Antimicrobial Resistance in Commensal Escherichia coli of the Porcine Gastrointestinal Tract
Abstract
1. Introduction
2. Antimicrobial Resistance in Escherichia coli of Porcine Origin: Surveillance Programmes
3. Resistance in Escherichia coli Isolated from Finisher Pigs: Data from Surveillance Programmes and Published Studies
4. Resistance in Escherichia coli Isolated from Pigs in Age Groups Other Than Finisher
5. Mechanisms of Antimicrobial Resistance in Escherichia coli
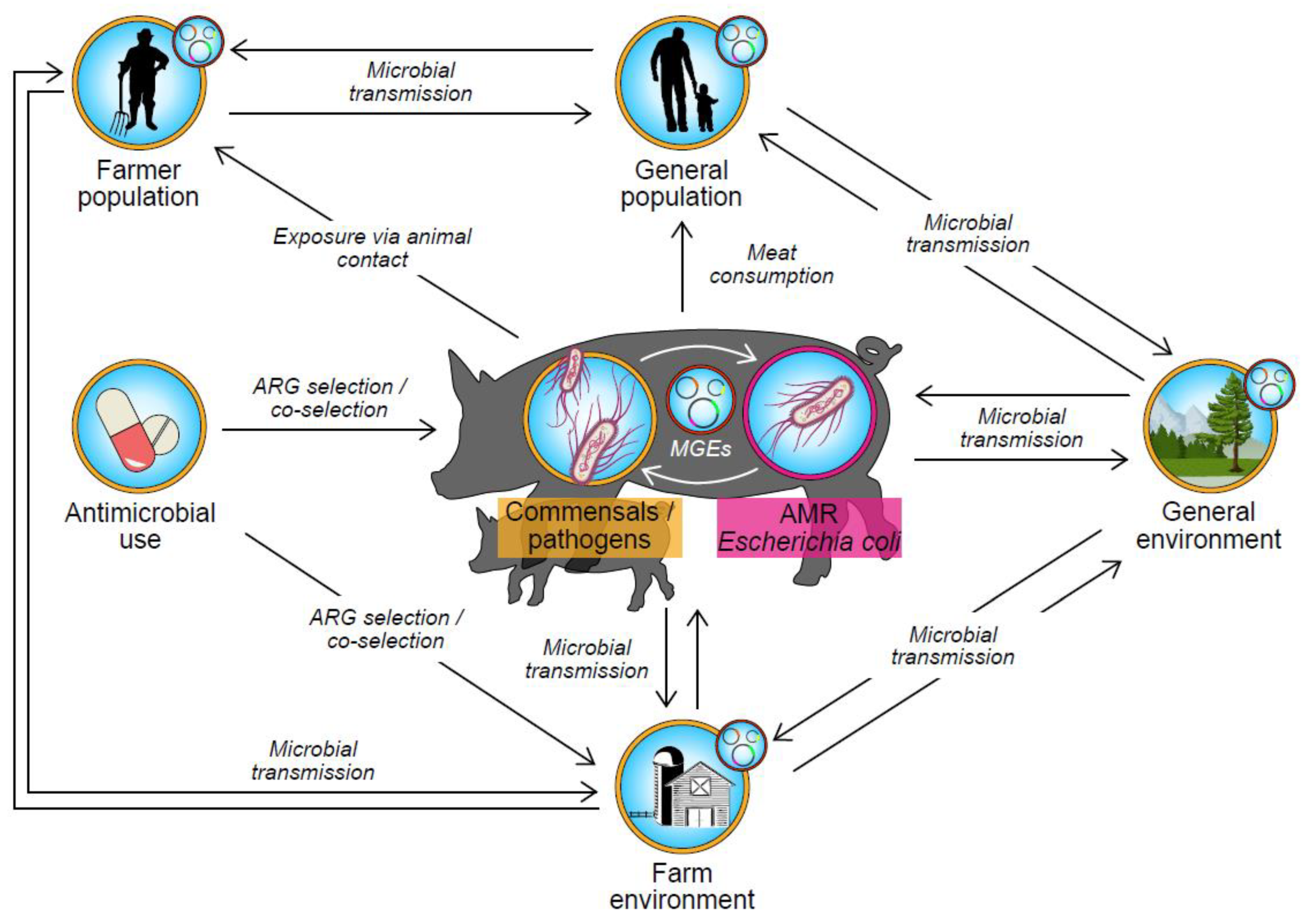
6. Relationship between AMU in Pig Farming and AMR in Escherichia coli
6.1. Ecological Associations between AMU and AMR in Escherichia coli
6.2. Intervention Studies Investigating Relationship between AMU and AMR
6.3. Observational Studies Investigating Relationship between AMU and AMR
7. Resistance to Extended Spectrum Cephalosporins, Fluoroquinolones, and Polymyxins
8. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Savageau, M.A. Escherichia coli Habitats, Cell Types, and Molecular Mechanisms of Gene Control. Am. Nat. 1983, 122, 732–744. [Google Scholar] [CrossRef]
- Tenaillon, O.; Skurnik, D.; Picard, B.; Denamur, E. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 2010, 8, 207–217. [Google Scholar] [CrossRef]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Nataro, J.P.; Kaper, J.B. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998, 11, 142–201. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority (EFSA) and European Centre for Disease Prevention and Control (ECDC). The European Union One Health 2021 Zoonoses Report. EFSA J. 2022, 20, 7666. [Google Scholar] [CrossRef]
- Tandogdu, Z.; Wagenlehner, F. Global epidemiology of urinary tract infections. Curr. Opin. Infect. Dis. 2016, 29, 73–79. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control (ECDC). Antimicrobial Resistance in the EU/EEA (EARS-Net)—Annual Epidemiological Report. 2021. Available online: https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2021 (accessed on 11 July 2023).
