Therapeutic Potential of an Azithromycin-Colistin Combination against XDR K. pneumoniae in a 3D Collagen-Based In Vitro Wound Model of a Biofilm Infection
Abstract
1. Introduction
2. Results
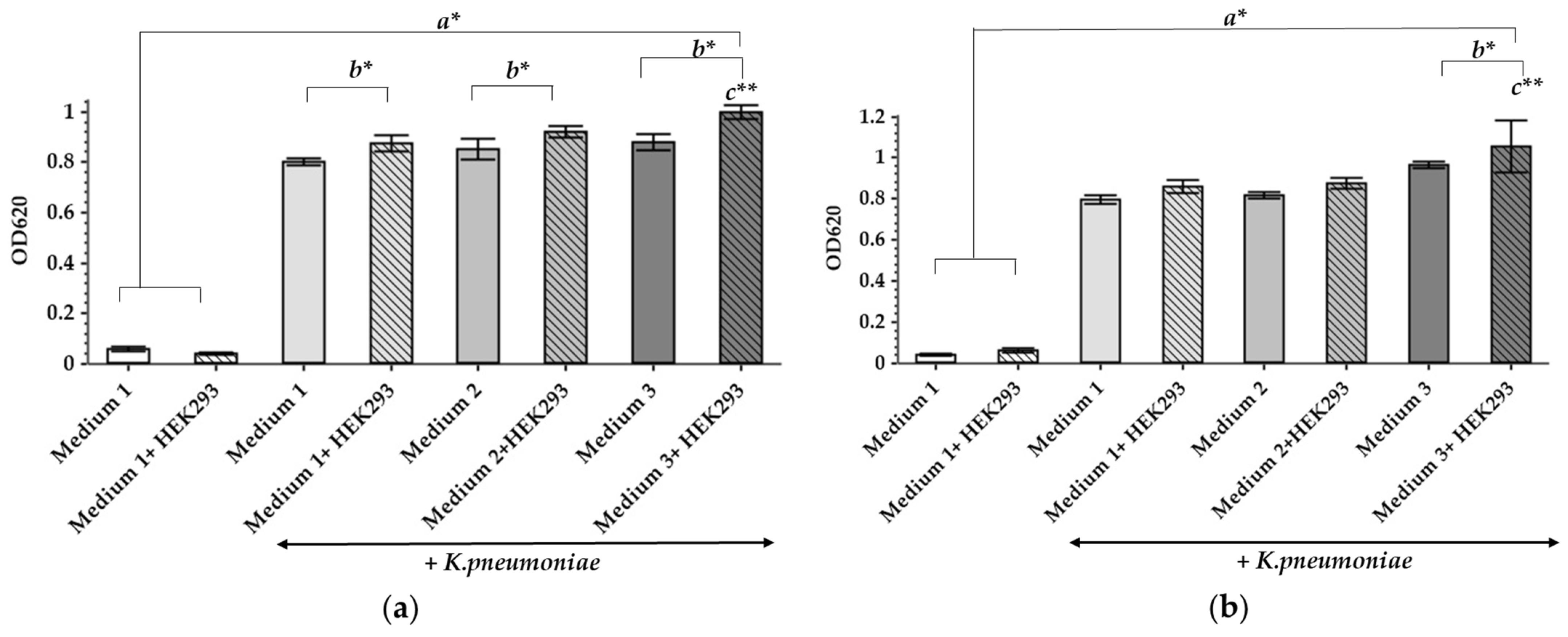
2.1. The Influence of Different Culture Media and the Presence of HEK293 on the Growth of K. pneumoniae UHI 1090
2.2. AZM and CMS Effects on the Viability of HEK293 Cells In Vitro
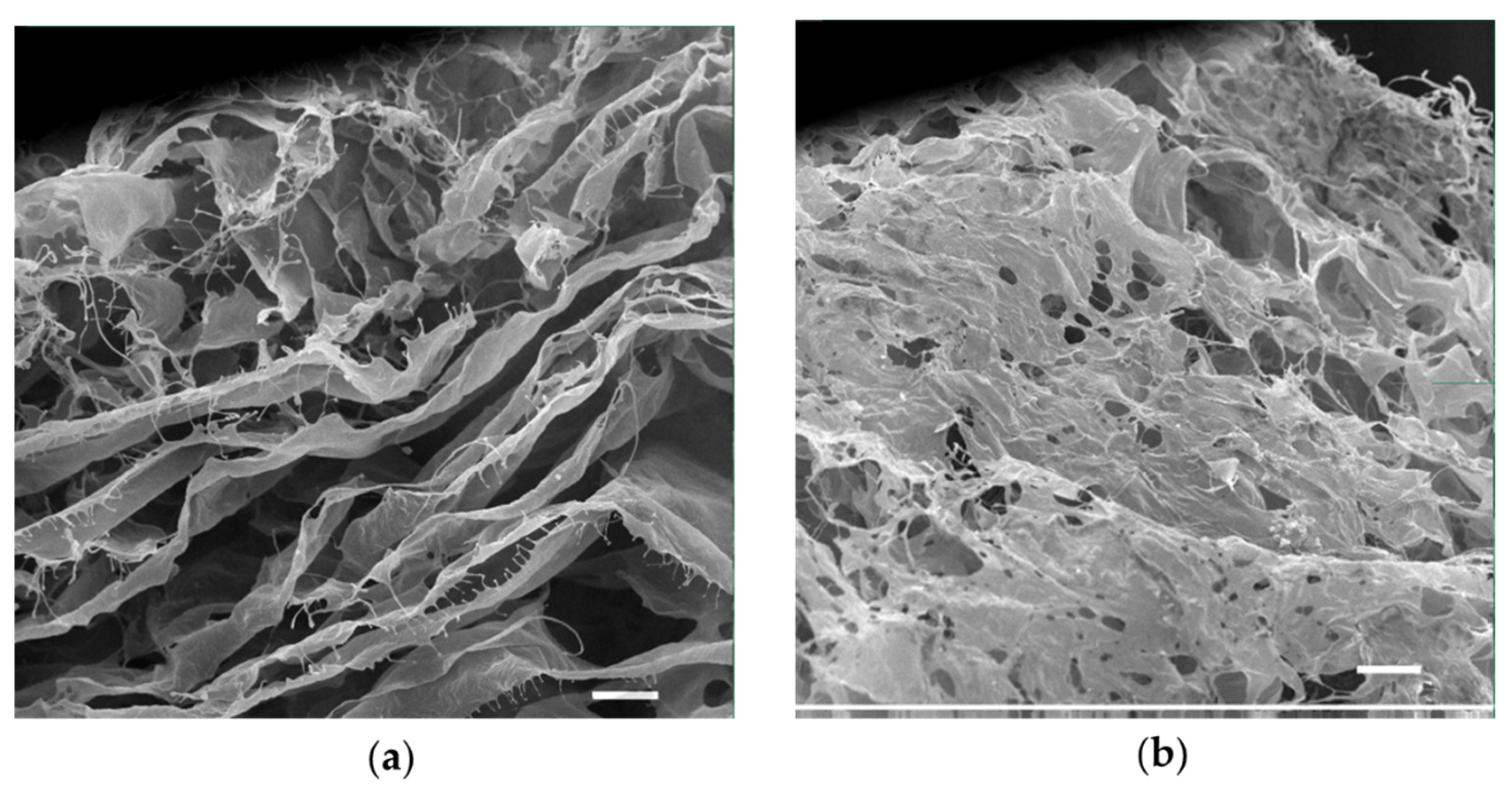
2.3. Development and Characterization of a Porous Collagen Scaffold for a 3D Collagen-Based Wound Model In Vitro
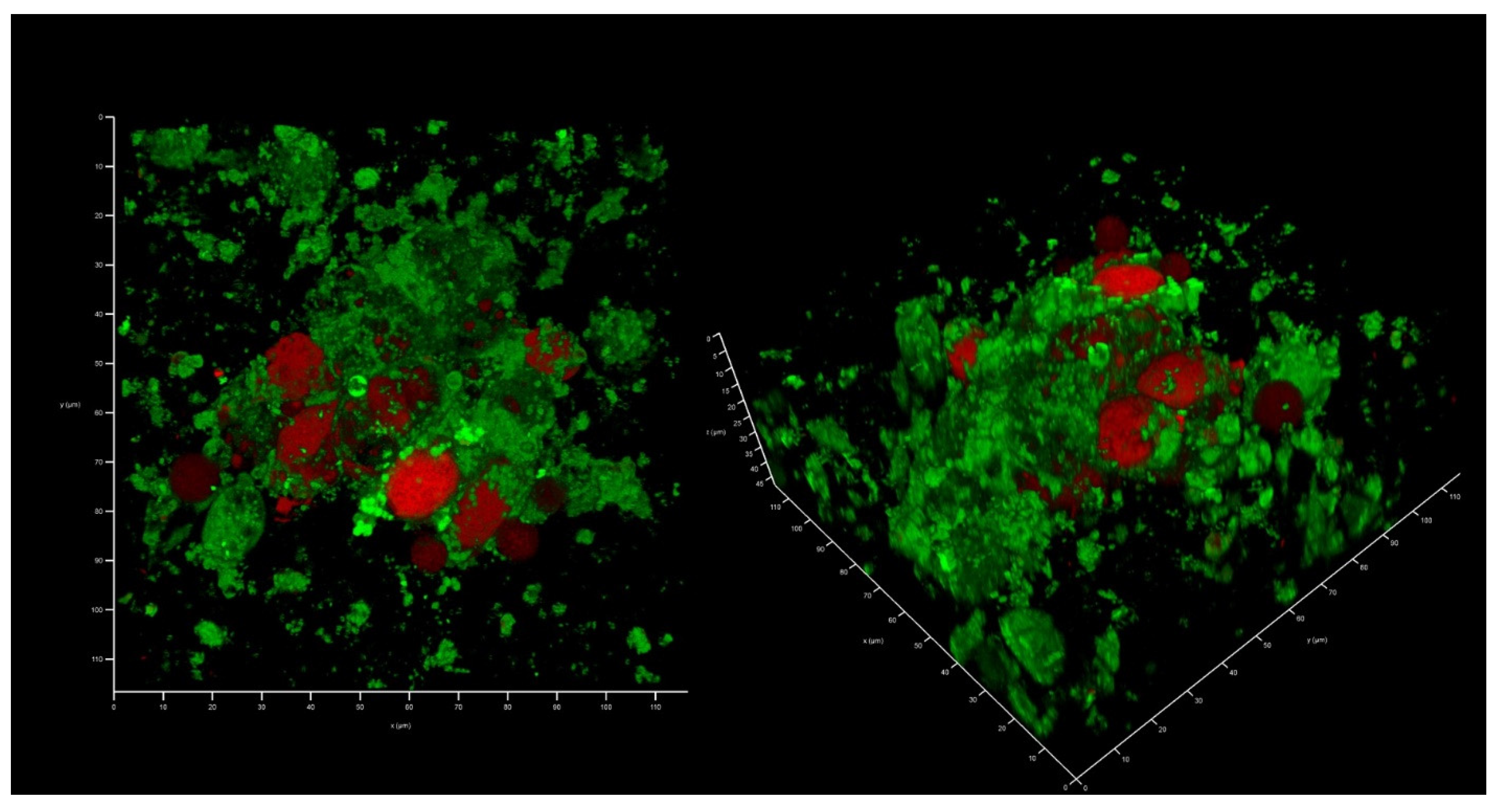
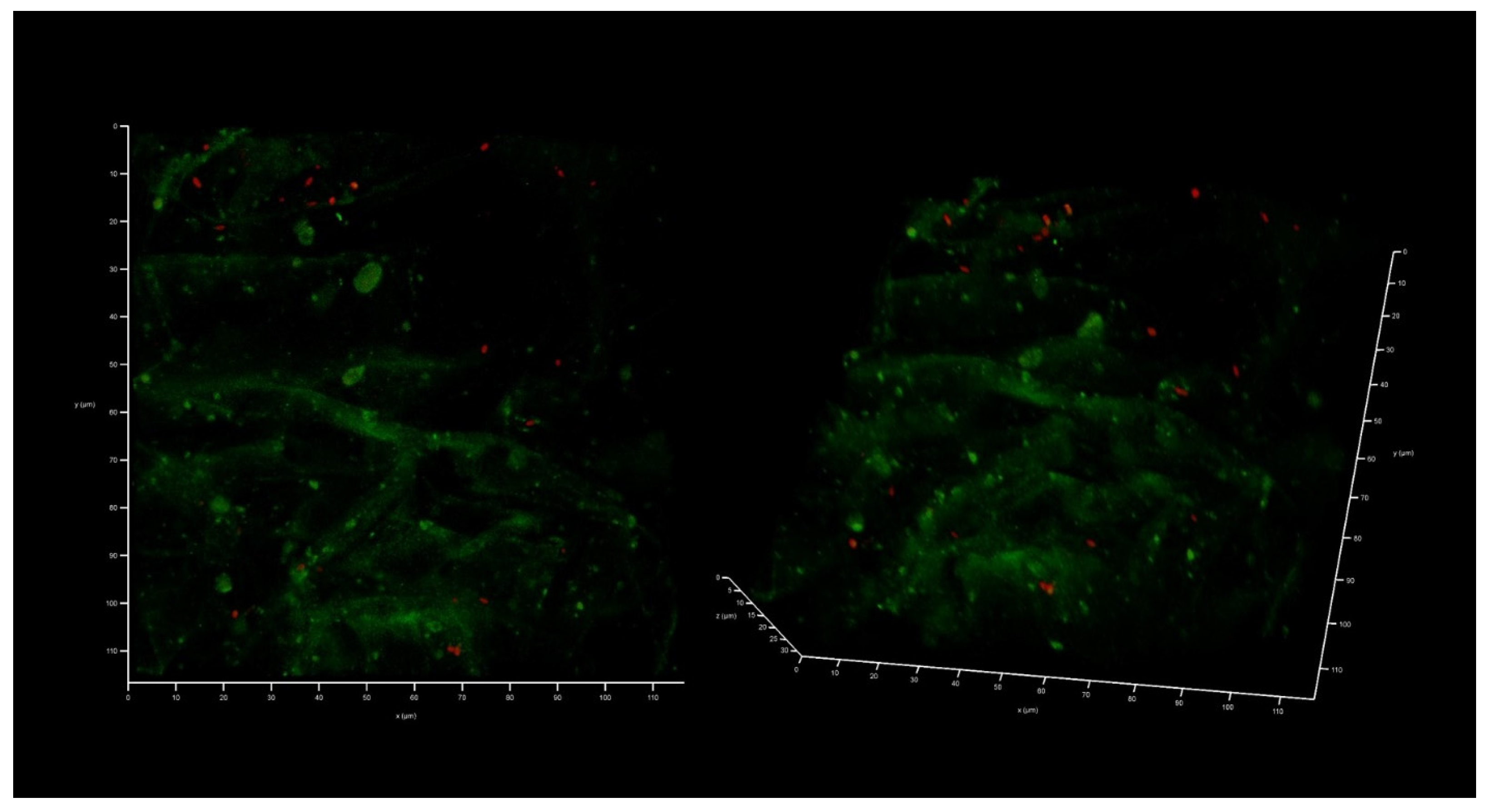
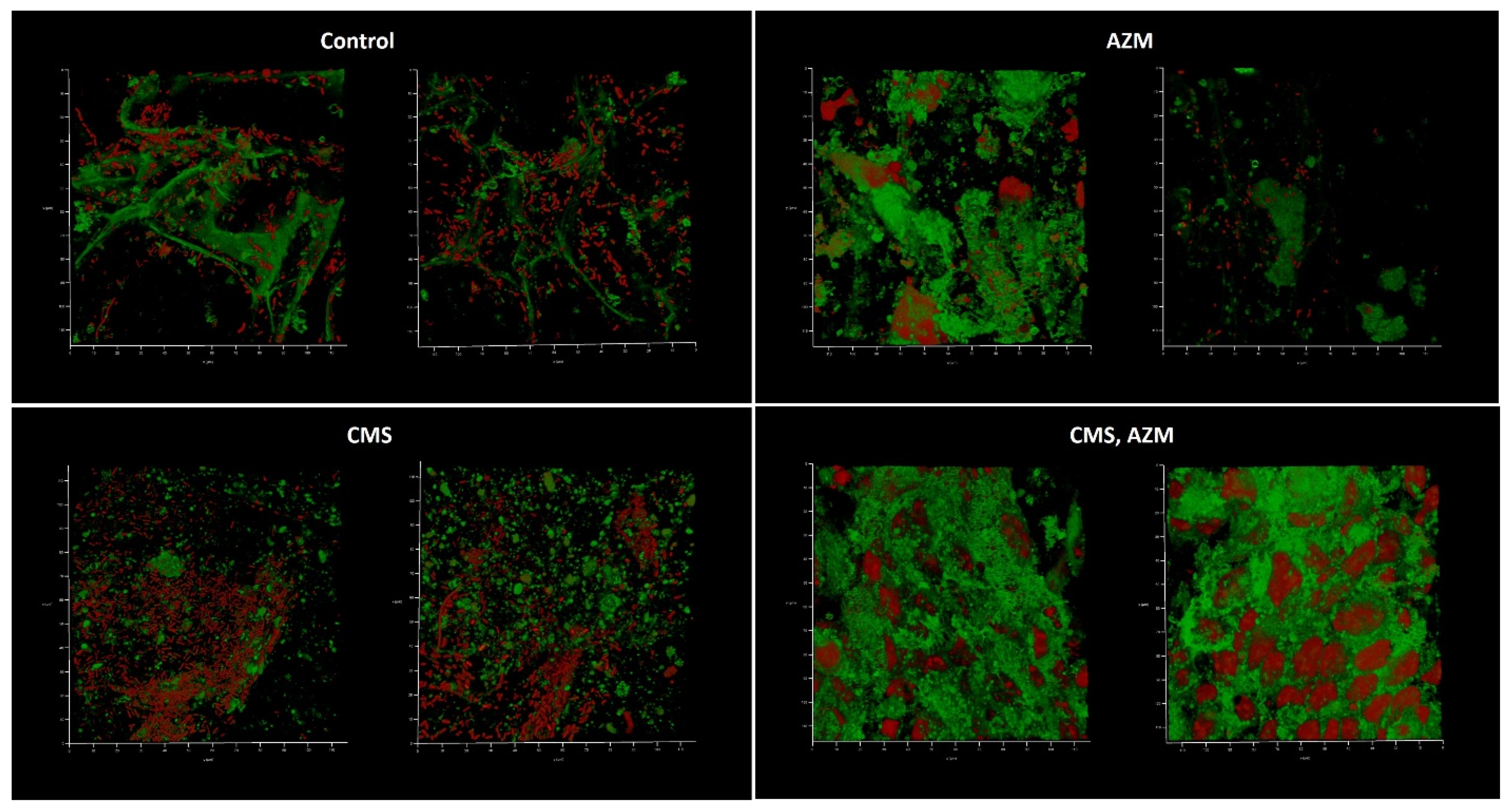
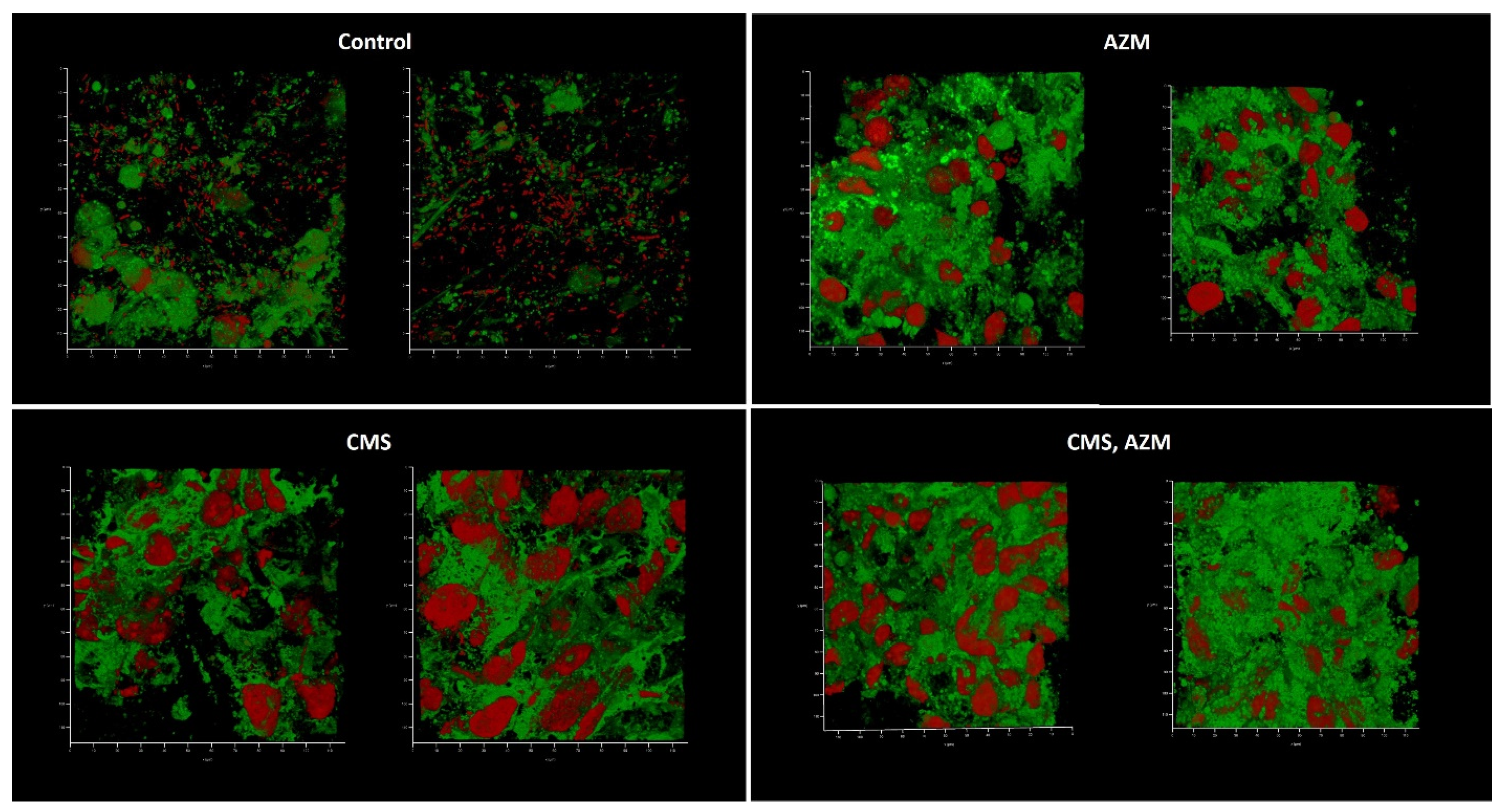
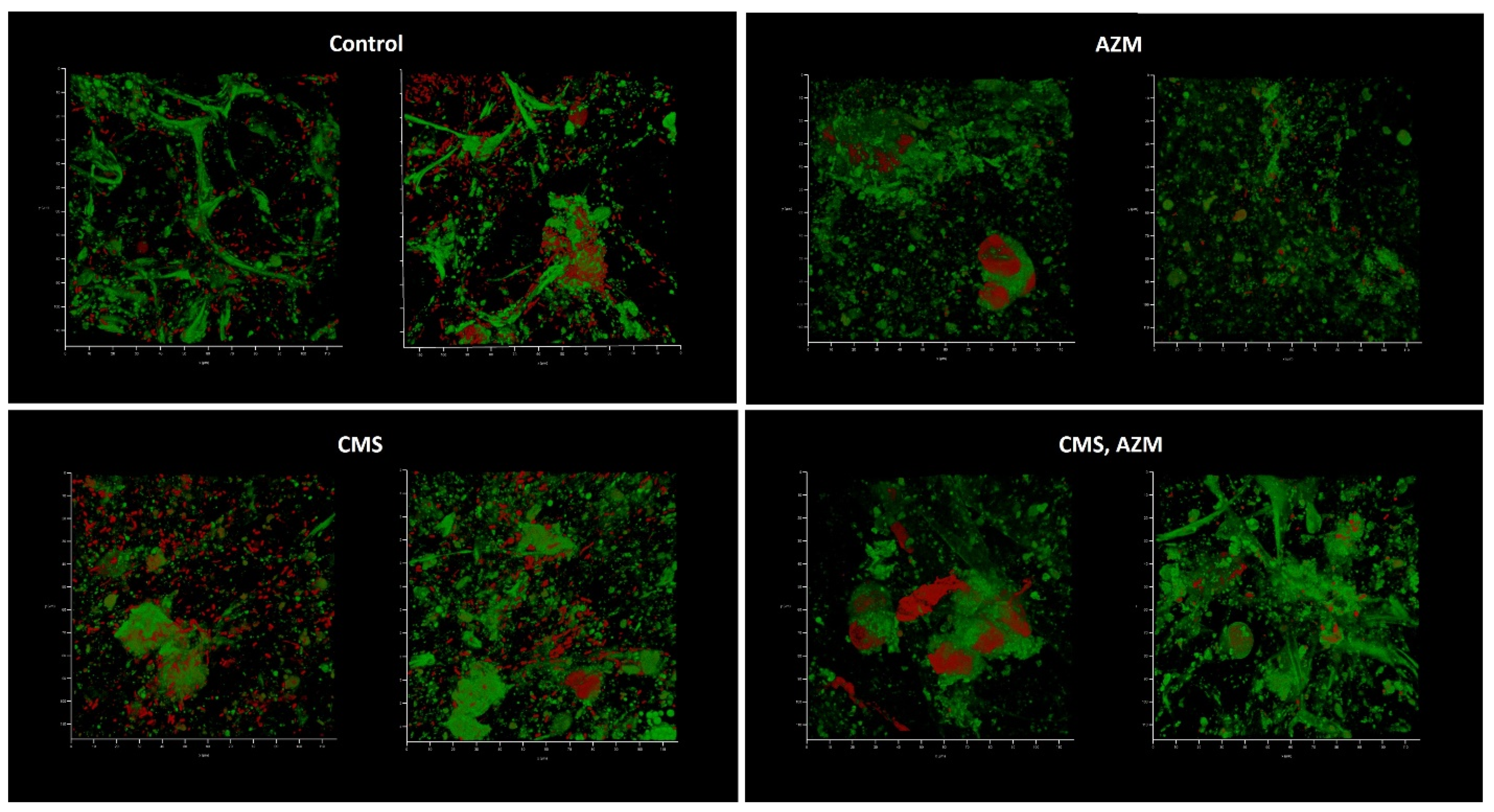
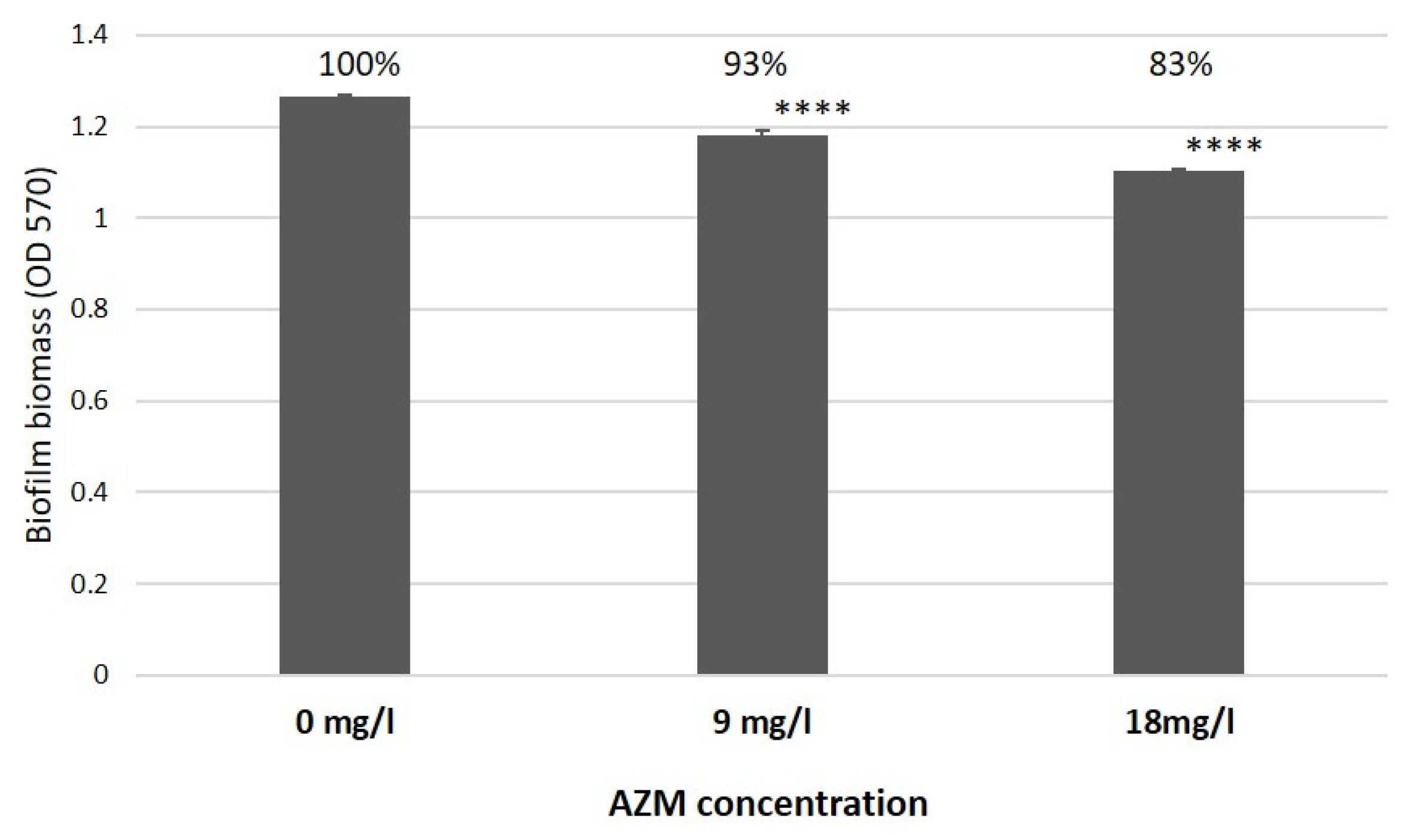
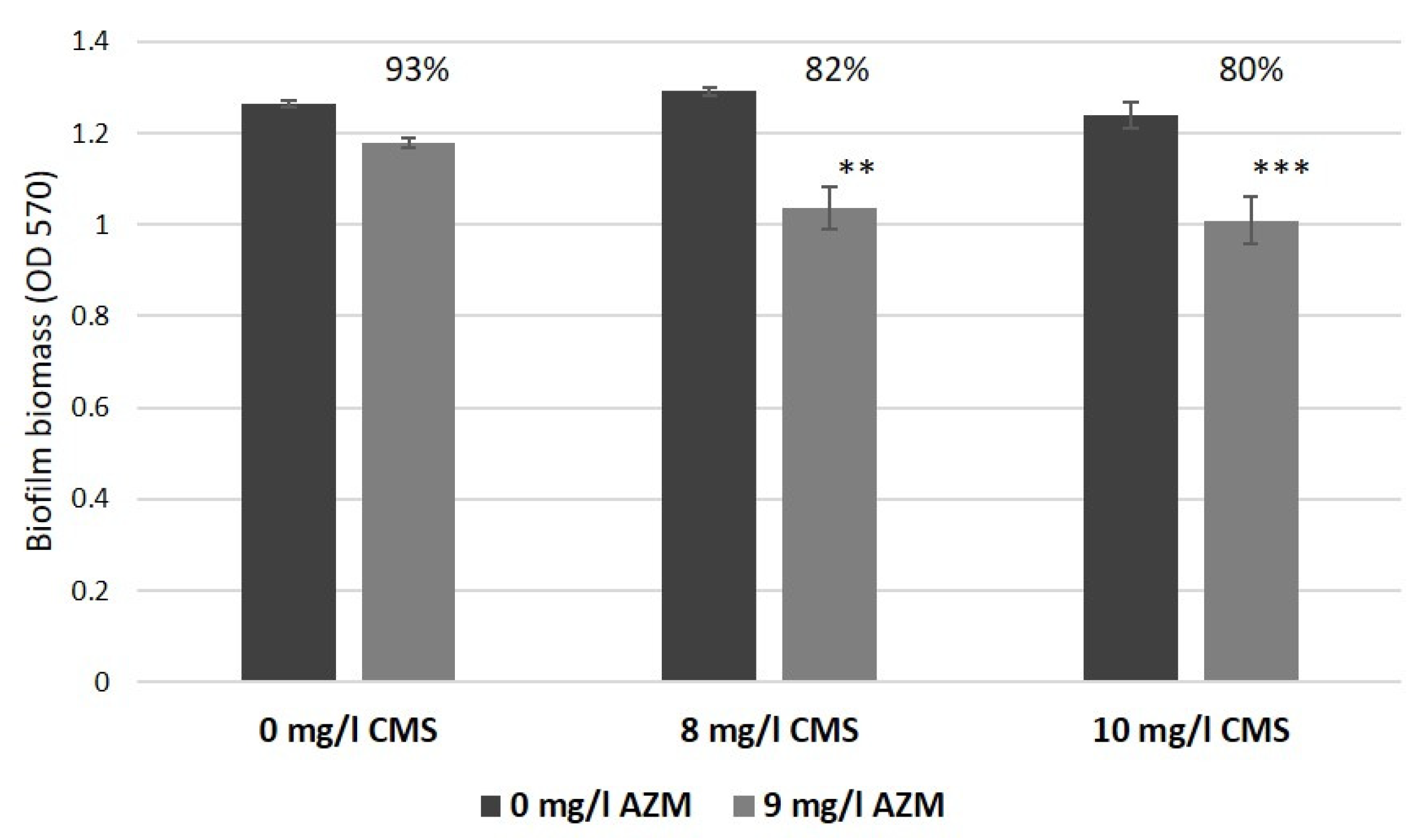
2.4. AZM and CMS Effects on Biofilm Development by K. pneumoniae UHI 1090 in a 3D Collagen-Based In Vitro Model
3. Discussion
4. Materials and Methods
4.1. Bacterial Strain and Culture Conditions
4.2. Mammalian Cell Lines
4.3. Viability of Eukaryotic Cells after Antibiotic Treatment
4.4. Effects of Different Culture Media and HEK293 Cells on the Growth of K. pneumoniae UHI 1090
4.5. Cultivation of Bacteria and Antibiotic Treatments
4.6. Collagen Scaffold Preparation
4.7. Preparation of the Wound Model
4.8. Confocal Laser Scanning Microscopy (CLSM)
4.9. Scanning Electron Microscopy (SEM)
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karakonstantis, S.; Kritsotakis, E.I.; Gikas, A. Pandrug-resistant Gram-negative bacteria: A systematic review of current epidemiology, prognosis and treatment options. J. Antimicrob. Chemother. 2019, 75, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Higgins, P.G.; Hagen, R.M.; Podbielski, A.; Frickmann, H.; Warnke, P. Molecular Epidemiology of Carbapenem-Resistant Acinetobacter baumannii Isolated from War-Injured Patients from the Eastern Ukraine. Antibiotics 2020, 9, 579. [Google Scholar] [CrossRef]
