Strategies to Name Metallo-β-Lactamases and Number Their Amino Acid Residues
Abstract
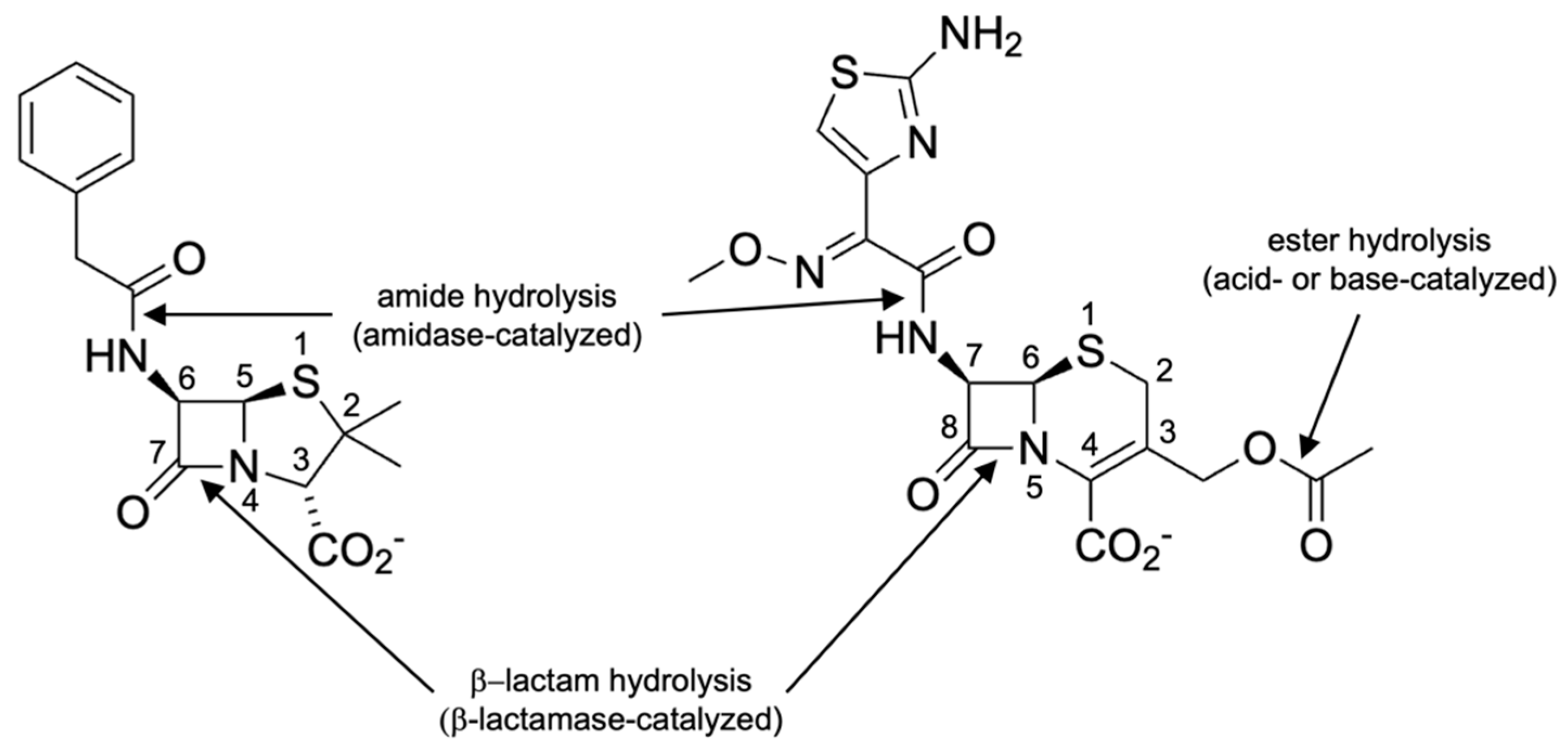
1. Introduction
2. The Challenge of Naming β-Lactamases and Numbering Their Amino Acid Residues
3. Naming of β-Lactamases
3.1. Naming of β-Lactamases Based on the Enzymatic Reaction Catalyzed
3.2. Naming of Metallo-β-Lactamase Families
3.3. Naming of Metallo-β-Lactamase Family Members
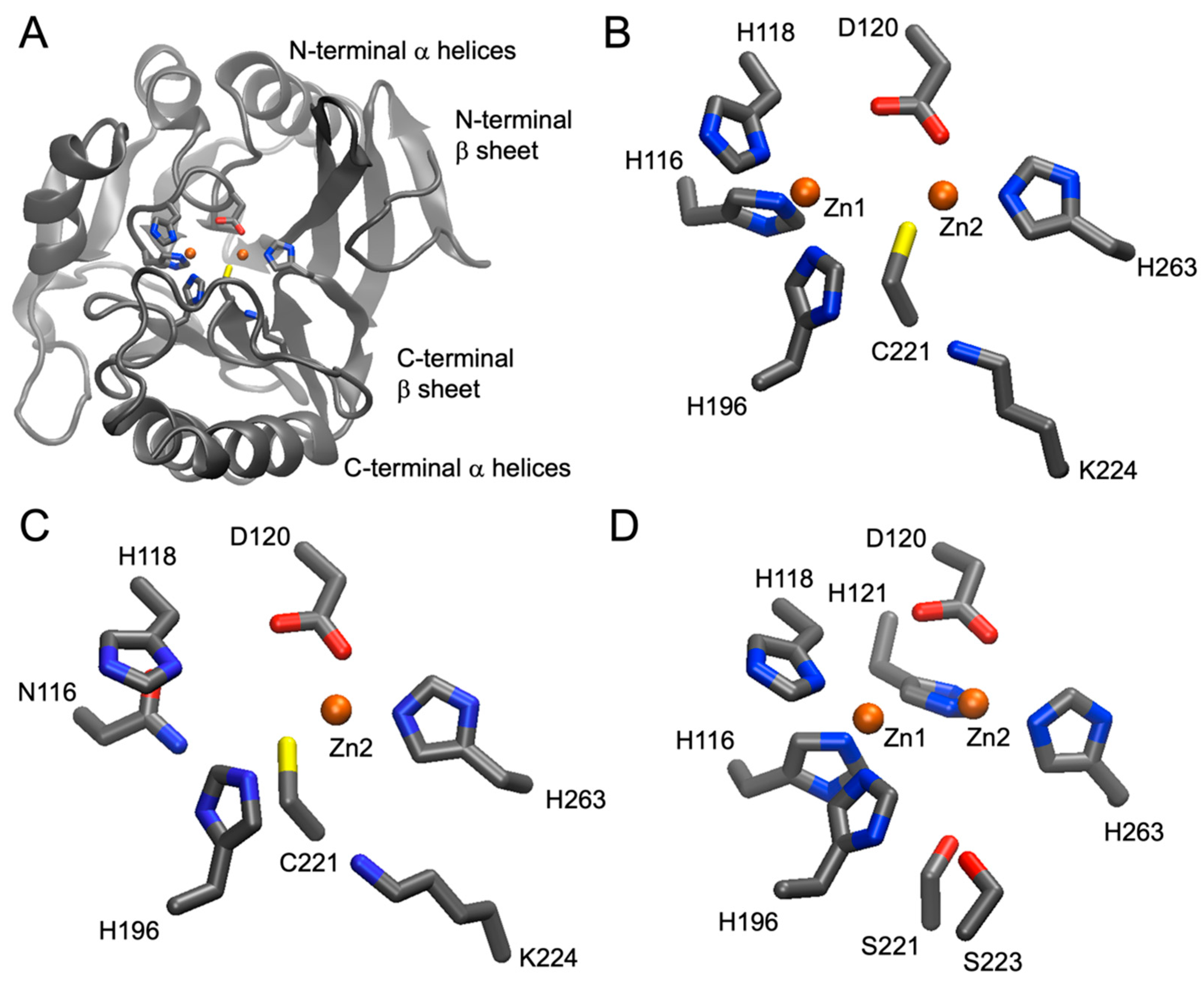
4. Numbering Amino Acid Residues in Metallo-β-Lactamases
4.1. The Class B (Metallo-)β-Lactamase Standard Numbering Scheme
4.2. Strategies for MBL Renumbering
4.2.1. Manual Renumbering
- H120, H122, and D124 become H116, H118, and D120 (−4), respectively;
- H189 becomes H196 (+7);
- C208 becomes C221 (+13);
- H250 becomes H263 (+13).
