Anti-Biofilm Effects of Rationally Designed Peptides against Planktonic Cells and Pre-Formed Biofilm of Pseudomonas aeruginosa
Abstract
1. Introduction
2. Results and Discussion
2.1. Antibacterial Activity of Peptides against Clinical Isolates of P. aeruginosa
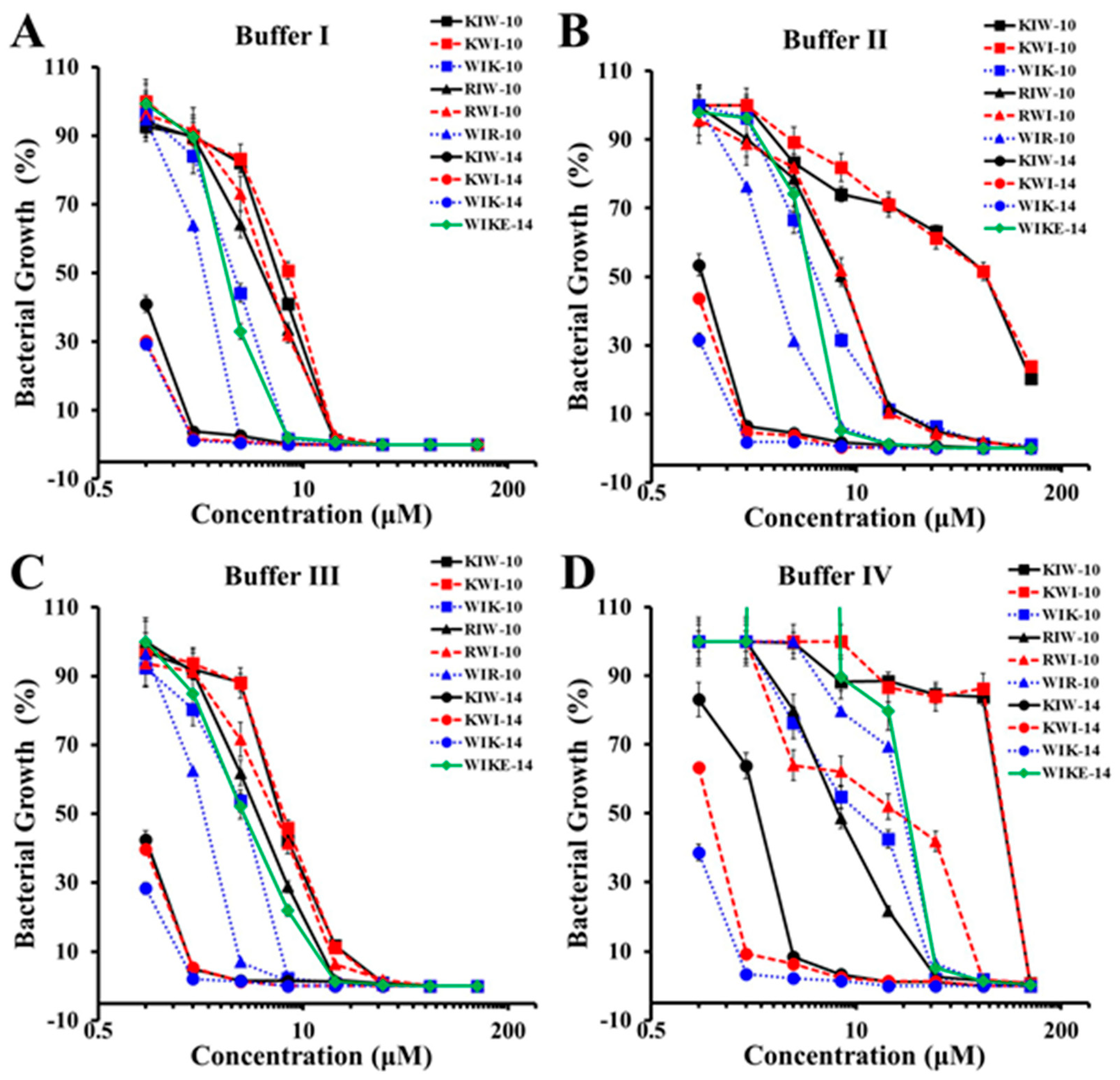
2.2. Growth Inhibition of Peptides in Various Ionic Strength
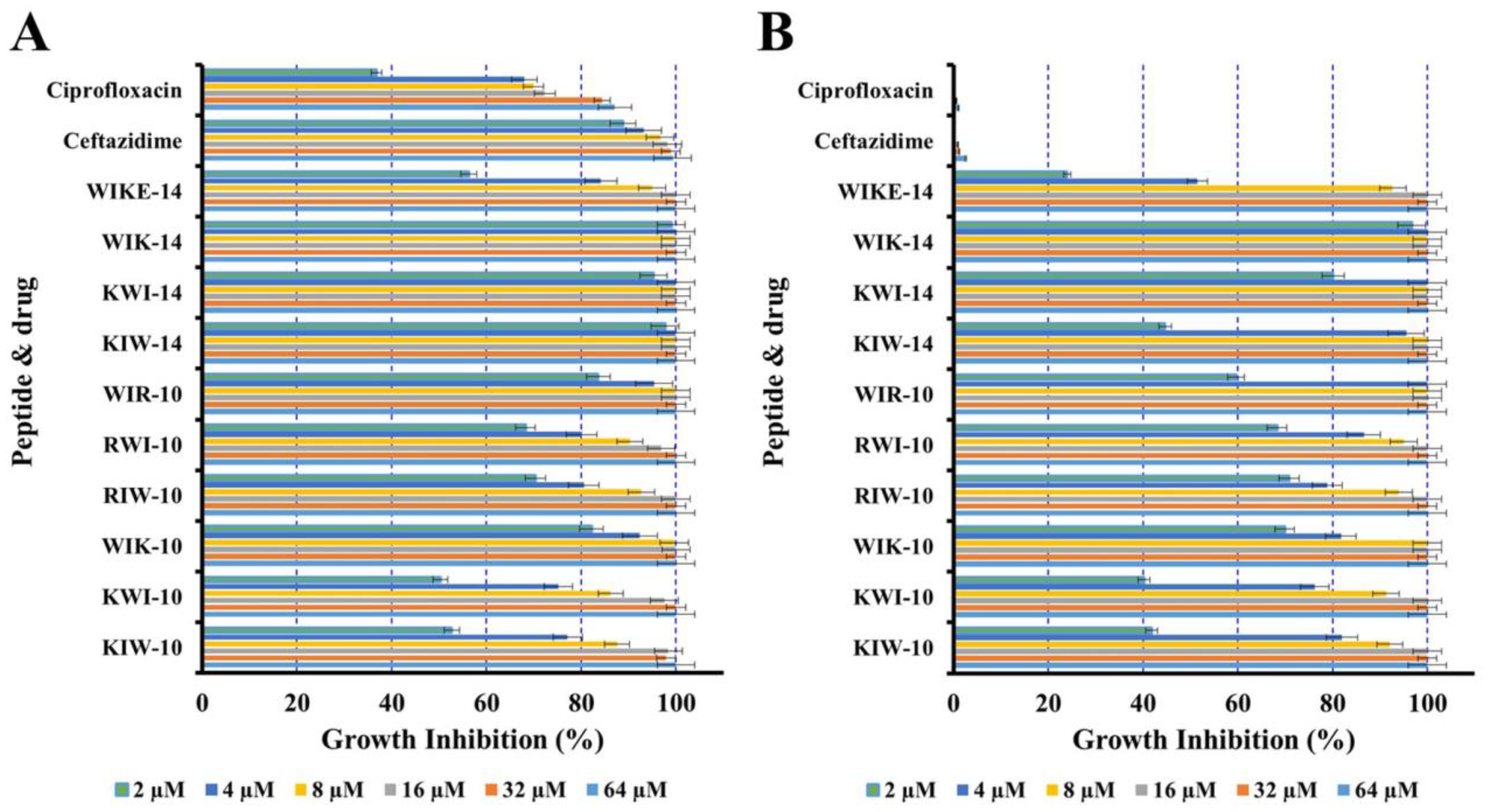
2.3. Growth Inhibition of P. aeruginosa Planktonic Cells
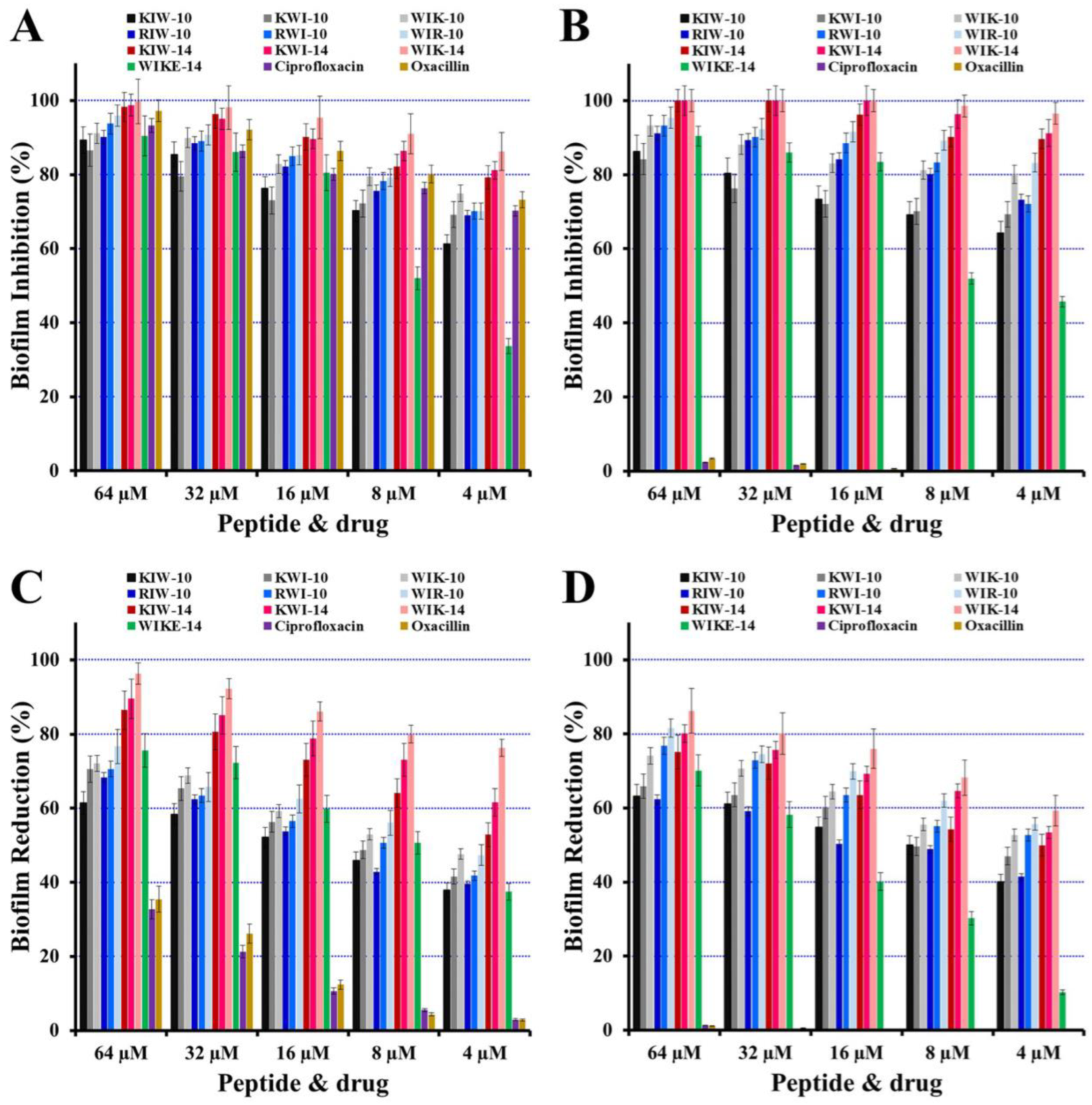
2.4. Inhibition of Biofilm Formation and Reduction on Preformed Biofilm
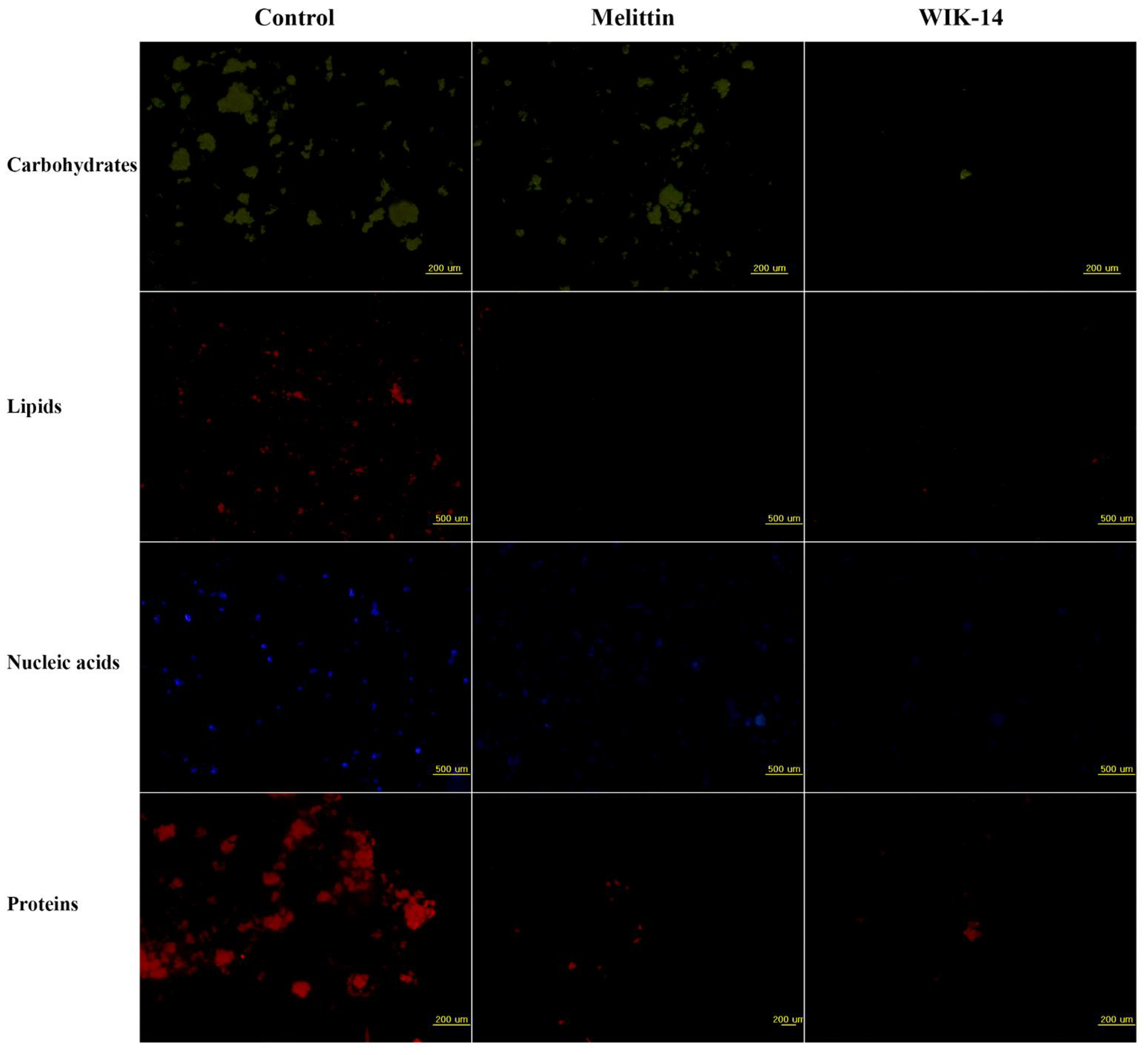
2.5. Effects of Peptides on Biofilm Components
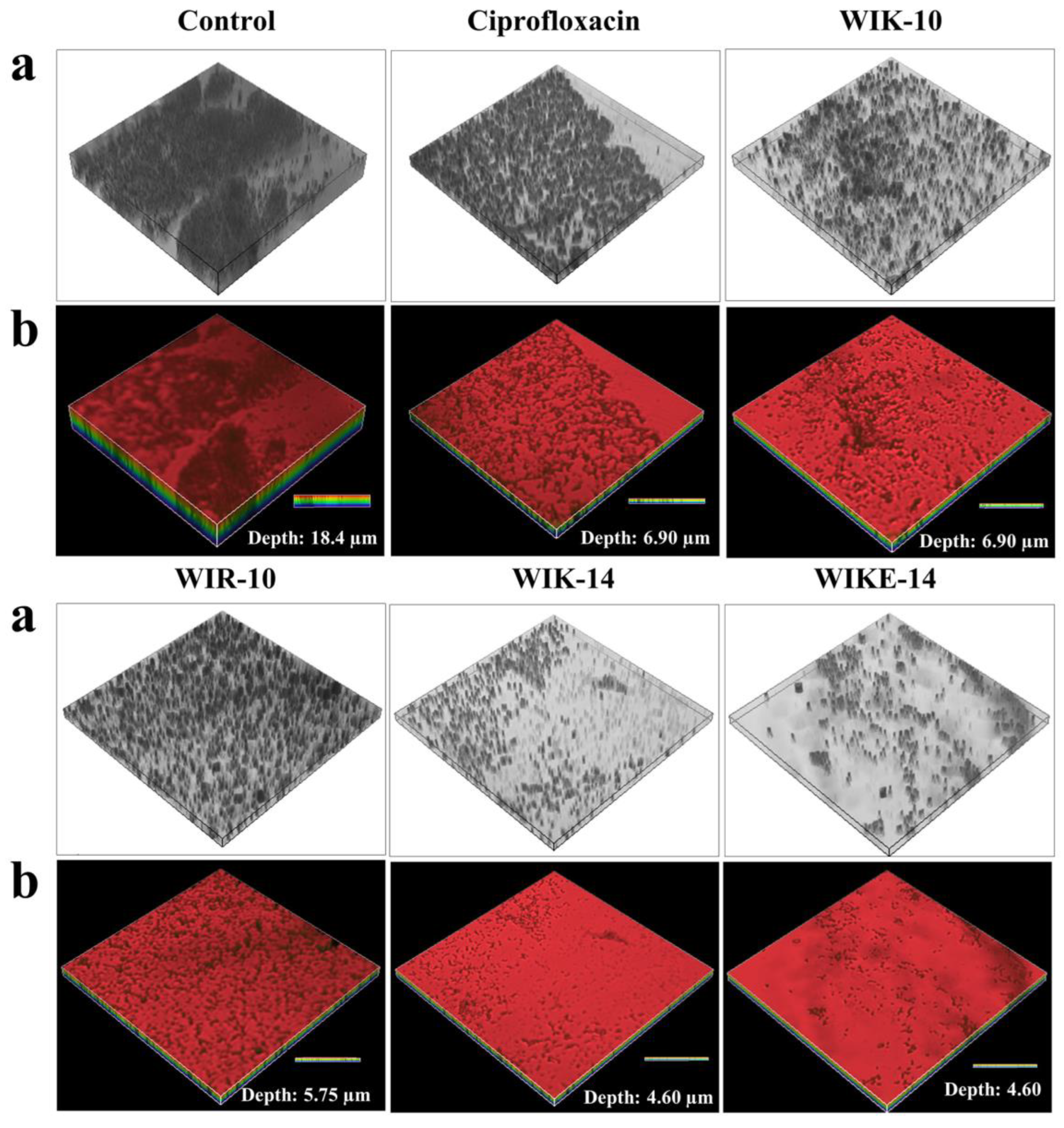
2.6. Ex Vivo Biofilm Eliminative Effects of Peptides on a Plastic Coverslip
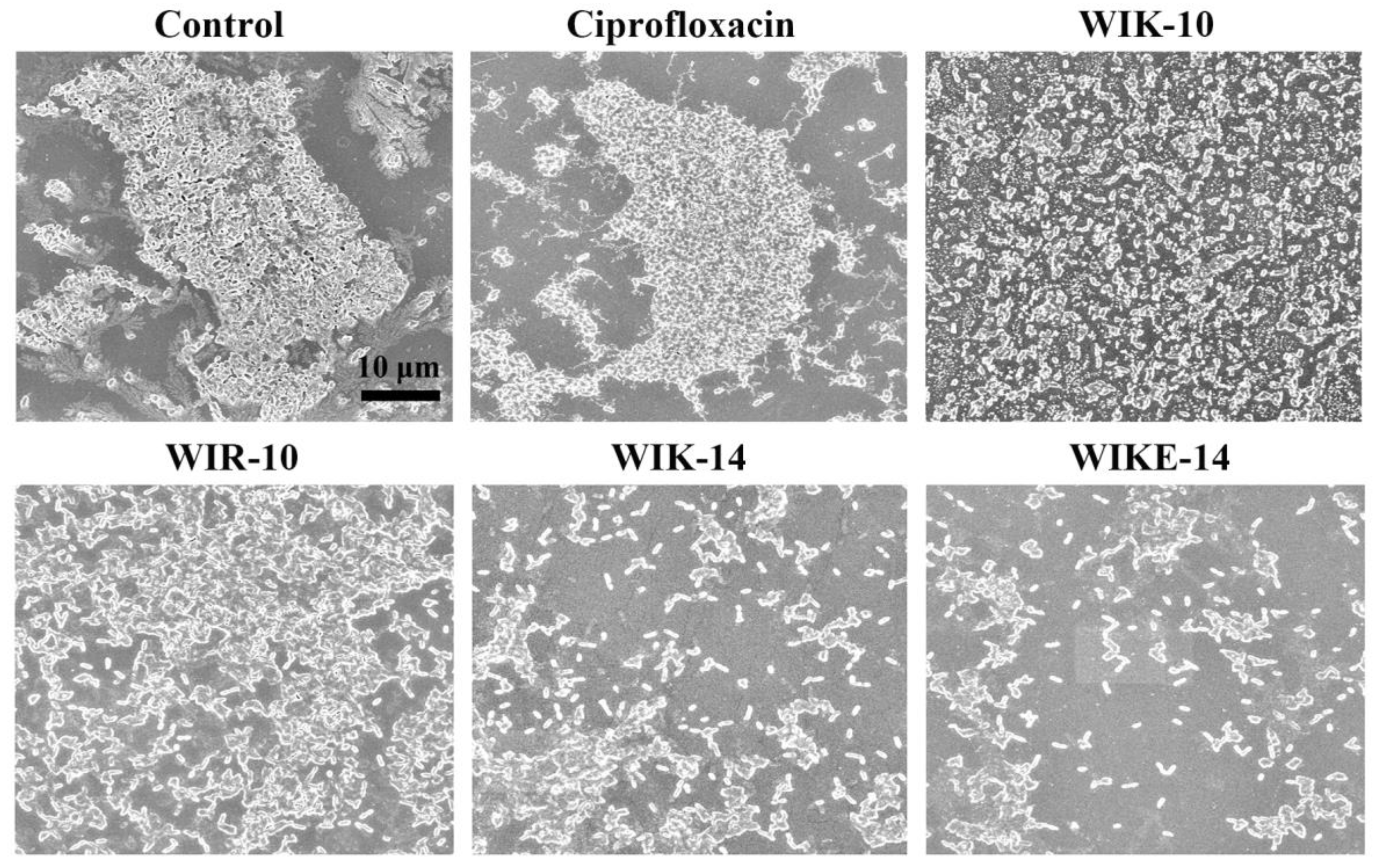
2.7. Morphological Analysis under Scanning Electron Microscopy (SEM)
3. Materials and Methods
3.1. Materials
3.2. Peptide Synthesis
3.3. Bacterial Strains
3.4. Antibacterial Assay
3.5. Growth Inhibition in Planktonic Bacteria
3.6. Inhibition Assay in Biofilm Formation
3.7. Reduction Assay on Preformed Biofilm
3.8. Reduction of EPS Components
3.9. SEM Analysis
3.10. CLSM Analysis
