1. Introduction
Antimicrobial resistance is currently one of the most critical issues in both human and animal health. Conservative estimates already suggest annual human deaths numbering 700,000 and economic losses in the trillions of dollars due to this issue [
1], with projected mortality figures reaching 10 million by 2050 [
2]. Antibiotics have been a cornerstone in treating infectious diseases since the 1940s, with the discovery of several new classes of antibiotics during the 1960s representing the golden age of antibiotics [
3]. Infectious diseases rank as the second leading cause of global mortality, with particular concern arising from panresistant strains of Gram-negative bacteria [
4]. Target-based antibiotic development has not yielded significant breakthroughs, necessitating a greater emphasis on target-based antibiotic research [
5].
From both animal and human health perspectives, it is crucial to delineate the precise mechanisms of resistance to various groups of active substances and explore potential correlations among them, particularly in terms of coresistance. Florfenicol is a broad-spectrum veterinary antibiotic belonging to the phenicol group, which functions as an inhibitor of bacterial protein synthesis, exerting bacteriostatic activity by targeting the 50S ribosome [
6]. Bacteria develop resistance to florfenicol through various mechanisms, ranging from antibiotic modification to efflux pumps. Currently, resistance to florfenicol is on the rise due to global trade and misuse [
6].
In our research, we adapted [
7] the MEGA-plate method [
8], for the first time in veterinary medicine, to investigate the phenotypic and genotypic expressions of induced resistance. Within the categorization established by the Antimicrobial Advice Ad Hoc Expert Group (AMEG)—encompassing categories A, B, C and D—AMEG B antibiotics are recognized as critically important for human medicine. Despite this designation, these antibiotics continue to be extensively utilized in farm animals, significantly contributing to the widespread emergence of antibiotic resistance. Notably, resistance is on the rise in the poultry sector, affecting AMEG B antibiotics such as third and fourth generation cephalosporins and fluoroquinolones. It is crucial to underscore that the use of cephalosporins in poultry is not authorized [
9,