4.2.2. Automated Renumbering Based on Conserved Motive Recognition
4.2.3. Automated Renumbering Based on Profile Hidden Markov Model
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ligon, B.L. Penicillin: Its discovery and early development. Semin. Pediatr. Infect. Dis. 2004, 15, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Chain, E.; Florey, H.W.; Gardner, A.D.; Heatley, N.G.; Jennings, M.A.; Orr-Ewing, J.; Sanders, A.G. Penicillin as a chemotherapeutic agent. Lancet 1940, 236, 226–228. [Google Scholar] [CrossRef]
- Abraham, E.P.; Chain, E.; Fletcher, C.M.; Gardner, A.D.; Heatley, N.G.; Jennings, M.A.; Florey, H.W. Further observations on penicillin. Lancet 1941, 238, 177–189. [Google Scholar] [CrossRef]
- Meyer, K.; Chaffee, E.; Hobby, G.L.; Dawson, M.H.; Schwenk, E.; Fleischer, G. On Penicillin. Science 1942, 96, 20–21. [Google Scholar] [CrossRef] [PubMed]
- Versporten, A.; Bolokhovets, G.; Ghazaryan, L.; Abilova, V.; Pyshnik, G.; Spasojevic, T.; Korinteli, I.; Raka, L.; Kambaralieva, B.; Cizmovic, L.; et al. Antibiotic use in eastern Europe: A cross-national database study in coordination with the WHO Regional Office for Europe. Lancet Infect. Dis. 2014, 14, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Versporten, A.; Zarb, P.; Caniaux, I.; Gros, M.F.; Drapier, N.; Miller, M.; Jarlier, V.; Nathwani, D.; Goossens, H.; Global, P.P.S.n. Antimicrobial consumption and resistance in adult hospital inpatients in 53 countries: Results of an internet-based global point prevalence survey. Lancet Glob. Health 2018, 6, e619–e629. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A. Penicillin. Nobel Lect. 1945. Available online: https://www.nobelprize.org/uploads/2018/06/fleming-lecture.pdf (accessed on 31 October 2023).
- Abraham, E.P.; Chain, E. An enzyme from bacteria able to destroy penicillin. Nature 1940, 3713, 837. [Google Scholar] [CrossRef]
- Bush, K.; Jacoby, G.A. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar] [CrossRef]
- Carfi, A.; Pares, S.; Duee, E.; Galleni, M.; Duez, C.; Frere, J.M.; Dideberg, O. The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 1995, 14, 4914–4921. [Google Scholar] [CrossRef]
- Garau, G.; Bebrone, C.; Anne, C.; Galleni, M.; Frere, J.M.; Dideberg, O. A metallo-beta-lactamase enzyme in action: Crystal structures of the monozinc carbapenemase CphA and its complex with biapenem. J. Mol. Biol. 2005, 345, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Spencer, J.; Read, J.; Sessions, R.B.; Howell, S.; Blackburn, G.M.; Gamblin, S.J. Antibiotic recognition by binuclear metallo-beta-lactamases revealed by X-ray crystallography. J. Am. Chem. Soc. 2005, 127, 14439–14444. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Galleni, M.; Lamotte-Brasseur, J.; Rossolini, G.M.; Spencer, J.; Dideberg, O.; Frere, J.M.; Metallo-beta-lactamases Working, G. Standard numbering scheme for class B beta-lactamases. Antimicrob. Agents Chemother. 2001, 45, 660–663. [Google Scholar] [CrossRef] [PubMed]
- Garau, G.; Garcia-Saez, I.; Bebrone, C.; Anne, C.; Mercuri, P.; Galleni, M.; Frere, J.M.; Dideberg, O. Update of the standard numbering scheme for class B beta-lactamases. Antimicrob. Agents Chemother. 2004, 48, 2347–2349. [Google Scholar] [CrossRef] [PubMed]
- Bahr, G.; Gonzalez, L.J.; Vila, A.J. Metallo-beta-lactamases in the Age of Multidrug Resistance: From Structure and Mechanism to Evolution, Dissemination, and Inhibitor Design. Chem. Rev. 2021, 121, 7957–8094. [Google Scholar] [CrossRef] [PubMed]
- Palzkill, T. Structural and Mechanistic Basis for Extended-Spectrum Drug-Resistance Mutations in Altering the Specificity of TEM, CTX-M, and KPC beta-lactamases. Front. Mol. Biosci. 2018, 5, 16. [Google Scholar] [CrossRef]
- Oelschlaeger, P.; Ai, N.; Duprez, K.T.; Welsh, W.J.; Toney, J.H. Evolving carbapenemases: Can medicinal chemists advance one step ahead of the coming storm? J. Med. Chem. 2010, 53, 3013–3027. [Google Scholar] [CrossRef]
- Bush, K. Proliferation and significance of clinically relevant beta-lactamases. Ann. N. Y. Acad. Sci. 2013, 1277, 84–90. [Google Scholar] [CrossRef]
- Tooke, C.L.; Hinchliffe, P.; Bragginton, E.C.; Colenso, C.K.; Hirvonen, V.H.A.; Takebayashi, Y.; Spencer, J. beta-Lactamases and beta-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431, 3472–3500. [Google Scholar] [CrossRef]
- Mojica, M.F.; Rossi, M.A.; Vila, A.J.; Bonomo, R.A. The urgent need for metallo-beta-lactamase inhibitors: An unattended global threat. Lancet Infect. Dis. 2022, 22, e28–e34. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Oelschlaeger, P.; Wang, D.; Xu, H.; Wang, Q.; Wang, C.; Zhao, A.; Yang, K.W. Structure and Mechanism-Guided Design of Dual Serine/Metallo-Carbapenemase Inhibitors. J. Med. Chem. 2022, 65, 5954–5974. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yan, Y.H.; Schofield, C.J.; McNally, A.; Zong, Z.; Li, G.B. Metallo-beta-lactamase-mediated antimicrobial resistance and progress in inhibitor discovery. Trends Microbiol. 2023, 31, 735–748. [Google Scholar] [CrossRef] [PubMed]
- Denakpo, E.; Naas, T.; Iorga, B.I. An updated patent review of metallo-beta-lactamase inhibitors (2020–2023). Expert. Opin. Ther. Pat. 2023, 33, 523–538. [Google Scholar] [CrossRef] [PubMed]
- Naas, T.; Oueslati, S.; Bonnin, R.A.; Dabos, M.L.; Zavala, A.; Dortet, L.; Retailleau, P.; Iorga, B.I. Beta-lactamase database (BLDB)—Structure and function. J. Enzym. Inhib. Med. Chem. 2017, 32, 917–919. [Google Scholar] [CrossRef] [PubMed]
- Sutcliffe, J.G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc. Natl. Acad. Sci. USA 1978, 75, 3737–3741. [Google Scholar] [CrossRef] [PubMed]
- Joris, B.; Ledent, P.; Dideberg, O.; Fonze, E.; Lamotte-Brasseur, J.; Kelly, J.A.; Ghuysen, J.M.; Frere, J.M. Comparison of the sequences of class A beta-lactamases and of the secondary structure elements of penicillin-recognizing proteins. Antimicrob. Agents Chemother. 1991, 35, 2294–2301. [Google Scholar] [CrossRef]
- Mack, A.R.; Barnes, M.D.; Taracila, M.A.; Hujer, A.M.; Hujer, K.M.; Cabot, G.; Feldgarden, M.; Haft, D.H.; Klimke, W.; van den Akker, F.; et al. A Standard Numbering Scheme for Class C beta-Lactamases. Antimicrob. Agents Chemother. 2020, 64, e01841-19. [Google Scholar] [CrossRef]
- Ambler, R.P.; Coulson, A.F.; Frere, J.M.; Ghuysen, J.M.; Joris, B.; Forsman, M.; Levesque, R.C.; Tiraby, G.; Waley, S.G. A standard numbering scheme for the class A beta-lactamases. Biochem. J. 1991, 276 Pt 1, 269–270. [Google Scholar] [CrossRef]
- Kang, J.S.; Zhang, A.L.; Faheem, M.; Zhang, C.J.; Ai, N.; Buynak, J.D.; Welsh, W.J.; Oelschlaeger, P. Virtual Screening and Experimental Testing of B1 Metallo-beta-lactamase Inhibitors. J. Chem. Inf. Model. 2018, 58, 1902–1914. [Google Scholar] [CrossRef]
- Nomenclature committee of the international union of biochemistry and molecular biology (NC-IUBMB), Enzyme Supplement 5 (1999). Eur. J. Biochem. 1999, 264, 610–650. Available online: https://pubmed.ncbi.nlm.nih.gov/10491110/ (accessed on 31 October 2023). [CrossRef]
- Schomburg, I.; Chang, A.; Ebeling, C.; Gremse, M.; Heldt, C.; Huhn, G.; Schomburg, D. BRENDA, the enzyme database: Updates and major new developments. Nucleic Acids Res. 2004, 32, D431–D433. [Google Scholar] [CrossRef] [PubMed]