3.11. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Y.; Bian, Z.; Wang, Y. Biofilm formation and inhibition mediated by bacterial quorum sensing. Appl. Microbiol. Biotechnol. 2022, 106, 6365–6381. [Google Scholar] [CrossRef] [PubMed]
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Hall-Stoodley, L.; Burmølle, M.; Stewart, P.S.; Bjarnsholt, T. The biofilm life cycle: Expanding the conceptual model of biofilm formation. Nat. Rev. Microbiol. 2022, 20, 608–620. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Wuertz, S. Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 2019, 17, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Gedefie, A.; Demsis, W.; Ashagrie, M.; Kassa, Y.; Tesfaye, M.; Tilahun, M.; Bisetegn, H.; Sahle, Z. Acinetobacter baumannii Biofilm Formation and Its Role in Disease Pathogenesis: A Review. Infect. Drug Resist. 2021, 14, 3711. [Google Scholar] [CrossRef]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An Emergent Form of Bacterial Life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Park, S.C.; Park, Y.; Hahm, K.S. The role of antimicrobial peptides in preventing multidrug-resistant bacterial infections and biofilm formation. Int. J. Mol. Sci. 2011, 12, 5971–5992. [Google Scholar] [CrossRef]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef]
- Percival, S.L.; Suleman, L.; Vuotto, C.; Donelli, G. Healthcare-associated infections, medical devices and biofilms: Risk, tolerance and control. J. Med. Microbiol. 2015, 64, 323–334. [Google Scholar] [CrossRef]
- Paranjpye, R.N.; Strom, M.S. A vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence. Infect. Immun. 2005, 73, 1411–1422. [Google Scholar] [CrossRef]
- Kanwar, I.; Sah, A.K.; Suresh, P.K. Biofilm-mediated Antibiotic-resistant Oral Bacterial Infections: Mechanism and Combat Strategies. Curr. Pharm. Des. 2017, 23, 2084–2095. [Google Scholar] [CrossRef] [PubMed]
- Wasfi, R.; Hamed, S.M.; Amer, M.A.; Fahmy, L.I. Proteus mirabilis biofilm: Development and therapeutic strategies. Front. Cell Infect. Microbiol. 2020, 10, 414. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef] [PubMed]
- Graf, A.C.; Leonard, A.; Schäuble, M.; Rieckmann, L.M.; Hoyer, J.; Maass, S.; Lalk, M.; Becher, D.; Pané-Farré, J.; Riedel, K. Virulence factors produced by staphylococcus aureus biofilms have a moonlighting function contributing to biofilm integrity. Mol. Cell Proteom. 2019, 18, 1036–1053. [Google Scholar] [CrossRef] [PubMed]