- Voidazan, S.; Albu, S.; Toth, R.; Grigorescu, B.; Rachita, A.; Moldovan, I. Healthcare Associated Infections-A New Pathology in Medical Practice? Int. J. Environ. Res. Public Health 2020, 17, 760. [Google Scholar] [CrossRef] [PubMed]
- Michalopoulos, A.S.; Falagas, M.E. Colistin: Resent data on pharmacodynamics properties and clinical efficacy in critically ill patients. Ann. Intensive Care 2011, 1, 30. [Google Scholar] [CrossRef] [PubMed]
- Michalopoulos, A.S.; Karatza, D.C.; Gregorakos, L. Pharmacokinetic evaluation of colistin sodium. Expert Opin. Drug Metab. Toxicol. 2011, 7, 245–255. [Google Scholar] [CrossRef]
- Li, J.; Turnidge, J.; Milne, R.; Nation, R.L.; Coulthard, K. In vitro pharmacodynamics properties of colistin and colistin methanesulfonate against Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob. Agents Chemother. 2001, 45, 781–785. [Google Scholar] [CrossRef]
- Paul, M.; Daikos, G.L.; Durante-Mangoni, E.; Yahav, D.; Carmeli, Y.; Benattar, Y.D.; Skiada, A.; Andini, R.; Eliakim-Raz, N.; Nutman, A.; et al. Colistin alone versus colistin plus meropenem for treatment of severe infections caused by carbapenem-resistant Gram-negative bacteria: An open-label, randomized controlled trial. Lancet Infect Dis. 2018, 18, 391–400. [Google Scholar] [CrossRef]
- Imberti, R.; Cusato, M.; Villani, P.; Carnavale, L.; Iotti, G.A.; Langer, M.; Regazzi, M. Steady-state pharmacokinetics and bronchoalveolar lavage concentration of colistin in critically ill patients after intravenous colistin methanesulphonate administration. Chest 2010, 138, 1333–1339. [Google Scholar] [CrossRef]
- Karakonstantis, S. Macrolides: An underappreciated option for treatment of Pseudomonas aeruginosa infections (and potentially other gram-negative pathogens)? Clin. Microbiol. Infect. 2022, 28, 1665–1666. [Google Scholar] [CrossRef]
- Moshynets, O.V.; Baranovskyi, T.P.; Cameron, S.; Iungin, O.S.; Pokholenko, I.; Jerdan, R.; Kamyshnyi, A.; Krikunov, A.A.; Potochilova, V.V.; Rudnieva, K.L.; et al. Azithromycin possesses biofilm-inhibitory activity and potentiates non-bactericidal colistin methanesulfonate (CMS) and polymyxin B against Klebsiella pneumonia. PLoS ONE 2022, 2022 17, e0270983. [Google Scholar] [CrossRef]