10], highlighting the potential for the concurrent development of coresistance. Considering the AMEG B category, it is not advisable to prioritize substances within this category as the primary choice in veterinary medicine. When selecting a treatment, it is essential to conduct sensitivity testing for an accurate determination of the appropriate substance. Based on the results, a judicious approach involves favoring substances from the less critical AMEG D or AMEG C categories. This strategy aligns with the objective of promoting responsible and sustainable use of antimicrobial agents in veterinary practices [
11].
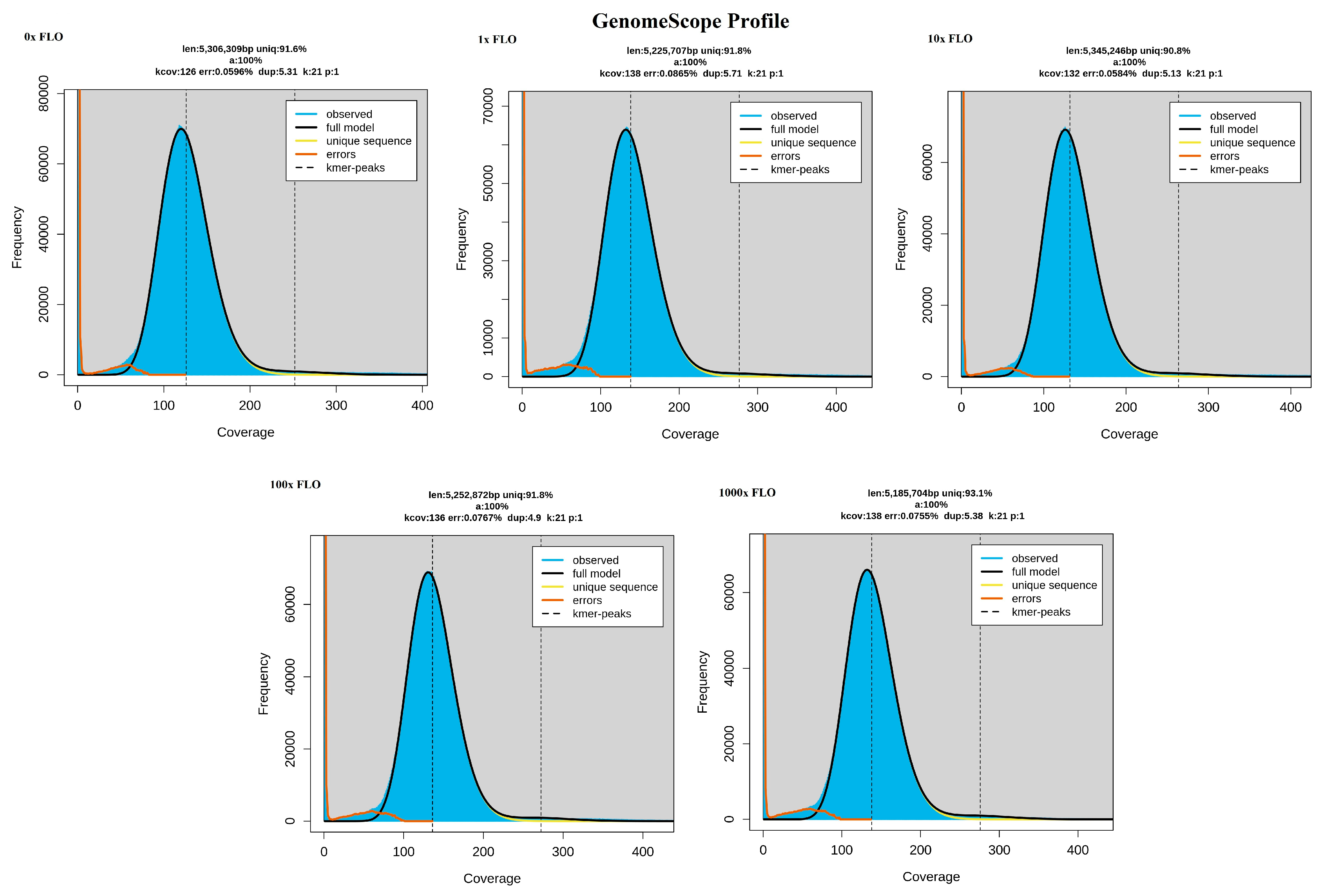
This system allows for the migration and adaptation of the bacteria under study in a large space, which is also a structured environment. The movement of the bacteria, subjected to continuous selection pressure, is enabled by the uppermost soft agar layer, which spreads by chemotaxis as it consumes nutrients. The size of the plate allows for the formation of mutations during the numerous generation changes and maintains the gradient of the antibiotic under study despite diffusion [
8,
12]. It is, however, a slower process than adaptation in a microenvironment [
13], which can be explained by the additional time of 10–12 days, on average, required for the plate to fully grow. Furthermore, a limitation of the method may be that among the development of several parallel lineages, it is not certain that the frontline is driven by those that are best adapted [
8]. The method is a population genetics study describable by Muller plots [
14]. Our objective was to examine the impact of florfenicol on the development of coresistance to specific active substances approved for poultry use, with particular emphasis on cephalosporins not authorized for poultry use, through evolutionary and co-selection studies.
3. Discussion
In this study, we employed the modified MEGA-plate method [
7] to investigate the development of antibiotic resistance in
E. coli strains under selection pressure by veterinary antibiotic florfenicol. The escalating resistance to florfenicol, a commonly used antibiotic in the swine industry, is concerning, with resistance levels rising from 2.1% in 2004 to 18.1% in 2017 [
27], Additionally, in broiler chickens, florfenicol administration during rearing has been linked to a significant increase in
E. coli resistance to phenicols [
28].