- Davies, D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2003, 2, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Yousif, A.; Jamal, M.A.; Raad, I. Biofilm-based central line-associated bloodstream infections. Adv. Exp. Med. Biol. 2015, 830, 157–179. [Google Scholar]
- Zhang, O.L.; Niu, J.Y.; Yin, I.X.; Yu, O.Y.; Mei, M.L.; Chu, C.H. Growing Global Research Interest in Antimicrobial Peptides for Caries Management: A Bibliometric Analysis. J. Funct. Biomater. 2022, 13, 210. [Google Scholar] [CrossRef]
- Dini, I.; De Biasi, M.G.; Mancusi, A. An Overview of the Potentialities of Antimicrobial Peptides Derived from Natural Sources. Antibiotics 2022, 11, 1483. [Google Scholar] [CrossRef]
- Talapko, J.; Meštrović, T.; Juzbašić, M.; Tomas, M.; Erić, S.; Horvat Aleksijević, L.; Bekić, S.; Schwarz, D.; Matić, S.; Neuberg, M.; et al. Antimicrobial Peptides-Mechanisms of Action, Antimicrobial Effects and Clinical Applications. Antibiotics 2022, 11, 1417. [Google Scholar] [CrossRef]
- Zhang, R.; Xu, L.; Dong, C. Antimicrobial Peptides: An Overview of their Structure, Function and Mechanism of Action. Protein Pept. Lett. 2022, 29, 641–650. [Google Scholar] [CrossRef]