- Holt, R.J.; Stewart, G.T. Production of Amidase and Beta-Lactamase by Bacteria. J. Gen. Microbiol. 1964, 36, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Volpato, G.; Rodrigues, R.C.; Fernandez-Lafuente, R. Use of enzymes in the production of semi-synthetic penicillins and cephalosporins: Drawbacks and perspectives. Curr. Med. Chem. 2010, 17, 3855–3873. [Google Scholar] [CrossRef] [PubMed]
- Srirangan, K.; Orr, V.; Akawi, L.; Westbrook, A.; Moo-Young, M.; Chou, C.P. Biotechnological advances on penicillin G acylase: Pharmaceutical implications, unique expression mechanism and production strategies. Biotechnol. Adv. 2013, 31, 1319–1332. [Google Scholar] [CrossRef] [PubMed]
- Buchholz, K. A breakthrough in enzyme technology to fight penicillin resistance-industrial application of penicillin amidase. Appl. Microbiol. Biotechnol. 2016, 100, 3825–3839. [Google Scholar] [CrossRef] [PubMed]
- Grulich, M.; Stepanek, V.; Kyslik, P. Perspectives and industrial potential of PGA selectivity and promiscuity. Biotechnol. Adv. 2013, 31, 1458–1472. [Google Scholar] [CrossRef]
- Yamana, T.; Tsuji, A. Comparative stability of cephalosporins in aqueous solution: Kinetics and mechanisms of degradation. J. Pharm. Sci. 1976, 65, 1563–1574. [Google Scholar] [CrossRef]
- Patel, K.B.; Nicolau, D.P.; Nightingale, C.H.; Quintiliani, R. Pharmacokinetics of cefotaxime in healthy volunteers and patients. Diagn. Microbiol. Infect. Dis. 1995, 22, 49–55. [Google Scholar] [CrossRef]
- Crowfoot, D. X-ray crystallographic studies of compounds of biochemical interest. Annu. Rev. Biochem. 1948, 17, 115–146. [Google Scholar] [CrossRef]
- Rasmussen, R.S. Infrared spectroscopy in structure determination and its application to penicillin. Fortschritte Chem. Org. Naturstoffe 1948, 5, 331–386. [Google Scholar]
- Bush, K. Classification for beta-lactamases: Historical perspectives. Expert. Rev. Anti Infect. Ther. 2023, 21, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Sabath, L.D.; Abraham, E.P. Cephalosporinase and penicillinase activity of Bacillus cereus. Antimicrob. Agents Chemother. 1965, 5, 392–397. [Google Scholar] [PubMed]
- Sabath, L.D.; Abraham, E.P. Zinc as a cofactor for cephalosporinase from Bacillus cereus 569. Biochem. J. 1966, 98, 11C–13C. [Google Scholar] [CrossRef] [PubMed]
- Madgwick, P.J.; Waley, S.G. beta-lactamase I from Bacillus cereus. Structure and site-directed mutagenesis. Biochem. J. 1987, 248, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Carlino, A.; Madonna, M.J.; Lampen, J.O. Cloning and sequencing of the metallothioprotein beta-lactamase II gene of Bacillus cereus 569/H in Escherichia coli. J. Bacteriol. 1985, 164, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.R.; Hall, L.; Assinder, S.J.; Nichols, W.W.; Cartwright, S.J.; MacGowan, A.P.; Bennett, P.M. Sequence analysis of the L1 metallo-beta-lactamase from Xanthomonas maltophilia. Biochim. Biophys. Acta 1994, 1218, 199–201. [Google Scholar] [CrossRef]
- Walsh, T.R.; MacGowan, A.P.; Bennett, P.M. Sequence analysis and enzyme kinetics of the L2 serine beta-lactamase from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 1997, 41, 1460–1464. [Google Scholar] [CrossRef]
- Ambler, R.P. The structure of beta-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1980, 289, 321–331. [Google Scholar] [CrossRef]
- Jacoby, G.A. Beta-lactamase nomenclature. Antimicrob. Agents Chemother. 2006, 50, 1123–1129. [Google Scholar] [CrossRef]
- Bradford, P.A.; Bonomo, R.A.; Bush, K.; Carattoli, A.; Feldgarden, M.; Haft, D.H.; Ishii, Y.; Jacoby, G.A.; Klimke, W.; Palzkill, T.; et al. Consensus on beta-Lactamase Nomenclature. Antimicrob. Agents Chemother. 2022, 66, e0033322. [Google Scholar] [CrossRef] [PubMed]
- Bush, K. The ABCD’s of beta-lactamase nomenclature. J. Infect. Chemother. 2013, 19, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Thai, Q.K.; Bos, F.; Pleiss, J. The Lactamase Engineering Database: A critical survey of TEM sequences in public databases. BMC Genom. 2009, 10, 390. [Google Scholar] [CrossRef] [PubMed]
- Widmann, M.; Pleiss, J.; Oelschlaeger, P. Systematic analysis of metallo-beta-lactamases using an automated database. Antimicrob. Agents Chemother. 2012, 56, 3481–3491. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Farrell, C.M.; Feldgarden, M.; Fine, A.M.; Funk, K.; et al. Database resources of the National Center for Biotechnology Information in 2023. Nucleic Acids Res. 2023, 51, D29–D38. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef]
- Keshri, V.; Diene, S.M.; Estienne, A.; Dardaillon, J.; Chabrol, O.; Tichit, L.; Rolain, J.M.; Raoult, D.; Pontarotti, P. An Integrative Database of beta-Lactamase Enzymes: Sequences, Structures, Functions, and Phylogenetic Trees. Antimicrob. Agents Chemother. 2019, 63, e02319-18. [Google Scholar] [CrossRef]
- Mao, Y.; Liu, X.; Zhang, N.; Wang, Z.; Han, M. NCRD: A non-redundant comprehensive database for detecting antibiotic resistance genes. iScience 2023, 26, 108141. [Google Scholar] [CrossRef]
- Chen, M.; Cai, H.; Li, Y.; Wang, N.; Zhang, P.; Hua, X.; Yu, Y.; Sun, R. Plasmid-Borne AFM Alleles in Pseudomonas aeruginosa Clinical Isolates from China. Microbiol. Spectr. 2022, 10, e0203522. [Google Scholar] [CrossRef]
- Berglund, F.; Marathe, N.P.; Osterlund, T.; Bengtsson-Palme, J.; Kotsakis, S.; Flach, C.F.; Larsson, D.G.J.; Kristiansson, E. Identification of 76 novel B1 metallo-beta-lactamases through large-scale screening of genomic and metagenomic data. Microbiome 2017, 5, 134. [Google Scholar] [CrossRef] [PubMed]
- Souza, C.O.; Cayo, R.; Lima, K.V.B.; Brasiliense, D.M.; Streling, A.P.; Siqueira, A.V.; Alberto-Lei, F.; Leal, J.T.; Nodari, C.S.; Perez-Chaparro, P.J.; et al. Genetic and biochemical characterization of BIM-1, a novel acquired subgroup B1 MBL found in a Pseudomonas sp. strain from the Brazilian Amazon region. J. Antimicrob. Chemother. 2023, 78, 1359–1366. [Google Scholar] [CrossRef] [PubMed]
- Bellais, S.; Aubert, D.; Naas, T.; Nordmann, P. Molecular and biochemical heterogeneity of class B carbapenem-hydrolyzing beta-lactamases in Chryseobacterium meningosepticum. Antimicrob. Agents Chemother. 2000, 44, 1878–1886. [Google Scholar] [CrossRef] [PubMed]
- Boyd, D.A.; Lisboa, L.F.; Rennie, R.; Zhanel, G.G.; Dingle, T.C.; Mulvey, M.R. Identification of a novel metallo-beta-lactamase, CAM-1, in clinical Pseudomonas aeruginosa isolates from Canada. J. Antimicrob. Chemother. 2019, 74, 1563–1567. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.S.; Malamy, M.H. Sequencing the gene for an imipenem-cefoxitin-hydrolyzing enzyme (CfiA) from Bacteroides fragilis TAL2480 reveals strong similarity between CfiA and Bacillus cereus beta-lactamase II. J. Bacteriol. 1990, 172, 2584–2593. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, B.A.; Gluzman, Y.; Tally, F.P. Cloning and sequencing of the class B beta-lactamase gene (ccrA) from Bacteroides fragilis TAL3636. Antimicrob. Agents Chemother. 1990, 34, 1590–1592. [Google Scholar] [CrossRef]