Our findings reveal a phenotypic decrease in florfenicol susceptibility, attributed to genome mutations mediating the upregulation of efflux pump expression. These genome mutations are a consequence of stress-induced changes in the bacterium, which contribute to the expression of resistance genes. Additionally, it is possible that clones with active efflux pumps were selectively favored during this process. Langsrud et al. previously reported a 1.5–20× increase in MIC for antibiotics due to stress-induced cross-resistance to benzalkonium chloride [
29]. In our study, we observed a 2–167× increase in MIC values against various antibiotics following treatment with 1000× concentrations of florfenicol. Cheng et al. also demonstrated an increased MIC against tigecycline in
Acinetobacter baumani strain [
30]. Ding et al. were able to achieve similarly elevated MIC values in
Streptoccus strain by passaging [
31]. The antimicrobial agents were categorized based on their public health significance by AMEG of European Medicines Agency (EMA). Notably, florfenicol caused a substantial MIC increase to a veterinary fluoroquinolone, enrofloxacin, classified as an AMEG B [
32], thus highlighting the importance of cross-resistance development to critically important antimicrobials, although florfenicol itself is an AMEG C substance. Chueca et al. exposed
E. coli strains to essential oils for 10 days, leading to increased MIC values against several antibiotics [
33]. The exposure of bacteria to antibiotics at subinhibitory concentrations can induce an SOS stress response, potentially causing a transient elevation in mutation rates [
34,
35]. Notably, no prior studies have reported such am extensive cross-resistance development across different antibiotic classes following exposure to varying doses of florfenicol. Resistance to penicillins and cephalosporins is presumed to be linked to efflux pump activation.
Among the 44 identified ARGs, the presence of genes responsible for enzymatic inactivation, particularly
ampC and
ampH, is of particular concern. These genes are associated with the overproduction of beta-lactamase enzymes, which can induce resistance to cephalosporins. Resistance to critically important agents includes the presence of genes, such as
bacA,
pmtF,
eptA and
ugd, which can confer resistance through target modification against colistin. We identified the presence of the
marA gene, which can induce multidrug resistance through reduced permeability. Additionally, we identified two genes responsible for developing resistance to drug classes (AMEG A) reserved for human healthcare (
marA and
tolC). Furthermore, 59.1% of all identified genes were found to be responsible for resistance to essential drug classes for both public health and animal health (
acrA,
acrB,
acrE,
acrF,
acrS,
ampC,
ampH,
bacA,
CRP,
emrA,
emrB,
emrR,
eptA,
evgA,
evgS,
gadW,
H-NS,
marA,
mdtE,
mdtF,
mdtH,
mdtM,
pmrF,
tolC,
ugd and
cojI). Notably, the
ampH and
bacA genes were encoded as MGEs, potentially enabling the transfer of these crucial ARGs to other bacterial strains. Surprisingly, we did not detect the
floR gene, responsible for florfenicol resistance in
Salmonella enterica, in our
E. coli samples [
36].
The activation of multidrug efflux pumps appears to underlie the elevated MIC values against most tested drugs. Previous research by Ma et al. elucidated the responsiveness of the
acrAB gene-encoded complex pump system to various environmental stresses [
37], In our study, we observed deletion mutations in the
marR gene for the 10× and 100× florfenicol samples, and it is well-documented that mutations in the
marR gene can upregulate the
acrAB operon [
37], a key transcriptional regulator of multiple antibiotic resistance genes [
38]. Additionally, Pourahmad et al. reported a reduced antibiotic efficacy in
E. coli due to the overactivation of the
acrAB-tolC efflux pump complex [
39]. Yaqoob et al. described the
acrR and
robA genes as regulators of this pump system [
40], and we identified mutations in these genes in the 1000× sample. Interestingly, when synthetic antibiotics, honey, and various plant alkaloids were applied, they enhanced pump system function and reduced oxidative stress in an
E. coli strain [
39]. Maslowska et al. identified two pivotal genes,
lexA and
recA, governing the SOS stress response, which involves the upregulation of at least 50 additional genes [
41]. In our study, we consistently detected both genes in all samples and identified most of the SOS-box genes under their regulation. Notably, single-point mutations in genes encoding multidrug efflux pumps were common, potentially accounting for the observed gene expression.