- Wang, L.; Qu, L.; Lin, S.; Yang, Q.; Zhang, X.; Jin, L.; Dong, H.; Sun, D. Biological Functions and Applications of Antimicrobial Peptides. Curr. Protein Pept. Sci. 2022, 23, 226–247. [Google Scholar] [PubMed]
- Li, X.; Zuo, S.; Wang, B.; Zhang, K.; Wang, Y. Antimicrobial Mechanisms and Clinical Application Prospects of Antimicrobial Peptides. Molecules 2022, 27, 2675. [Google Scholar] [CrossRef]
- Pontes, J.T.C.; Toledo Borges, A.B.; Roque-Borda, C.A.; Pavan, F.R. Antimicrobial Peptides as an Alternative for the Eradication of Bacterial Biofilms of Multi-Drug Resistant Bacteria. Pharmaceutics 2022, 14, 642. [Google Scholar] [CrossRef]
- Park, S.C.; Lee, M.Y.; Kim, J.Y.; Kim, H.; Jung, M.; Shin, M.K.; Lee, W.K.; Cheong, G.W.; Lee, J.R.; Jang, M.K. Anti-Biofilm Effects of Synthetic Antimicrobial Peptides Against Drug-Resistant Pseudomonas aeruginosa and Staphylococcus aureus Planktonic Cells and Biofilm. Molecules 2019, 24, 4560. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Park, S.; Kim, Y.M.; Guk, T.; Lee, M.Y.; Park, S.C.; Lee, J.R.; Jang, M.K. Candidacidal and Antibiofilm Activity of PS1-3 Peptide against Drug-Resistant Candida albicans on Contact Lenses. Pharmaceutics 2022, 14, 1602. [Google Scholar] [CrossRef] [PubMed]