- Moshynets, O.V.; Spiers, A.J. Viewing biofilms within the larger context of bacterial aggregations. In Microbial Biofilms-Importance and Applications; Dhanasekaran, D., Thajuddin, N., Eds.; InTech Press: London, UK, 2016; pp. 3–22. [Google Scholar] [CrossRef]
- Vyas, H.K.N.; Xia, B.; Mai-Prochnow, A. Clinically relevant in vitro biofilm models: A need to mimic and recapitulate the host environment. Biofilm 2022, 4, 100069. [Google Scholar] [CrossRef] [PubMed]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef] [PubMed]
- Stein, H.; Wilensky, M.; Tsafrir, Y.; Rosenthal, M.; Amir, R.; Avraham, T.; Ofir, K.; Dgany, O.; Yayon, A.; Shoseyov, O. Production of Bioactive, Post-Translationally Modified, Heterotrimeric, Human Recombinant Type-I Collagen in Transgenic Tobacco. Biomacromolecules 2009, 10, 2640–2645. [Google Scholar] [CrossRef] [PubMed]
- Ramshaw, J.A.M.; Werkmeister, J.A.; Glattauer, V. Recent progress with recombinant collagens produced in Escherichia coli. Curr. Opin. Biomed. Eng. 2019, 10, 149–155. [Google Scholar] [CrossRef]
- Matinong, A.M.E.; Chisti, Y.; Pickering, K.L.; Haverkamp, R.G. Collagen Extraction from Animal Skin. Biology 2022, 11, 905. [Google Scholar] [CrossRef]
- Maistrenko, L.; Iungin, O.; Pikus, P.; Pokholenko, I.; Gorbatiuk, O.; Moshynets, O.; Okhmat, O.; Kolesnyk, T.; Potters, G.; Mokrousova, O. Collagen Obtained from Leather Production Waste Provides Suitable Gels for Biomedical Applications. Polymers 2022, 14, 4749. [Google Scholar] [CrossRef]
- Slade, E.A.; Thorn, R.M.S.; Young, A.; Reynolds, D.M. An in vitro collagen perfusion wound biofilm model; with applications for antimicrobial studies and microbial metabolomics. BMC Microbiol. 2019, 19, 310. [Google Scholar] [CrossRef]
- Buyck, J.M.; Plésiat, P.; Traore, H.; Vanderbist, F.; Tulkens, P.M.; Van Bambeke, F. Increased susceptibility of Pseudomonas aeruginosa to macrolides and ketolides in eukaryotic cell culture media and biological fluids due to decreased expression of oprM and increased outer-membrane permeability. Clin. Infect. Dis. 2012, 55, 534–542. [Google Scholar] [CrossRef]