- Bellais, S.; Naas, T.; Nordmann, P. Genetic and biochemical characterization of CGB-1, an Ambler class B carbapenem-hydrolyzing beta-lactamase from Chryseobacterium gleum. Antimicrob. Agents Chemother. 2002, 46, 2791–2796. [Google Scholar] [CrossRef]
- Klimkaite, L.; Ragaisis, I.; Krasauskas, R.; Ruzauskas, M.; Suziedeliene, E.; Armalyte, J. Novel Antibiotic Resistance Genes Identified by Functional Gene Library Screening in Stenotrophomonas maltophilia and Chryseobacterium spp. Bacteria of Soil Origin. Int. J. Mol. Sci. 2023, 24, 6037. [Google Scholar] [CrossRef]
- Alvarez-Marin, M.T.; Zarzuela, L.; Camacho, E.M.; Santero, E.; Flores, A. Detection by metagenomic functional analysis and improvement by experimental evolution of beta-lactams resistance genes present in oil contaminated soils. Sci. Rep. 2022, 12, 10059. [Google Scholar] [CrossRef]
- Soki, J.; Lang, U.; Schumacher, U.; Nagy, I.; Berenyi, A.; Feher, T.; Burian, K.; Nagy, E. A novel Bacteroides metallo-beta-lactamase (MBL) and its gene (crxA) in Bacteroides xylanisolvens revealed by genomic sequencing and functional analysis. J. Antimicrob. Chemother. 2022, 77, 1553–1556. [Google Scholar] [CrossRef]
- Zhang, L.; Calvo-Bado, L.; Murray, A.K.; Amos, G.C.A.; Hawkey, P.M.; Wellington, E.M.; Gaze, W.H. Novel clinically relevant antibiotic resistance genes associated with sewage sludge and industrial waste streams revealed by functional metagenomic screening. Environ. Int. 2019, 132, 105120. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Rodriguez-Martinez, J.M.; Al Naiemi, N.; Debets-Ossenkopp, Y.J.; Nordmann, P. Characterization of DIM-1, an integron-encoded metallo-beta-lactamase from a Pseudomonas stutzeri clinical isolate in the Netherlands. Antimicrob. Agents Chemother. 2010, 54, 2420–2424. [Google Scholar] [CrossRef] [PubMed]
- Bellais, S.; Girlich, D.; Karim, A.; Nordmann, P. EBR-1, a novel Ambler subclass B1 beta-lactamase from Empedobacter brevis. Antimicrob. Agents Chemother. 2002, 46, 3223–3227. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Tan, S.; Gao, J.; Han, H.; Liu, J.; Lu, G.; Liu, D.; Yi, Y.; Zhu, B.; Gao, G.F. An unexpected similarity between antibiotic-resistant NDM-1 and beta-lactamase II from Erythrobacter litoralis. Protein Cell 2011, 2, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Pollini, S.; Maradei, S.; Pecile, P.; Olivo, G.; Luzzaro, F.; Docquier, J.D.; Rossolini, G.M. FIM-1, a new acquired metallo-beta-lactamase from a Pseudomonas aeruginosa clinical isolate from Italy. Antimicrob. Agents Chemother. 2013, 57, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Toleman, M.A.; Jones, R.N.; Schmidt, F.J.; Walsh, T.R. Molecular characterization of a beta-lactamase gene, blaGIM-1, encoding a new subclass of metallo-beta-lactamase. Antimicrob. Agents Chemother. 2004, 48, 4654–4661. [Google Scholar] [CrossRef]
- Schauer, J.; Gatermann, S.G.; Eisfeld, J.; Hans, J.B.; Ziesing, S.; Schluter, D.; Pfennigwerth, N. Characterization of GMB-1, a novel metallo-beta-lactamase (MBL) found in three different Enterobacterales species. J. Antimicrob. Chemother. 2022, 77, 1247–1253. [Google Scholar] [CrossRef]
- Gudeta, D.D.; Bortolaia, V.; Pollini, S.; Docquier, J.D.; Rossolini, G.M.; Amos, G.C.; Wellington, E.M.; Guardabassi, L. Expanding the Repertoire of Carbapenem-Hydrolyzing Metallo-ss-Lactamases by Functional Metagenomic Analysis of Soil Microbiota. Front. Microbiol. 2016, 7, 1985. [Google Scholar] [CrossRef]
- Chertkov, O.; Brown, P.J.; Kysela, D.T.; de Pedro, M.A.; Lucas, S.; Copeland, A.; Lapidus, A.; Del Rio, T.G.; Tice, H.; Bruce, D.; et al. Complete genome sequence of Hirschia baltica type strain (IFAM 1418(T)). Stand. Genomic. Sci. 2011, 5, 287–297. [Google Scholar] [CrossRef]
- Pfennigwerth, N.; Lange, F.; Belmar Campos, C.; Hentschke, M.; Gatermann, S.G.; Kaase, M. Genetic and biochemical characterization of HMB-1, a novel subclass B1 metallo-beta-lactamase found in a Pseudomonas aeruginosa clinical isolate. J. Antimicrob. Chemother. 2017, 72, 1068–1073. [Google Scholar] [CrossRef][Green Version]
- Osano, E.; Arakawa, Y.; Wacharotayankun, R.; Ohta, M.; Horii, T.; Ito, H.; Yoshimura, F.; Kato, N. Molecular characterization of an enterobacterial metallo beta-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 1994, 38, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Bellais, S.; Leotard, S.; Poirel, L.; Naas, T.; Nordmann, P. Molecular characterization of a carbapenem-hydrolyzing beta-lactamase from Chryseobacterium (Flavobacterium) indologenes. FEMS Microbiol. Lett. 1999, 171, 127–132. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Naas, T.; Bellais, S.; Nordmann, P. Molecular and biochemical characterization of a carbapenem-hydrolysing beta-lactamase from Flavobacterium johnsoniae. J. Antimicrob. Chemother. 2003, 51, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Sekiguchi, J.; Morita, K.; Kitao, T.; Watanabe, N.; Okazaki, M.; Miyoshi-Akiyama, T.; Kanamori, M.; Kirikae, T. KHM-1, a novel plasmid-mediated metallo-beta-lactamase from a Citrobacter freundii clinical isolate. Antimicrob. Agents Chemother. 2008, 52, 4194–4197. [Google Scholar] [CrossRef] [PubMed]
- Mammeri, H.; Bellais, S.; Nordmann, P. Chromosome-encoded beta-lactamases TUS-1 and MUS-1 from Myroides odoratus and Myroides odoratimimus (formerly Flavobacterium odoratum), new members of the lineage of molecular subclass B1 metalloenzymes. Antimicrob. Agents Chemother. 2002, 46, 3561–3567. [Google Scholar] [CrossRef]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef]
- Kieffer, N.; Poirel, L.; Fournier, C.; Haltli, B.; Kerr, R.; Nordmann, P. Characterization of PAN-1, a Carbapenem-Hydrolyzing Class B beta-Lactamase From the Environmental Gram-Negative Pseudobacteriovorax antillogorgiicola. Front. Microbiol. 2019, 10, 1673. [Google Scholar] [CrossRef]
- Gudeta, D.D.; Bortolaia, V.; Amos, G.; Wellington, E.M.; Brandt, K.K.; Poirel, L.; Nielsen, J.B.; Westh, H.; Guardabassi, L. The Soil Microbiota Harbors a Diversity of Carbapenem-Hydrolyzing beta-Lactamases of Potential Clinical Relevance. Antimicrob. Agents Chemother. 2016, 60, 151–160. [Google Scholar] [CrossRef]
- Dai, J.; Dai, W.; Qiu, C.; Yang, Z.; Zhang, Y.; Zhou, M.; Zhang, L.; Fang, C.; Gao, Q.; Yang, Q.; et al. Unraveling adaptation of Pontibacter korlensis to radiation and infertility in desert through complete genome and comparative transcriptomic analysis. Sci. Rep. 2015, 5, 10929. [Google Scholar] [CrossRef]
- Poirel, L.; Heritier, C.; Nordmann, P. Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella livingstonensis (SLB-1) and Shewanella frigidimarina (SFB-1). J. Antimicrob. Chemother. 2005, 55, 680–685. [Google Scholar] [CrossRef]
- Lee, K.; Yum, J.H.; Yong, D.; Lee, H.M.; Kim, H.D.; Docquier, J.D.; Rossolini, G.M.; Chong, Y. Novel acquired metallo-beta-lactamase gene, bla(SIM-1), in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob. Agents Chemother. 2005, 49, 4485–4491. [Google Scholar] [CrossRef] [PubMed]