- Park, S.C.; Son, H.; Kim, Y.M.; Lee, J.K.; Park, S.; Lim, H.S.; Lee, J.R.; Jang, M.K. Design of Antimicrobial Peptides with Cell-Selective Activity and Membrane-Acting Mechanism against Drug-Resistant Bacteria. Antibiotics 2022, 11, 1619. [Google Scholar] [CrossRef] [PubMed]
- Khandelia, H.; Kaznessis, Y.N. Cation-pi interactions stabilize the structure of the antimicrobial peptide indolicidin near membranes: Molecular dynamics simulations. J. Phys. Chem. B 2007, 111, 242–250. [Google Scholar] [CrossRef]
- Bouffartigues, E.; Duchesne, R.; Bazire, A.; Simon, M.; Maillot, O.; Dufour, A.; Feuilloley, M.; Orange, N.; Chevalier, S. Sucrose favors Pseudomonas aeruginosa pellicle production through the extracytoplasmic function sigma factor SigX. FEMS Microbiol. Lett. 2014, 356, 193–200. [Google Scholar] [CrossRef]
- Cai, J.N.; Jung, J.E.; Lee, M.H.; Choi, H.M.; Jeon, J.G. Sucrose challenges to Streptococcus mutans biofilms and the curve fitting for the biofilm changes. FEMS Microbiol. Ecol. 2018, 94, fiy091. [Google Scholar] [CrossRef]
