF474 of the prolate-head phage JCL1032 is implicated in specificity of adsorption of phage to its host, Lactobacillus delbrueckii. J. Bacteriol. 2002, 184, 2455–2459. [Google Scholar] [CrossRef]
- Scanlan, P.D.; Hall, A.R.; Lopez-Pascua, L.D.; Buckling, A. Genetic basis of infectivity evolution in a bacteriophage. Mol. Ecol. 2011, 20, 981–989. [Google Scholar] [CrossRef] [PubMed]


| Specific Systems | Bacteria | Phage Resistance Mechanism (Anti-Phage Defense Strategies) | References |
|---|---|---|---|
| ToxN, RNase activity, destroying both host and phage transcripts | Pectobacterium atrosepticum | TA systems | [44] |
| MazF/MazE TA system | E. coli | TA systems | [43] |
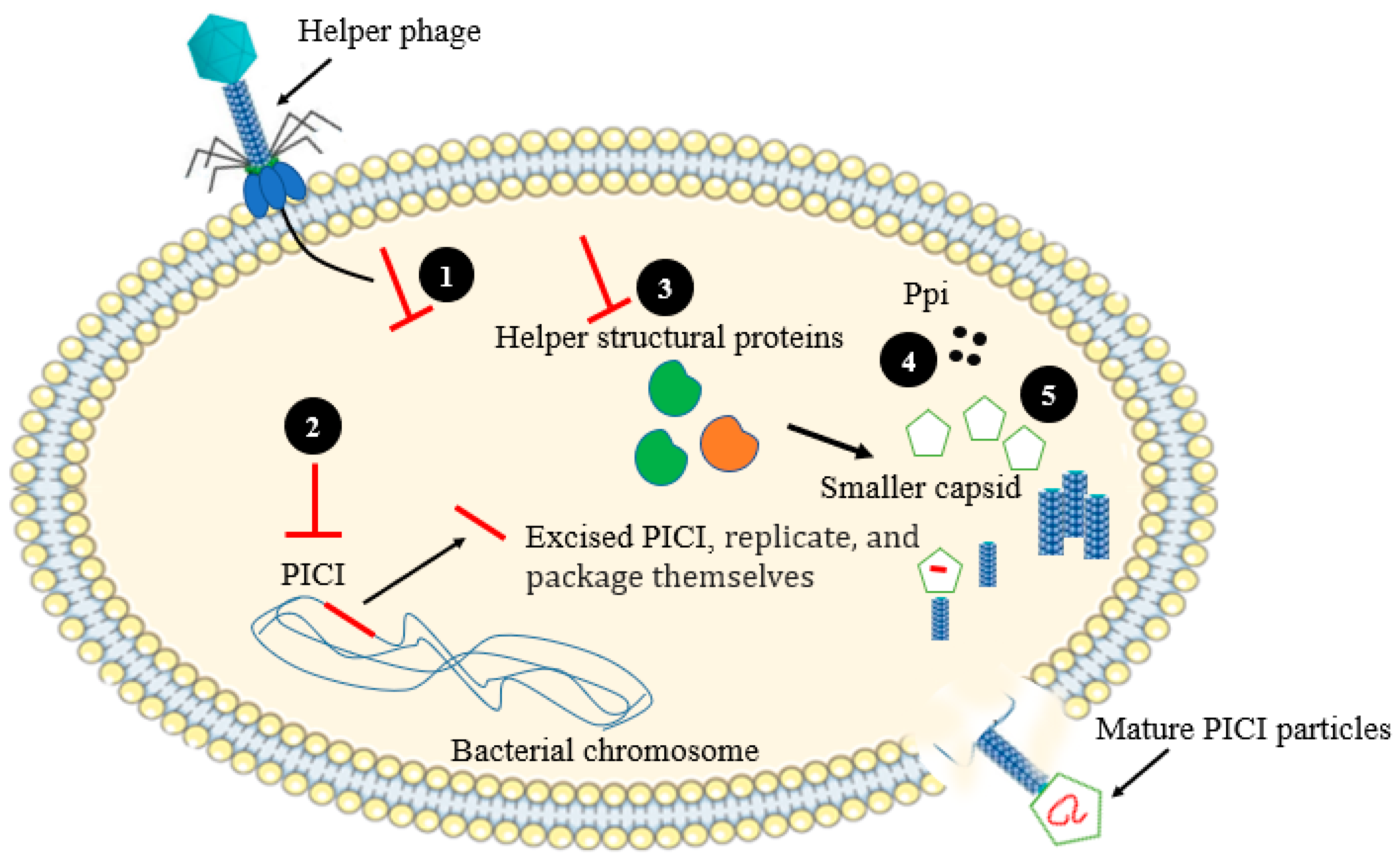
| Phage-inducible chromosomal islands (PICIs) | Staphylococcus aureus | Assembly Interference | [45] |
| Ppi protein prevent phage packaging process | Staphylococcus spp. | Assembly interference | [46] |
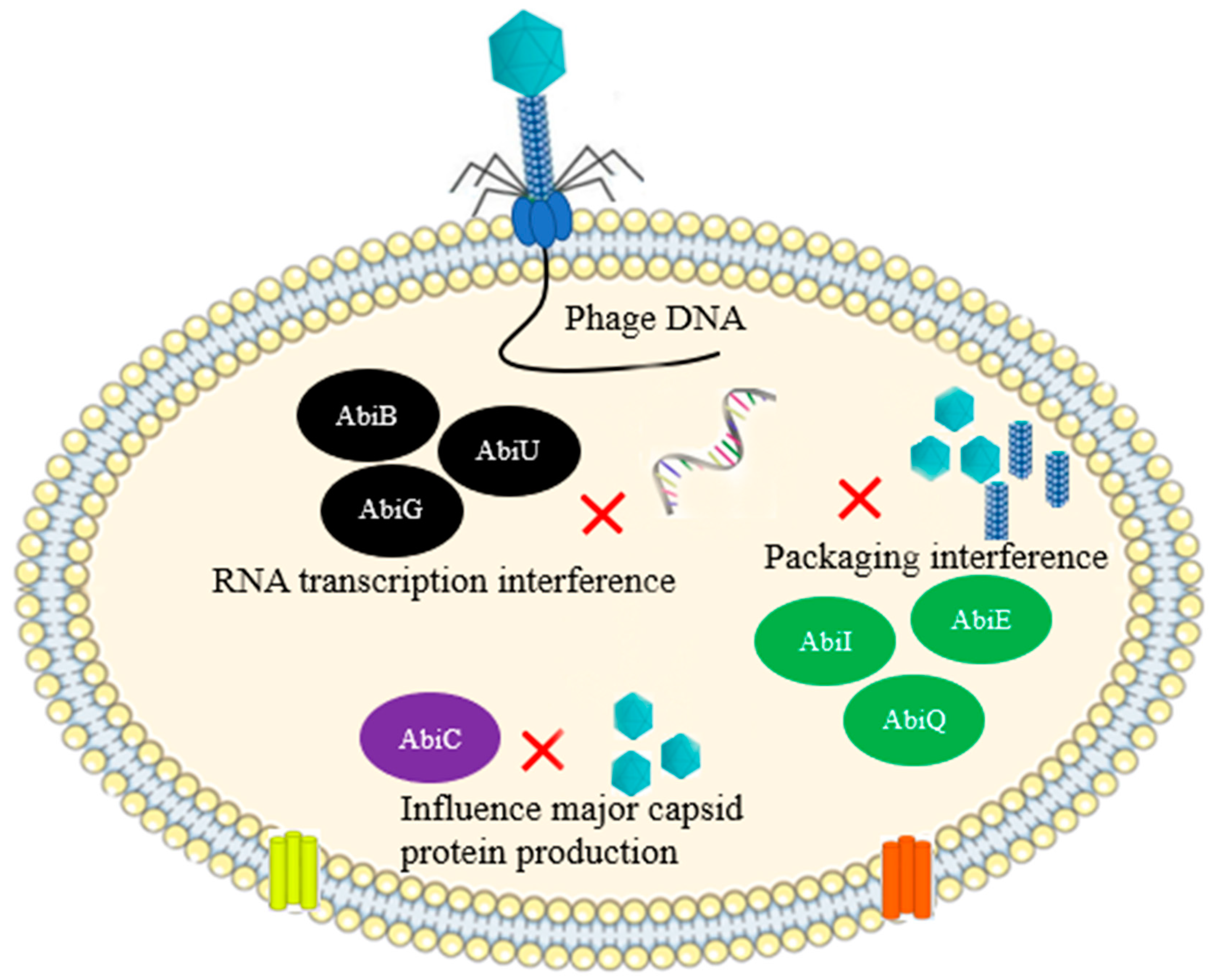
| abiK system | L. lactis | Abi | [47] |
| AbiZ (100-fold reduction of the burst size of phage Φ31) | L. lactis | Abi | [37] |
| Stk2 Abi System (kinase-mediated Abi mechanism) | Staphylococcus epidermidis | Abi | [36] |
| Inhibiting the protein translation system using the peptide Lit and the anticodon nuclease (PrrC) | E. coli | Abi | [48] |
| Stp protein of phage T4 affects the interaction of PrrC and EcoprrI, freeing triggered PrrC protein and leading to abortion | E. coli | Abi | [33] |
| “inverted” RM systems | Streptomyces coelicolor A2(3) | R–M systems | [49] |
| Type IV pili | Pseudomonas aeruginosa | Preventing phage adsorption | [50] |