- Toleman, M.A.; Simm, A.M.; Murphy, T.A.; Gales, A.C.; Biedenbach, D.J.; Jones, R.N.; Walsh, T.R. Molecular characterization of SPM-1, a novel metallo-beta-lactamase isolated in Latin America: Report from the SENTRY antimicrobial surveillance programme. J. Antimicrob. Chemother. 2002, 50, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Liu, Z.; Lu, Z.; Huang, R.; Xiang, R. Identification and characterization of a novel metallo beta-lactamase, SZM-1, in Shenzhen Bay, South China. Front. Microbiol. 2022, 13, 996834. [Google Scholar] [CrossRef] [PubMed]
- El Salabi, A.; Borra, P.S.; Toleman, M.A.; Samuelsen, O.; Walsh, T.R. Genetic and biochemical characterization of a novel metallo-beta-lactamase, TMB-1, from an Achromobacter xylosoxidans strain isolated in Tripoli, Libya. Antimicrob. Agents Chemother. 2012, 56, 2241–2245. [Google Scholar] [CrossRef]
- Cheng, Q.; Zheng, Z.; Ye, L.; Chen, S. Identification of a Novel Metallo-beta-Lactamase, VAM-1, in a Foodborne Vibrio alginolyticus Isolate from China. Antimicrob. Agents Chemother. 2021, 65, e0112921. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, W.; Peng, K.; Wang, Z.; Li, R. Identification of a Novel Plasmid-Mediated Carbapenemase-Encoding Gene, bla(VMB-2), in Vibrio diabolicus. Antimicrob. Agents Chemother. 2021, 65, e0020621. [Google Scholar] [CrossRef]
- Lauretti, L.; Riccio, M.L.; Mazzariol, A.; Cornaglia, G.; Amicosante, G.; Fontana, R.; Rossolini, G.M. Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 1999, 43, 1584–1590. [Google Scholar] [CrossRef]
- Zheng, Z.; Cheng, Q.; Chan, E.W.; Chen, S. Genetic and Biochemical Characterization of VMB-1, a Novel Metallo-beta-Lactamase Encoded by a Conjugative, Broad-Host Range IncC Plasmid from Vibrio spp. Adv. Biosyst. 2020, 4, e1900221. [Google Scholar] [CrossRef]
- Lu, W.J.; Hsu, P.H.; Lin, H.V. A Novel Cooperative Metallo-beta-Lactamase Fold Metallohydrolase from Pathogen Vibrio vulnificus Exhibits beta-Lactam Antibiotic-Degrading Activities. Antimicrob. Agents Chemother. 2021, 65, e0032621. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, L.; Feng, C.; Zhu, J.; Li, A.; Zhao, J.; Zhang, Y.; Gao, M.; Shi, W.; Li, Q.; et al. Characterization and Identification of a novel chromosome-encoded metallo-beta-lactamase WUS-1 in Myroides albus P34. Front. Microbiol. 2022, 13, 1059997. [Google Scholar] [CrossRef]
- Kieffer, N.; Guzman-Puche, J.; Poirel, L.; Kang, H.J.; Jeon, C.O.; Nordmann, P. ZHO-1, an intrinsic MBL from the environmental Gram-negative species Zhongshania aliphaticivorans. J. Antimicrob. Chemother. 2019, 74, 1568–1571. [Google Scholar] [CrossRef] [PubMed]
- Massidda, O.; Rossolini, G.M.; Satta, G. The Aeromonas hydrophila cphA gene: Molecular heterogeneity among class B metallo-beta-lactamases. J. Bacteriol. 1991, 173, 4611–4617. [Google Scholar] [CrossRef] [PubMed]
- Gomez, S.A.; Sanz, M.B.; Rapoport, M.; Sucin, G.; Corallo, T.A.; Poklepovich, T.; Campos, J.; Ceriana, P.; de Mendieta, J.M.; Prieto, M.; et al. Novel Metallo-beta-Lactamase bla(CVI-1) Isolated from a Chromobaterium violaceum Clinical Strain Resistant to Colistin. Pathogens 2023, 12, 961. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Palmieri, M.; Brilhante, M.; Masseron, A.; Perreten, V.; Nordmann, P. PFM-Like Enzymes Are a Novel Family of Subclass B2 Metallo-beta-Lactamases from Pseudomonas synxantha Belonging to the Pseudomonas fluorescens Complex. Antimicrob. Agents Chemother. 2020, 64, e01700-19. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, M.J.; Peixe, L.; Sousa, J.C.; Henriques, I.; Alves, A.; Correia, A. Sfh-I, a subclass B2 metallo-beta-lactamase from a Serratia fonticola environmental isolate. Antimicrob. Agents Chemother. 2003, 47, 2330–2333. [Google Scholar] [CrossRef] [PubMed]
- Mercuri, P.S.; Esposito, R.; Bletard, S.; Di Costanzo, S.; Perilli, M.; Kerff, F.; Galleni, M. Mutational Effects on Carbapenem Hydrolysis of YEM-1, a New Subclass B2 Metallo-beta-Lactamase from Yersinia mollaretii. Antimicrob. Agents Chemother. 2020, 64, e00105-20. [Google Scholar] [CrossRef] [PubMed]
- Yong, D.; Toleman, M.A.; Bell, J.; Ritchie, B.; Pratt, R.; Ryley, H.; Walsh, T.R. Genetic and biochemical characterization of an acquired subgroup B3 metallo-beta-lactamase gene, blaAIM-1, and its unique genetic context in Pseudomonas aeruginosa from Australia. Antimicrob. Agents Chemother. 2012, 56, 6154–6159. [Google Scholar] [CrossRef] [PubMed]
- Stoczko, M.; Frere, J.M.; Rossolini, G.M.; Docquier, J.D. Postgenomic scan of metallo-beta-lactamase homologues in rhizobacteria: Identification and characterization of BJP-1, a subclass B3 ortholog from Bradyrhizobium japonicum. Antimicrob. Agents Chemother. 2006, 50, 1973–1981. [Google Scholar] [CrossRef]
- Au, S.X.; Dzulkifly, N.S.; Muhd Noor, N.D.; Matsumura, H.; Raja Abdul Rahman, R.N.Z.; Normi, Y.M. Dual Activity BLEG-1 from Bacillus lehensis G1 Revealed Structural Resemblance to B3 Metallo-beta-Lactamase and Glyoxalase II: An Insight into Its Enzyme Promiscuity and Evolutionary Divergence. Int. J. Mol. Sci. 2021, 22, 9377. [Google Scholar] [CrossRef]
- Stoczko, M.; Frere, J.M.; Rossolini, G.M.; Docquier, J.D. Functional diversity among metallo-beta-lactamases: Characterization of the CAR-1 enzyme of Erwinia carotovora. Antimicrob. Agents Chemother. 2008, 52, 2473–2479. [Google Scholar] [CrossRef]
- Docquier, J.D.; Pantanella, F.; Giuliani, F.; Thaller, M.C.; Amicosante, G.; Galleni, M.; Frere, J.M.; Bush, K.; Rossolini, G.M. CAU-1, a subclass B3 metallo-beta-lactamase of low substrate affinity encoded by an ortholog present in the Caulobacter crescentus chromosome. Antimicrob. Agents Chemother. 2002, 46, 1823–1830. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pedroso, M.M.; Waite, D.W.; Melse, O.; Wilson, L.; Mitic, N.; McGeary, R.P.; Antes, I.; Guddat, L.W.; Hugenholtz, P.; Schenk, G. Broad spectrum antibiotic-degrading metallo-beta-lactamases are phylogenetically diverse. Protein Cell 2020, 11, 613–617. [Google Scholar] [CrossRef] [PubMed]
- Girlich, D.; Poirel, L.; Nordmann, P. Diversity of naturally occurring Ambler class B metallo-beta-lactamases in Erythrobacter spp. J. Antimicrob. Chemother. 2012, 67, 2661–2664. [Google Scholar] [CrossRef] [PubMed]
- Boschi, L.; Mercuri, P.S.; Riccio, M.L.; Amicosante, G.; Galleni, M.; Frere, J.M.; Rossolini, G.M. The Legionella (Fluoribacter) gormanii metallo-beta-lactamase: A new member of the highly divergent lineage of molecular-subclass B3 beta-lactamases. Antimicrob. Agents Chemother. 2000, 44, 1538–1543. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lange, F.; Pfennigwerth, N.; Hartl, R.; Kerschner, H.; Achleitner, D.; Gatermann, S.G.; Kaase, M. LMB-1, a novel family of class B3 MBLs from an isolate of Enterobacter cloacae. J. Antimicrob. Chemother. 2018, 73, 2331–2335. [Google Scholar] [CrossRef]
- Allen, H.K.; Moe, L.A.; Rodbumrer, J.; Gaarder, A.; Handelsman, J. Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME J. 2009, 3, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Miraula, M.; Schenk, G.; Mitic, N. Promiscuous metallo-beta-lactamases: MIM-1 and MIM-2 may play an essential role in quorum sensing networks. J. Inorg. Biochem. 2016, 162, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Suzuki, S.; Matsui, M.; Hiraki, Y.; Kawano, F.; Shibayama, K. A subclass B3 metallo-beta-lactamase found in Pseudomonas alcaligenes. J. Antimicrob. Chemother. 2014, 69, 1430–1432. [Google Scholar] [CrossRef][Green Version]