- Powell, L.C.; Pritchard, M.F.; Ferguson, E.L.; Powell, K.A.; Patel, S.U.; Rye, P.D.; Sakellakou, S.M.; Buurma, N.J.; Brilliant, C.D.; Copping, J.M.; et al. Targeted disruption of the extracellular polymeric network of Pseudomonas aeruginosa biofilms by alginate oligosaccharides. NPJ Biofilms Microbiomes 2018, 4, 13. [Google Scholar] [CrossRef]
- Gaglione, R.; Cesaro, A.; Dell’Olmo, E.; Di Girolamo, R.; Tartaglione, L.; Pizzo, E.; Arciello, A. Cryptides Identified in Human Apolipoprotein B as New Weapons to Fight Antibiotic Resistance in Cystic Fibrosis Disease. Int. J. Mol. Sci. 2020, 21, 2049. [Google Scholar] [CrossRef] [PubMed]
| Name | MIC (µM) | |||||
|---|---|---|---|---|---|---|
| Drug-Sensitive P. aeruginosa (ATCC 27853) | Drug-Resistant P. aeruginosa (DRPa) | |||||
| 3904 | 4830 | 4891 | 4007 | 5018 | ||
| KIW-10 | 16 | 16 | 16 | 8 | 16 | 16 |
| KWI-10 | 16 | 16 | 8 | 8 | 8 | 8 |
| WIK-10 | 8 | 4 | 8 | 8 | 8 | 8 |
| RIW-10 | 16 | 16 | 16 | 8 | 8 | 8 |
| RWI-10 | 16 | 16 | 4 | 4 | 8 | 8 |
| WIR-10 | 4 | 4 | 4 | 4 | 2 | 4 |
| KIW-14 | 2 | 2 | 2 | 2 | 2 | 2 |
| KWI-14 | 2 | 2 | 2 | 1 | 1 | 2 |
| WIK-14 | 2 | 2 | 2 | 1 | 1 | 1 |
| WIKE-14 | 8 | 16 | 8 | 8 | 8 | 16 |
| Melittin | 2 | 2 | 4 | 4 | 4 | 4 |
| Ciprofloxacin | 1 | >256 | 64 | >256 | >256 | >256 |
| Ceftazidime | 1 | >256 | 128 | >256 | >256 | >256 |