However, it appears that the action of these multidrug efflux pumps may not be adequate to effectively expel peptide and aminoglycoside antibiotics from
E. coli. Babosan et al. demonstrated that the presence of
qnr genes and deletion of the
recB gene contributed to SOS stress induction by aminoglycosides [
42]. In our study, we identified the
recB gene in our
E. coli strain but observed no mutations. This might explain the lack of an increase in MIC against neomycin. The minor increase in MIC for beta-lactam antibiotics is likely attributed to efflux pump activity, although we do not suspect the expression of genes responsible for enzymatic inactivation. Such gene expression would have resulted in a much greater increase in MIC values, and we did not detect mutations in these genes.
In summary, our findings suggest that reduced susceptibility to florfenicol and other antimicrobials may be linked to mutations affecting three types of multidrug efflux pump systems encoded by the
mdtABC-tolC,
emrAB-tolC and
acrAB-tolC genes. Furthermore, mutations in the
acrR,
emrR and
rpbA genes that regulate these efflux pumps enhance their functionality. While our study unveils valuable insights into the mechanisms driving antimicrobial resistance in
E. coli, further research is needed to fully comprehend the intricate interplay between stress-induced gene activation, efflux pump systems and genomic mutations in the context of antibiotic resistance. Our results support the need for future in silico studies targeting the expression of genes associated with established resistance. A deletion of the
marR gene was shown to occur in response to 10× and 100× concentrations of florfenicol, a deletion that Notka et al. have previously shown to act as a positive regulator of the
acrAB efflux pump system and to result in a 16× increase in MIC with fluoroquinolones [
43]; in our studies, we observed similar MIC increases of an order of magnitude not only for enrofloxacin but also for florfenicol, cephalosporin and oxytetracycline. The point mutations and deletions identified underscore the importance of resistance induced by previously demonstrated repressor genes in a cascade-like SOS stress response, but the association of individual agents in increasing resistance has not been previously investigated in a comprehensive manner like our study. We need to highlight the fact that certain active substances used in veterinary medicine may induce co-selection against other agents, such as cephalosporins, which may be of public health importance. Our results confirmed the value of continuing the study with transcriptomic methods to support the phenotypic expression of genotypically occurring mutations.
In veterinary practice, understanding and managing antibiotic resistance to antibiotics is of paramount importance for the preservation of animal health and the safety of animal populations. Our research has shed light on the fact that antibiotic resistance not only poses a significant threat to animals but can also have profound implications for human health. The spread of antibiotic resistance in the E. coli bacterium is of particular concern for both animal and human populations. The results of our studies indicate that the development of antibiotic resistance is closely tied to genetic mutations occurring in the genomes of bacteria exposed to antibiotics. Antibiotic resistance evolves as bacteria undergo genetic changes and activate genes that confer resistance to various antibiotics. The rise in resistance to critically important antibiotics, such as fluoroquinolones, is especially worrisome. These antibiotics are of great importance to human health, and the emergence of resistance poses a serious health risk to human populations.
5. Conclusions
Our results show that the development of antimicrobial resistance is closely linked to the activation of bacterial multidrug efflux pumps. Mutations in genes that determine these pumps may increase antibiotic resistance, which may explain the observed increase in MIC.
Stress factors (such as antibiotic use) may trigger SOS mechanisms that lead to mutations in the genome. Our study supports the hypothesis that the presence of florfenicol induces significant gene expression through mutations, the resulting multidrug resistance is mainly due to efflux pumps, and the expression of these pumps is affected by a single nucleic acid substitution. On the basis of these results, it may be particularly important to consider the administration of antioxidants alongside antibiotics, which may attenuate SOS processes. As a more far-reaching hypothesis, which needs to be tested, these processes may contribute not only to the spread of strains’ resistance to first-line agents but may also lead to the emergence of resistance to second-line agents, and, therefore, in this context, when selecting a second-line agent following unsuccessful treatment, it may be worthwhile to be careful to choose an agent to which sensitivity may still be maintained.
Understanding and monitoring antibiotic resistance to antibiotics used in veterinary practice is key to protecting animal health and, through this, human health. Our results can help to improve the use of antibiotics and contribute to preventing the spread of antibiotic resistance in both animal and human populations. The intertwined role of veterinary medicine and human health underlines the global importance of science and health in safeguarding our well-being.