- Yamada, K.; Ishii, Y.; Tateda, K. Biochemical Characterization of the Subclass B3 Metallo-beta-Lactamase PJM-1 from Pseudoxanthomonas japonensis. Antimicrob. Agents Chemother. 2022, 66, e0069122. [Google Scholar] [CrossRef]
- Viana, A.T.; Caetano, T.; Covas, C.; Santos, T.; Mendo, S. Environmental superbugs: The case study of Pedobacter spp. Environ. Pollut. 2018, 241, 1048–1055. [Google Scholar] [CrossRef]
- Thaller, M.C.; Borgianni, L.; Di Lallo, G.; Chong, Y.; Lee, K.; Dajcs, J.; Stroman, D.; Rossolini, G.M. Metallo-beta-lactamase production by Pseudomonas otitidis: A species-related trait. Antimicrob. Agents Chemother. 2011, 55, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Park, K.S.; Kim, T.Y.; Kim, J.H.; Lee, J.H.; Jeon, J.H.; Karim, A.M.; Malik, S.K.; Lee, S.H. PNGM-1, a novel subclass B3 metallo-beta-lactamase from a deep-sea sediment metagenome. J. Glob. Antimicrob. Resist. 2018, 14, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Salimraj, R.; Zhang, L.; Hinchliffe, P.; Wellington, E.M.; Brem, J.; Schofield, C.J.; Gaze, W.H.; Spencer, J. Structural and Biochemical Characterization of Rm3, a Subclass B3 Metallo-beta-Lactamase Identified from a Functional Metagenomic Study. Antimicrob. Agents Chemother. 2016, 60, 5828–5840. [Google Scholar] [CrossRef]
- Wilson, L.A.; Knaven, E.G.; Morris, M.T.; Monteiro Pedroso, M.; Schofield, C.J.; Bruck, T.B.; Boden, M.; Waite, D.W.; Hugenholtz, P.; Guddat, L.; et al. Kinetic and Structural Characterization of the First B3 Metallo-beta-Lactamase with an Active-Site Glutamic Acid. Antimicrob. Agents Chemother. 2021, 65, e0093621. [Google Scholar] [CrossRef] [PubMed]
- Wachino, J.; Yoshida, H.; Yamane, K.; Suzuki, S.; Matsui, M.; Yamagishi, T.; Tsutsui, A.; Konda, T.; Shibayama, K.; Arakawa, Y. SMB-1, a novel subclass B3 metallo-beta-lactamase, associated with ISCR1 and a class 1 integron, from a carbapenem-resistant Serratia marcescens clinical isolate. Antimicrob. Agents Chemother. 2011, 55, 5143–5149. [Google Scholar] [CrossRef] [PubMed]
- Vella, P.; Miraula, M.; Phelan, E.; Leung, E.W.; Ely, F.; Ollis, D.L.; McGeary, R.P.; Schenk, G.; Mitic, N. Identification and characterization of an unusual metallo-beta-lactamase from Serratia proteamaculans. J. Biol. Inorg. Chem. 2013, 18, 855–863. [Google Scholar] [CrossRef] [PubMed]
- Rossolini, G.M.; Condemi, M.A.; Pantanella, F.; Docquier, J.D.; Amicosante, G.; Thaller, M.C. Metallo-beta-lactamase producers in environmental microbiota: New molecular class B enzyme in Janthinobacterium lividum. Antimicrob. Agents Chemother. 2001, 45, 837–844. [Google Scholar] [CrossRef]
- Poirel, L.; Naas, T.; Nicolas, D.; Collet, L.; Bellais, S.; Cavallo, J.D.; Nordmann, P. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-beta-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 2000, 44, 891–897. [Google Scholar] [CrossRef]
- Walsh, T.R.; Toleman, M.A. The emergence of pan-resistant Gram-negative pathogens merits a rapid global political response. J. Antimicrob. Chemother. 2012, 67, 1–3. [Google Scholar] [CrossRef]
- Haruta, S.; Yamaguchi, H.; Yamamoto, E.T.; Eriguchi, Y.; Nukaga, M.; O’Hara, K.; Sawai, T. Functional analysis of the active site of a metallo-beta-lactamase proliferating in Japan. Antimicrob. Agents Chemother. 2000, 44, 2304–2309. [Google Scholar] [CrossRef]
- Oelschlaeger, P.; Mayo, S.L.; Pleiss, J. Impact of remote mutations on metallo-beta-lactamase substrate specificity: Implications for the evolution of antibiotic resistance. Protein Sci. 2005, 14, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Yano, H.; Kuga, A.; Okamoto, R.; Kitasato, H.; Kobayashi, T.; Inoue, M. Plasmid-encoded metallo-beta-lactamase (IMP-6) conferring resistance to carbapenems, especially meropenem. Antimicrob. Agents Chemother. 2001, 45, 1343–1348. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.M.; Pegg, K.M.; Oelschlaeger, P. The sequence-activity relationship between metallo-beta-lactamases IMP-1, IMP-6, and IMP-25 suggests an evolutionary adaptation to meropenem exposure. Antimicrob. Agents Chemother. 2012, 56, 6403–6406. [Google Scholar] [CrossRef] [PubMed]
- Riccio, M.L.; Franceschini, N.; Boschi, L.; Caravelli, B.; Cornaglia, G.; Fontana, R.; Amicosante, G.; Rossolini, G.M. Characterization of the metallo-beta-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of bla(IMP) allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 2000, 44, 1229–1235. [Google Scholar] [CrossRef] [PubMed]
- Bebrone, C.; Delbruck, H.; Kupper, M.B.; Schlomer, P.; Willmann, C.; Frere, J.M.; Fischer, R.; Galleni, M.; Hoffmann, K.M. The structure of the dizinc subclass B2 metallo-beta-lactamase CphA reveals that the second inhibitory zinc ion binds in the histidine site. Antimicrob. Agents Chemother. 2009, 53, 4464–4471. [Google Scholar] [CrossRef] [PubMed]
- Horsfall, L.E.; Garau, G.; Lienard, B.M.; Dideberg, O.; Schofield, C.J.; Frere, J.M.; Galleni, M. Competitive inhibitors of the CphA metallo-beta-lactamase from Aeromonas hydrophila. Antimicrob. Agents Chemother. 2007, 51, 2136–2142. [Google Scholar] [CrossRef] [PubMed]
- Lienard, B.M.; Garau, G.; Horsfall, L.; Karsisiotis, A.I.; Damblon, C.; Lassaux, P.; Papamicael, C.; Roberts, G.C.; Galleni, M.; Dideberg, O.; et al. Structural basis for the broad-spectrum inhibition of metallo-beta-lactamases by thiols. Org. Biomol. Chem. 2008, 6, 2282–2294. [Google Scholar] [CrossRef]
- Lassaux, P.; Hamel, M.; Gulea, M.; Delbruck, H.; Mercuri, P.S.; Horsfall, L.; Dehareng, D.; Kupper, M.; Frere, J.M.; Hoffmann, K.; et al. Mercaptophosphonate compounds as broad-spectrum inhibitors of the metallo-beta-lactamases. J. Med. Chem. 2010, 53, 4862–4876. [Google Scholar] [CrossRef]
- Fonseca, F.; Bromley, E.H.; Saavedra, M.J.; Correia, A.; Spencer, J. Crystal structure of Serratia fonticola Sfh-I: Activation of the nucleophile in mono-zinc metallo-beta-lactamases. J. Mol. Biol. 2011, 411, 951–959. [Google Scholar] [CrossRef]
- Hinchliffe, P.; Moreno, D.M.; Rossi, M.A.; Mojica, M.F.; Martinez, V.; Villamil, V.; Spellberg, B.; Drusano, G.L.; Banchio, C.; Mahler, G.; et al. 2-Mercaptomethyl Thiazolidines (MMTZs) Inhibit All Metallo-beta-Lactamase Classes by Maintaining a Conserved Binding Mode. ACS Infect. Dis. 2021, 7, 2697–2706. [Google Scholar] [CrossRef]
- Nauton, L.; Kahn, R.; Garau, G.; Hernandez, J.F.; Dideberg, O. Structural insights into the design of inhibitors for the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia. J. Mol. Biol. 2008, 375, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Crisp, J.; Conners, R.; Garrity, J.D.; Carenbauer, A.L.; Crowder, M.W.; Spencer, J. Structural basis for the role of Asp-120 in metallo-beta-lactamases. Biochemistry 2007, 46, 10664–10674. [Google Scholar] [CrossRef] [PubMed]
- Hinchliffe, P.; Tanner, C.A.; Krismanich, A.P.; Labbe, G.; Goodfellow, V.J.; Marrone, L.; Desoky, A.Y.; Calvopina, K.; Whittle, E.E.; Zeng, F.; et al. Structural and Kinetic Studies of the Potent Inhibition of Metallo-beta-lactamases by 6-Phosphonomethylpyridine-2-carboxylates. Biochemistry 2018, 57, 1880–1892. [Google Scholar] [CrossRef] [PubMed]