- Lin, L.; Nonejuie, P.; Munguia, J.; Hollands, A.; Olson, J.; Dam, Q.; Kumaraswamy, M.; Rivera, H., Jr.; Corriden, R.; Rohde, M.; et al. Azithromycin Synergizes with Cationic Antimicrobial Peptides to Exert Bactericidal and Therapeutic Activity Against Highly Multidrug-Resistant Gram-Negative Bacterial Pathogens. eBioMedicine 2015, 2, 690–698. [Google Scholar] [CrossRef]
- Dillon, N.; Holland, M.; Tsunemoto, H.; Hancock, B.; Cornax, I.; Pogliano, J.; Sakoulas, G.; Nizet, V. Surprising synergy of dual translation inhibition vs. Acinetobacter baumannii and other multidrug-resistant bacterial pathogens. eBioMedicine 2019, 46, 193–201. [Google Scholar] [CrossRef]
- Kadam, S.; Nadkarni, S.; Lele, J.; Sakhalkar, S.; Mokashi, P.; Kaushik, K.S. Bioengineered Platforms for Chronic Wound Infection Studies: How Can We Make Them More Human-Relevant? Front. Bioeng. Biotechnol. 2019, 7, 418. [Google Scholar] [CrossRef]
- Cano, V.; Moranta, D.; Llobet-Brossa, E.; Bengoechea, J.A.; Garmendia, J. Klebsiella pneumoniae triggers a cytotoxic effect on airway epithelial cells. BMC Microbiol. 2009, 9, 156. [Google Scholar] [CrossRef]
- Kalghatgi, S.; Spina, C.S.; Costello, J.C.; Liesa, M.; Morones-Ramirez, J.R.; Slomovic, S.; Molina, A.; Shirihai, O.S.; Collins, J.J. Bactericidal antibiotics induce mitochondrial dysfunction and oxidative damage in mammalian cells. Sci. Transl. Med. 2013, 5, 192ra85. [Google Scholar] [CrossRef]
- Zavascki, A.P.; Nation, R.L. Nephrotoxicity of Polymyxins: Is There Any Difference between Colistimethate and Polymyxin B? Antimicrob. Agents Chemother. 2017, 61, e02319-16. [Google Scholar] [CrossRef]
- Nation, R.L.; Rigatto, M.H.P.; Falci, D.R.; Zavascki, A.P. Polymyxin Acute Kidney Injury: Dosing and Other Strategies to Reduce Toxicity. Antibiotics 2019, 8, 24. [Google Scholar] [CrossRef]