| Lipopolysaccharide (LPS) | E. coli K1 | Preventing phage adsorption | [12] |
| Mediated by imm and sp | E. coli | Sie systems | [17] |
| Sie2009 | L. lactis and lactococcal prophages | Sie systems | [51,52] |
| Using the signal peptide lipoprotein prophage TP-J34 (LTP) | Prophage of Streptococcus thermophilus | Sie systems | [53] |
| Using MDS enzymes (DpnI for Streptococcus pneumoniae, McrBC, McrA, and Mrr for E. coli) | Streptococcus pneumoniae, E. coli | Modification-dependent systems (MDSs), which detect the modified phage DNA | [26,54] |
| RNA-guided DNA silencing and DNA-guided DNA silencing | It is available in some bacterial spp. | Argonautes (pAgos) | [55] |
| Specific System | Anti-Phage Resistance Strategies | Example of Phages | References |
|---|---|---|---|
| Absence of endonuclease recognition sites (Lack of Sau 3A regions in its dsDNA) | Anti-R-M system | Staphylococcus phage K | [77] |
| Phage DNA modification comprised the rare base hydroxymethylcytosine (HMC) in place of the cytosine | Anti-R-M system | Phage T4 | [26] |
| Ocr protein prevent restriction activity | Anti-R-M system | Coliphage T7 | [67] |
| Using anti-restriction protein | Anti-R-M system | Phage P1 | [65] |
| Using protein RIIA and RIIB | Anti-Abi mechanism | Phage T4 | [33] |
| The toxin effect of LsoA and RnlA neutralized by Dmd during replication of phage | Anti-Abi system | Coliphage T4 | [71] |
| Polysaccharide-degradation using hydrolases and lyases | Accessing the receptors | Depolymerase producing phages | [78] |
| Anti-CRISPR proteins | Interfere with CRISPR–Cas system | P. aeruginosa prophages | [75] |
| Protospacer mutation | Interfere with CRISPR–Cas system | Streptococcus thermophilus phages | [73] |