- Leiros, H.K.; Borra, P.S.; Brandsdal, B.O.; Edvardsen, K.S.; Spencer, J.; Walsh, T.R.; Samuelsen, O. Crystal structure of the mobile metallo-beta-lactamase AIM-1 from Pseudomonas aeruginosa: Insights into antibiotic binding and the role of Gln157. Antimicrob. Agents Chemother. 2012, 56, 4341–4353. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Saez, I.; Mercuri, P.S.; Papamicael, C.; Kahn, R.; Frere, J.M.; Galleni, M.; Rossolini, G.M.; Dideberg, O. Three-dimensional structure of FEZ-1, a monomeric subclass B3 metallo-beta-lactamase from Fluoribacter gormanii, in native form and in complex with D-captopril. J. Mol. Biol. 2003, 325, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.M.; Herman, R.; Ghiglione, B.; Kerff, F.; D’Amico Gonzalez, G.; Bouillenne, F.; Galleni, M.; Handelsman, J.; Charlier, P.; Gutkind, G.; et al. Crystal structure and kinetic analysis of the class B3 di-zinc metallo-beta-lactamase LRA-12 from an Alaskan soil metagenome. PLoS ONE 2017, 12, e0182043. [Google Scholar] [CrossRef] [PubMed]
- Selleck, C.; Pedroso, M.M.; Wilson, L.; Krco, S.; Knaven, E.G.; Miraula, M.; Mitic, N.; Larrabee, J.A.; Bruck, T.; Clark, A.; et al. Structure and mechanism of potent bifunctional beta-lactam- and homoserine lactone-degrading enzymes from marine microorganisms. Sci. Rep. 2020, 10, 12882. [Google Scholar] [CrossRef]
- Kumarasamy, K.K.; Toleman, M.A.; Walsh, T.R.; Bagaria, J.; Butt, F.; Balakrishnan, R.; Chaudhary, U.; Doumith, M.; Giske, C.G.; Irfan, S.; et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010, 10, 597–602. [Google Scholar] [CrossRef]
- Zhang, H.; Hao, Q. Crystal structure of NDM-1 reveals a common beta-lactam hydrolysis mechanism. FASEB J. 2011, 25, 2574–2582. [Google Scholar] [CrossRef]
- Kim, Y.; Tesar, C.; Mire, J.; Jedrzejczak, R.; Binkowski, A.; Babnigg, G.; Sacchettini, J.; Joachimiak, A. Structure of apo- and monometalated forms of NDM-1--a highly potent carbapenem-hydrolyzing metallo-beta-lactamase. PLoS ONE 2011, 6, e24621. [Google Scholar] [CrossRef]
- King, D.; Strynadka, N. Crystal structure of New Delhi metallo-beta-lactamase reveals molecular basis for antibiotic resistance. Protein Sci. 2011, 20, 1484–1491. [Google Scholar] [CrossRef] [PubMed]
- Palacios, A.R.; Mojica, M.F.; Giannini, E.; Taracila, M.A.; Bethel, C.R.; Alzari, P.M.; Otero, L.H.; Klinke, S.; Llarrull, L.I.; Bonomo, R.A.; et al. The Reaction Mechanism of Metallo-beta-Lactamases Is Tuned by the Conformation of an Active-Site Mobile Loop. Antimicrob. Agents Chemother. 2019, 63, 10–1128. [Google Scholar] [CrossRef]
- Spyrakis, F.; Santucci, M.; Maso, L.; Cross, S.; Gianquinto, E.; Sannio, F.; Verdirosa, F.; De Luca, F.; Docquier, J.D.; Cendron, L.; et al. Virtual screening identifies broad-spectrum beta-lactamase inhibitors with activity on clinically relevant serine- and metallo-carbapenemases. Sci. Rep. 2020, 10, 12763. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.A.; Martinez, V.; Hinchliffe, P.; Mojica, M.F.; Castillo, V.; Moreno, D.M.; Smith, R.; Spellberg, B.; Drusano, G.L.; Banchio, C.; et al. 2-Mercaptomethyl-thiazolidines use conserved aromatic-S interactions to achieve broad-range inhibition of metallo-beta-lactamases. Chem. Sci. 2021, 12, 2898–2908. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, L.J.; Bahr, G.; Nakashige, T.G.; Nolan, E.M.; Bonomo, R.A.; Vila, A.J. Membrane anchoring stabilizes and favors secretion of New Delhi metallo-beta-lactamase. Nat. Chem. Biol. 2016, 12, 516–522. [Google Scholar] [CrossRef]
- Oelschlaeger, P.; Dhungana, R. Automated class B b-lactamase (BBL) renumbering of NDM enzymes. In Proceedings of the ASM Microbe, Washington, DC, USA, 9–13 June 2022. [Google Scholar]
- Vogel, C.; Widmann, M.; Pohl, M.; Pleiss, J. A standard numbering scheme for thiamine diphosphate-dependent decarboxylases. BMC Biochem. 2012, 13, 24. [Google Scholar] [CrossRef]
- Davies, D.T.; Everett, M. Designing Inhibitors of beta-Lactamase Enzymes to Overcome Carbapenem Resistance in Gram-Negative Bacteria. Acc. Chem. Res. 2021, 54, 2055–2064. [Google Scholar] [CrossRef]
- Oelschlaeger, P. beta-Lactamases: Sequence, Structure, Function, and Inhibition. Biomolecules 2021, 11, 986. [Google Scholar] [CrossRef]


| Name | Derivation | Pattern | Pattern Explanation | Reference |
|---|---|---|---|---|
| AFM | Alcaligenes faecalis MBL | GsM | Genus species MBL | 36000902 [60] |
| ANA | Anaeromyxobacter spp. | Gen | Genus | 29020980 [61] |
| BcII | Bacillus cereus type II BL | Gs | Genus species | 3930467 [46] |
| BIM | Belém imipenemase | LSu | Location Substrate Specificity | 37038995 [62] |
| BlaB | β-lactamase class B | Other * | 10858348 [63] | |
| CAM | Central Alberta MBL | LoM | Location MBL | 30789204 [64] |
| CfiA (CcrA) | Cefoxitin and imipenem-resistant A Cefoxitin and carbapenem-resistant A | Other Other | 2110145 [65] 2121094 [66] | |
| CGB | Chryseobacterium gleum class B | GsB | Genus species BBL | 12183230 [67] |
| CHM | Chryseobacterium MBL | GeM | Genus MBL | 37047008 [68] |
| CEMC19 | Cefixime (cem) resistance | Other | 35768448 [69] | |
| CrxA | Carbapenem-resistant Bacteroides xylanisolvens A | Other | 35296904 [70] | |
| CX1 | Isolated from clone CX1 | Other | 31487611 [71] | |
| DIM | Dutch imipenemase | LSu | Location Substrate Specificity | 20308383 [72] |
| EBR | Empedobacter brevis | Gsp | Genus species | 12234848 [73] |
| ECV | Echinicola vietnamensis | Ges | Genus species | 29020980 [61] |
| ElBla2 | Erythrobacter litoralis β-lactamase 2 | Other | 21468894 [74] | |
| FIA | Fibrella aestuarina | Ges | Genus species | 29020980 [61] |
| FIM | Florence imipenemase | LSu | Location Substrate Specificity | 23114762 [75] |
| GIM | German imipenemase | LSu | Location Substrate Specificity | 15561840 [76] |
| GMB | German MBL | LMB | Location MBL | 35257174 [77] |
| GRD23 | Gemmatimonadetes resistant Denmark | Other | 28082950 [78] | |
| HBA | Hirschia baltica | Gsp | Genus species | 22675580 [79] |
| HMB | Hamburg MBL | LMB | Location MBL | 28065891 [80] |
| IMP | Imipenemase | Sub | Substrate Specificity | 8141584 [81] |
| IND | Chryseobacterium indologenes | spe | species | 10077836 [82] |
| JOHN | Chryseobacterium johnsoniae | spec | species | 12562690 [83] |
| KHM | Kyorin University Hospital MBL | LoM | Location MBL | 18765691 [84] |
| MOC | Myroides odoratus carbapenemase | GsM | Genus species MBL | |
| MUS | Myroides odoratimimus | spe | species | 12384365 [85] |
| MYO | Myroides odoratimimus | Ges | Genus species | 29020980 [61] |
| MYX | Myxococcus xanthus | Ges | Genus species | 29020980 [61] |
| NDM | New Delhi MBL | LoM | Location MBL | 19770275 [86] |
| ORR | Ornithobacterium rhinotracheale | Ges | Genus species | 29020980 [61] |
| PAN | Pseudobacteriovorax antillogorgiicola | Gsp | Genus species | 31396187 [87] |
| PEDO | Pedobacter roseus | Genu | Genus | 26482314 [88] |
| PKB | Pontibacter korlensis class B | GsB | Genus species BBL | 26057562 [89] |
| PST | Pseudomonas stutzeri | Gsp | Genus species | 29020980 [61] |
| SFB | Shewanella frigidimarina class B | GsB | Genus species BBL | 15772146 [90] |