- Karvanen, M.; Plachouras, D.; Friberg, L.E.; Paramythiotou, E.; Papadomichelakis, E.; Karaiskos, I.; Tsangaris, I.; Armaganidis, A.; Cars, O.; Giamarellou, H. Colistin methanesulfonate and colistin pharmacokinetics in critically ill patients receiving continuous venovenous hemodiafiltration. Antimicrob. Agents Chemother. 2013, 57, 668–671. [Google Scholar] [CrossRef]
- Instruction for Colomycin. Available online: https://compendium.com.ua/dec/266344/378710/ (accessed on 27 December 2022).
- Foulds, G.; Shepard, R.M.; Johnson, R.B. The pharmacokinetics of azithromycin in human serum and tissues. J. Antimicrob. Chemother. 1990, 25, 73–82. [Google Scholar] [CrossRef]
- Instruction for Azithromycin. Available online: https://compendium.com.ua/dec/337677/ (accessed on 27 December 2022).
- Moshynets, O.; Chernii, S.; Chernii, V.; Losytskyy, M.Y.; Karakhim, S.; Czerwieniec, R.; Pekhnyo, V.; Yarmoluk, S.; Kovalska, V. Fluorescent β-ketoenole AmyGreen dye for visualization of amyloid components of bacterial biofilms. Methods Appl. Fluoresc. 2020, 8, 035006. [Google Scholar] [CrossRef]
- Moshynets, O.V.; Pokholenko, I.; Iungin, O.; Potters, G.; Spiers, A.J. eDNA, Amyloid fibers and membrane vesicles identified in Pseudomonas fluorescens SBW25 biofilms. Int. J. Mol. Sci. 2022, 23, 15096. [Google Scholar] [CrossRef]
- Haseeb, A.; Faidah, H.S.; Alghamdi, S.; Alotaibi, A.F.; Elrggal, M.E.; Mahrous, A.J.; Almarzoky Abuhussain, S.S.; Obaid, N.A.; Algethamy, M.; AlQarni, A.; et al. Dose Optimization of Colistin: A Systematic Review. Antibiotics 2021, 10, 1454. [Google Scholar] [CrossRef]
- Fekety, F.R.; Norman, P.S.; Cluff, L.E. The treatment of gram-negative bacillary infections with colistin. Ann. Intern. Med. 1962, 57, 214–229. [Google Scholar] [CrossRef] [PubMed]
- Plachouras, D.; Karvanen, M.; Friberg, L.E.; Papadomichelakis, E.; Antoniadou, A.; Tsangaris, I.; Karaiskos, I.; Poulakou, G.; Kontopidou, F.; Armaganidis, A.; et al. Population pharmacokinetic analysis of colistin methanesulfonate and colistin after intravenous administration in critically ill patients with infections caused by gram-negative bacteria. Antimicrob. Agents Chemother. 2009, 53, 3430–3436. [Google Scholar] [CrossRef] [PubMed]