| Tobramycin | 0.25 | >256 | >256 | >256 | >256 | >256 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.-M.; Son, H.; Park, S.-C.; Lee, J.-K.; Jang, M.-K.; Lee, J.R. Anti-Biofilm Effects of Rationally Designed Peptides against Planktonic Cells and Pre-Formed Biofilm of Pseudomonas aeruginosa. Antibiotics 2023, 12, 349. https://doi.org/10.3390/antibiotics12020349
Kim Y-M, Son H, Park S-C, Lee J-K, Jang M-K, Lee JR. Anti-Biofilm Effects of Rationally Designed Peptides against Planktonic Cells and Pre-Formed Biofilm of Pseudomonas aeruginosa. Antibiotics. 2023; 12(2):349. https://doi.org/10.3390/antibiotics12020349
Chicago/Turabian StyleKim, Young-Min, Hyosuk Son, Seong-Cheol Park, Jong-Kook Lee, Mi-Kyeong Jang, and Jung Ro Lee. 2023. "Anti-Biofilm Effects of Rationally Designed Peptides against Planktonic Cells and Pre-Formed Biofilm of Pseudomonas aeruginosa" Antibiotics 12, no. 2: 349. https://doi.org/10.3390/antibiotics12020349
APA StyleKim, Y.-M., Son, H., Park, S.-C., Lee, J.-K., Jang, M.-K., & Lee, J. R. (2023). Anti-Biofilm Effects of Rationally Designed Peptides against Planktonic Cells and Pre-Formed Biofilm of Pseudomonas aeruginosa. Antibiotics, 12(2), 349. https://doi.org/10.3390/antibiotics12020349