| SHD | Shewanella denitrificans | Ges | Genus species | 29020980 [61] |
| SHN | Shewanella denitrificans | Ges | Genus species | 29020980 [61] |
| SIM | Seoul imipenemase | LSS | Location Substrate Specificity | 16251286 [91] |
| SLB | Shewanella livinstonensis class B | GsB | Genus species BBL | 15772146 [90] |
| SPM | Sao Paulo MBL | LoM | Location MBL | 12407123 [92] |
| SPN79 | Spain | Loc | Location | 28082950 [78] |
| SPS | Sediminispirochaeta smaragdinae | Ges | Genus species | 29020980 [61] |
| STA | Stigmatella aurantiaca | Ges | Genus species | 29020980 [61] |
| SZM | Shenzhen MBL | LoM | Location MBL | 36225370 [93] |
| TMB | Tripoli MBL | LMB | Location MBL | 22290947 [94] |
| TTU | Teredinibacter turnerae | Gsp | Genus species | 29020980 [61] |
| TUS | Myroides odoratus | spe | species | 12384365 [85] |
| VAM (VMB) | Vibrio alginolyticus MBL Vibrio alginolyticus MBL | GsM GMB | Genus species MBL Genus MBL | 34424042 [95] 34097496 [96] |
| VIM | Verona integron-borne MBL | LIM | Location integron-borne MBL | 10390207 [97] |
| VMB | Vibrio MBL | GMB | Genus MBL | 32293144 [98] |
| VMH | Vibrio vulnificus metallohydrolase | GMH | Genus metallohydrolase | 34228542 [99] |
| WUS | Wenzhou Monopterus albus | Lsp | Location species | 36532482 [100] |
| ZHO | Zhongshania aliphaticivorans | Gen | Genus | 30778547 [101] |
| ZOG | Zobellia galactanivorans | Ges | Genus species | 29020980 [61] |
| Name | Derivation | Pattern | Pattern Explanation | Reference |
|---|---|---|---|---|
| CphA | Carbapenem-hydrolyzing enzyme A | Other | 1856163 [102] | |
| CVI | Chromobacterium violaceum | Gsp | Genus species | 37513808 [103] |
| PFM | Pseudomonas fluorescens MBL | GsM | Genus species MBL | 31685461 [104] |
| Sfh | Serratia fonticola carbapenem hydrolase | GsH | Genus species hydrolase | 12821491 [105] |
| YEM | Yersinia mollaretii | Ges | Genus species | 32540974 [106] |
| Name | Derivation | Pattern | Pattern Explanation | Reference |
|---|---|---|---|---|
| AIM | Adelaide imipenemase | LSu | Location Substrate Specificity | 22985886 [107] |
| ALG6 | Algeria | Loc | Location | 28082950 [78] |
| ALG11 | Algeria | Loc | Location | 28082950 [78] |
| AM1 | Isolated from clone AM1 | Other | 31487611 [71] | |
| B3SU1 | B3 subclass uncultured 1 | Other | ||
| B3SU2 | B3 subclass uncultured 2 | Other | ||
| BJP | Bradyrhizobium japonicum | Gsp | Genus species | 16723554 [108] |
| BLEG | Bacillus lehensis G | GspG | Genus species | 34502284 [109] |
| CAR | Erwinia caratovora | spe | species | 18443127 [110] |
| CAU | Caulobacter crescentus | Gen | Genus | 12019096 [111] |
| CHI | Chitinophaga pinensis | Gen | Genus | |
| CPS | Chryseobacterium piscium Stok-1 | Gss | Genus species strain | 26482314 [88] |
| CRD3 | CRUCIAL Denmark | Other | 28082950 [78] | |
| CSR | Chronobacter sakazakii resistant | Gsr | Genus species resistant | 32542533 [112] |
| DHT2 | Dossenheim plantomycin treated | Other | 28082950 [78] | |
| EAM | Erythrobacter aquimaris MBL | GsM | Genus species MBL | 22850693 [113] |
| ECM | Erythrobacter citreus MBL | GsM | Genus species MBL | 22850693 [113] |
| EFM | Erythrobacter flavus MBL | GsM | Genus species MBL | 22850693 [113] |
| ELM | Erythrobacter longus MBL | GsM | Genus species MBL | 22850693 [113] |
| ESP | Extended-spectrum BL | Other | 26482314 [88] | |
| EVM | Erythrobacter vulgaris MBL | GsM | Genus species MBL | 22850693 [113] |
| FEZ | Fluoribacter gormanii endogenous zinc BL | Other | 10817705 [114] | |
| GOB | Chryseobacterium meningosepticum class B | spB | species BBL | 10858348 [63] |
| L1 | Labile enzyme 1 from Stenotrophomonas maltophilia | Other | 8018721 [47] | |
| LMB | Linz MBL | LMB | Location MBL | 29897538 [115] |
| LRA2 | Lactam resistant from Alaskan soil | Other | 18843302 [116] | |
| LRA3 | Lactam resistant from Alaskan soil | Other | 18843302 [116] | |
| LRA7 | Lactam resistant from Alaskan soil | Other | 18843302 [116] | |
| LRA8 | Lactam resistant from Alaskan soil | Other | 18843302 [116] | |
| LRA12 | Lactam resistant from Alaskan soil | Other | 18843302 [116] | |
| LRA17 | Lactam resistant from Alaskan soil | Other | 18843302 [116] | |
| LRA17 | Lactam resistant from Alaskan soil | Other | 18843302 [116] | |
| MIM | Maynooth imipenemase | LSS | Location Substrate Specificity | 26775612 [117] |
| MSI | Massilia oculi | Gen | Genus | 26482314 [88] |
| NWM | North Rhine-Westphalia MBL | LoM | Location MBL | |
| PAM | Pseudomonas alcaligenes MBL | GsM | Genus species MBL | 24356301 [118] |
| PEDO | Pedobacter roseus | Genu | Genus | 26482314 [88] |
| PJM | Pseudoxanthomonas japonensis MBL | GsM | Genus species MBL | 35943258 [119] |
| PLN | Pedobacter lusitanus NL19 | Gss | Genus species strain | 30029312 [120] |
| POM | Pseudomonas otitidis MBL | GsM | Genus species MBL | 21060106 [121] |
| PNGM | Papua New Guinea MBL | LocM | Location MBL | 29842976 [122] |
| RM3 | Isolated from clone RM3 | Other | 27431213 [123] | |
| SAM | Simiduia agarivorans MBL | GsM | Genus species MBL | |
| SER | Salmonella enterica resistance | Gsr | Genus species resistance | 32542533 [112] |
| SIE | Sphingobium indicum B3-E (E116) | GsE | Genus species B3-E | 34310207 [124] |
| SIQ | Sphingobium indicum B3-Q (Q116) | GsQ | Genus species B3-Q | 34310207 [124] |
| SMB | Serratia marcescens class B | GsB | Genus species BBL | 21876060 [125] |
| SPG | Sphingomonas | Gen | Genus | 26482314 [88] |
| SPR | Serratia proteamaculans | Gsp | Genus species | 23982345 [126] |
| SSE | Sphingopyxis sp. Enzyme? | GsE | Genus species Enzyme | 32542533 [112] |
| THIN-B | Janthinobacterium lividum class B | GenB | Genus BBL | 11181369 [127] |
| MBL Subclass | Zn1 Ligands | Zn2 Ligands | ||||
|---|---|---|---|---|---|---|
| B1 | H116 | H118 | H196 | D120 | C221 | H263 |
| B2 | N116 | H118 | H196 | D120 | C221 | H263 |
| B3 | H/Q/E116 | H/R118 | H196 | D120 | H/Q121 | H/K263 |
| Original Number (FASTA Position) | Modification | BBL Number |
|---|---|---|
| 1 to 110 | −6 | −5 to 104 |
| 111 to 134 | −4 | 107 to 130 |
| 135 to 153 | −3 | 132 to 150 |
| 154 | −4 + a | 150 a |
| 155 to 201 | +7 | 162 to 208 |
| 202 to 225 | +13 | 215 to 238 |
| 226 to 239 | +15 | 241 to 254 |
| 240 | +14 + a | 254 a |
| 241 | +13 + b | 254 b |
| 242 to 253 | +13 | 255 to 266 |
| 254 to 270 | +41 | 295 to 311 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oelschlaeger, P.; Kaadan, H.; Dhungana, R. Strategies to Name Metallo-β-Lactamases and Number Their Amino Acid Residues. Antibiotics 2023, 12, 1746. https://doi.org/10.3390/antibiotics12121746
Oelschlaeger P, Kaadan H, Dhungana R. Strategies to Name Metallo-β-Lactamases and Number Their Amino Acid Residues. Antibiotics. 2023; 12(12):1746. https://doi.org/10.3390/antibiotics12121746
Chicago/Turabian StyleOelschlaeger, Peter, Heba Kaadan, and Rinku Dhungana. 2023. "Strategies to Name Metallo-β-Lactamases and Number Their Amino Acid Residues" Antibiotics 12, no. 12: 1746. https://doi.org/10.3390/antibiotics12121746
APA StyleOelschlaeger, P., Kaadan, H., & Dhungana, R. (2023). Strategies to Name Metallo-β-Lactamases and Number Their Amino Acid Residues. Antibiotics, 12(12), 1746. https://doi.org/10.3390/antibiotics12121746