- Daikos, G.L.; Skiada, A.; Pavleas, J.; Vafiadi, C.; Salatas, K.; Tofas, P.; Tzanetou, K.; Markogiannakis, A.; Thomopoulos, G.; Vafiadi, I.; et al. Serum bactericidal activity of three different dosing regimens of colistin with implications for optimum clinical use. J. Chemother. 2010, 22, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Vardakas, K.Z.; Mavroudis, A.D.; Georgiou, M.; Falagas, M.E. Intravenous colistin combination antimicrobial treatment vs. monotherapy: A systematic review and meta-analysis. Int. J. Antimicrob. Agents 2018, 51, 535–547. [Google Scholar] [CrossRef]
- Katip, W.; Uitrakul, S.; Oberdorfer, P. Clinical Efficacy and Nephrotoxicity of the Loading Dose Colistin for the Treatment of Carbapenem-Resistant Acinetobacter baumannii in Critically Ill Patients. Pharmaceutics 2022, 14, 31. [Google Scholar] [CrossRef]
- Saiman, L.; Marshall, B.C.; Mayer-Hamblett, N.; Burns, J.L.; Quittner, A.L.; Cibene, D.A.; Coquillette, S.; Feiberg, A.Y.; Accurso, F.J.; Campbell, P.W. Azithromycin in patients with cystic fibrosis chronically infected with Pseudomonas aeruginosa. JAMA 2003, 290, 1749–1756. [Google Scholar] [CrossRef]
- Gillis, R.J.; Iglewski, B.H. Azithromycin retards Pseudomonas aeruginosa biofilm formation. J. Clin. Microbiol. 2004, 42, 5842–5845. [Google Scholar] [CrossRef]
- Imperi, F.; Leoni, L.; Visca, P. Antivirulence activity of azithromycin in Pseudomonas aeruginosa. Front. Microbiol. 2014, 5, 178. [Google Scholar] [CrossRef]
- Swatton, J.E.; Davenport, P.W.; Maunders, E.A.; Griffin, J.L.; Lilley, K.S.; Welch, M. Impact of azithromycin on the quorum sensing-controlled proteome of Pseudomonas aeruginosa. PLoS ONE 2016, 1, e0147698. [Google Scholar] [CrossRef]
- Kohler, T.; Dumac, J.L.; Van Delden, C. Ribosome protection prevents azithromycin-mediated quorum-sensing modulation and stationary-phase killing of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2007, 51, 4243–4248. [Google Scholar] [CrossRef]