| Mutations in the RBP-encoding gene mutation | New receptors adaptation | Coliphages T7 and ϕX174 | [79] |
| Tail protein modification | New receptors adaptation | Pseudomonas fluorescens phage ϕ2, L. lactis phage LL-H | [80,81] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teklemariam, A.D.; Al-Hindi, R.R.; Qadri, I.; Alharbi, M.G.; Ramadan, W.S.; Ayubu, J.; Al-Hejin, A.M.; Hakim, R.F.; Hakim, F.F.; Hakim, R.F.; et al. The Battle between Bacteria and Bacteriophages: A Conundrum to Their Immune System. Antibiotics 2023, 12, 381. https://doi.org/10.3390/antibiotics12020381
Teklemariam AD, Al-Hindi RR, Qadri I, Alharbi MG, Ramadan WS, Ayubu J, Al-Hejin AM, Hakim RF, Hakim FF, Hakim RF, et al. The Battle between Bacteria and Bacteriophages: A Conundrum to Their Immune System. Antibiotics. 2023; 12(2):381. https://doi.org/10.3390/antibiotics12020381
Chicago/Turabian StyleTeklemariam, Addisu D., Rashad R. Al-Hindi, Ishtiaq Qadri, Mona G. Alharbi, Wafaa S. Ramadan, Jumaa Ayubu, Ahmed M. Al-Hejin, Raghad F. Hakim, Fanar F. Hakim, Rahad F. Hakim, and et al. 2023. "The Battle between Bacteria and Bacteriophages: A Conundrum to Their Immune System" Antibiotics 12, no. 2: 381. https://doi.org/10.3390/antibiotics12020381
APA StyleTeklemariam, A. D., Al-Hindi, R. R., Qadri, I., Alharbi, M. G., Ramadan, W. S., Ayubu, J., Al-Hejin, A. M., Hakim, R. F., Hakim, F. F., Hakim, R. F., Alseraihi, L. I., Alamri, T., & Harakeh, S. (2023). The Battle between Bacteria and Bacteriophages: A Conundrum to Their Immune System. Antibiotics, 12(2), 381. https://doi.org/10.3390/antibiotics12020381