- Tateda, K.; Ishii, Y.; Matsumoto, T.; Kobayashi, T.; Miyazaki, S.; Yamaguchi, K. Potential of macrolide antibiotics to inhibit protein synthesis of Pseudomonas aeruginosa: Suppression of virulence factors and stress response. J. Infect. Chemother. 2000, 6, 1–7. [Google Scholar] [CrossRef]
- Hirakata, Y.; Kaku, M.; Mizukane, R.; Ishida, K.; Furuya, N.; Matsumoto, T.; Tateda, K.; Yamaguchi, K. Potential effects of erythromycin on host defense systems and virulence of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1992, 36, 1922–1927. [Google Scholar] [CrossRef]
- Kawamura-Sato, K.; Iinuma, Y.; Hasegawa, T.; Horii, T.; Yamashino, T.; Ohta, M. Effect of subinhibitory concentrations of macrolides on expression of flagellin in Pseudomonas aeruginosa and Proteus mirabilis. Antimicrob. Agents Chemother. 2000, 44, 2869–2872. [Google Scholar] [CrossRef]
- Wolter, J.; Seeney, S.; Bell, S.; Bowler, S.; Masel, P.; McCormack, J. Effect of long term treatment with azithromycin on disease parameters in cystic fibrosis: A randomized trial. Thorax 2002, 57, 212–216. [Google Scholar] [CrossRef]
- Yanagihara, K.; Tomono, K.; Imamura, Y.; Kaneko, Y.; Kuroki, M.; Sawai, T.; Miyazaki, Y.; Hirakata, Y.; Mukae, H.; Kadota, J.I.; et al. Effect of clarithromycin on chronic respiratory infection caused by Pseudomonas aeruginosa with biofilm formation in an experimental murine model. J. Antimicrob. Chemother. 2002, 49, 867–870. [Google Scholar] [CrossRef]
- Tateda, K.; Ishii, Y.; Kimura, S.; Horikawa, M.; Miyairi, S.; Yamaguchi, K. Suppression of Pseudomonas aeruginosa quorum-sensing systems by macrolides: A promising strategy or an oriental mystery? J. Infect. Chemother. 2007, 13, 357–367. [Google Scholar] [CrossRef]
- Andrade, F.F.; Silva, D.; Rodrigues, A.; Pina-Vaz, C. Colistin Update on Its Mechanism of Action and Resistance; Present and Future Challenges. Microorganisms 2020, 8, 1716. [Google Scholar] [CrossRef]
- Yun, B.; Zhang, T.; Azad, M.A.K.; Wang, J.; Nowell, C.J.; Kalitsis, P.; Velkov, T.; Hudson, D.F.; Li, J. Polymyxin B causes DNA damage in HK-2 cells and mice. Arch. Toxicol. 2018, 92, 2259–2271. [Google Scholar] [CrossRef]
- Shenkutie, A.M.; Zhang, J.; Yao, M.; Asrat, D.; Chow, F.W.N.; Leung, P.H.M. Effects of sub-minimum inhibitory concentrations of imipenem and colistin on expression of biofilm-specific antibiotic resistance and virulence genes in Acinetobacter baumannii Sequence Type 1894. Int. J. Mol. Sci. 2022, 23, 12705. [Google Scholar] [CrossRef]
- Denizot, F.; Lang, R. Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J. Immunol. Methods 1986, 89, 271–277. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moshynets, O.V.; Baranovskyi, T.P.; Iungin, O.S.; Krikunov, A.A.; Potochilova, V.V.; Rudnieva, K.L.; Potters, G.; Pokholenko, I. Therapeutic Potential of an Azithromycin-Colistin Combination against XDR K. pneumoniae in a 3D Collagen-Based In Vitro Wound Model of a Biofilm Infection. Antibiotics 2023, 12, 293. https://doi.org/10.3390/antibiotics12020293
Moshynets OV, Baranovskyi TP, Iungin OS, Krikunov AA, Potochilova VV, Rudnieva KL, Potters G, Pokholenko I. Therapeutic Potential of an Azithromycin-Colistin Combination against XDR K. pneumoniae in a 3D Collagen-Based In Vitro Wound Model of a Biofilm Infection. Antibiotics. 2023; 12(2):293. https://doi.org/10.3390/antibiotics12020293
Chicago/Turabian StyleMoshynets, Olena V., Taras P. Baranovskyi, Olga S. Iungin, Alexey A. Krikunov, Viktoria V. Potochilova, Kateryna L. Rudnieva, Geert Potters, and Ianina Pokholenko. 2023. "Therapeutic Potential of an Azithromycin-Colistin Combination against XDR K. pneumoniae in a 3D Collagen-Based In Vitro Wound Model of a Biofilm Infection" Antibiotics 12, no. 2: 293. https://doi.org/10.3390/antibiotics12020293
APA StyleMoshynets, O. V., Baranovskyi, T. P., Iungin, O. S., Krikunov, A. A., Potochilova, V. V., Rudnieva, K. L., Potters, G., & Pokholenko, I. (2023). Therapeutic Potential of an Azithromycin-Colistin Combination against XDR K. pneumoniae in a 3D Collagen-Based In Vitro Wound Model of a Biofilm Infection. Antibiotics, 12(2), 293. https://doi.org/10.3390/antibiotics12020293