- Shane, A.L.; Mody, R.K.; Crump, J.A.; Tarr, P.I.; Steiner, T.S.; Kotloff, K.; Langley, J.M.; Wanke, C.; Warren, C.A.; Cheng, A.C.; et al. 2017 Infectious Diseases Society of America Clinical Practice Guidelines for the Diagnosis and Management of Infectious Diarrhea. Clin. Infect. Dis. 2017, 65, e45–e80. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Critically Important Antimicrobials for Human Medicine, 6th Revision. Geneva, Switzerland, 2019; Licence: CC BY-NC-SA 3.0 IGO. Available online: https://www.who.int/publications/i/item/9789241515528 (accessed on 21 July 2021).
- World Health Organization (WHO). Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. 2017. Available online: https://remed.org/wp-content/uploads/2017/03/lobal-priority-list-of-antibiotic-resistant-bacteria-2017.pdf (accessed on 21 September 2023).
- Bélanger, L.; Garenaux, A.; Harel, J.; Boulianne, M.; Nadeau, E.; Dozois, C.M. Escherichia coli from Animal Reservoirs as a Potential Source of Human Extraintestinal Pathogenic E. coli. FEMS Immunol. Med. Microbiol. 2011, 62, 1–10. [Google Scholar] [CrossRef]
- Manges, A.R.; Johnson, J.R. Food-Borne Origins of Escherichia coli Causing Extraintestinal Infections. Clin. Infect. Dis. 2012, 55, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Manges, A.R.; Johnson, J.R. Reservoirs of Extraintestinal Pathogenic Escherichia coli. Microbiol. Spectr. 2015, 3, 159–177. [Google Scholar] [CrossRef]
- Jørgensen, S.L.; Stegger, M.; Kudirkiene, E.; Lilje, B.; Poulsen, L.L.; Ronco, T.; Santos, T.; Kiil, K.; Bisgaard, M.; Pedersen, K.; et al. Diversity and Population Overlap between Avian and Human Escherichia coli Belonging to Sequence Type 95. mSphere 2019, 4, e00333-18. [Google Scholar] [CrossRef]
- Manges, A.R.; Smith, S.P.; Lau, B.J.; Nuval, C.J.; Eisenberg, J.; Dietrich, P.S.; Riley, L.W. Retail Meat Consumption and the Acquisition of Anti-microbial Resistant Escherichia coli Causing Urinary Tract Infections: A Case Control Study. Foodborne Pathog. Dis. 2007, 4, 419–431. [Google Scholar] [CrossRef]
- Manges, A.R. Escherichia coli and urinary tract infections: The role of poultry-meat. Clin. Microbiol. Infect. 2016, 22, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Aarestrup, F.M. The livestock reservoir for antimicrobial resistance: A personal view on changing patterns of risks, effects of interventions and the way forward. Phil. Trans. R Soc B. 2015, 370, 20140085. [Google Scholar] [CrossRef]
- Winokur, P.; Vonstein, D.; Hoffman, L.; Uhlenhopp, E.; Doe, G. Evidence for Transfer of CMY-2 AmpC β-Lactamase Plasmids between Escherichia coli and Salmonella Isolates from Food Animals and Humans. Antimicrob. Agents Chemother. 2001, 45, 2716–2722. [Google Scholar] [CrossRef]
- Mathew, A.G.; Liamthong, S.; Lin, J.; Hong, Y. Evidence of Class 1 Integron Transfer Between Escherichia coli and Salmonella spp. on Livestock Farms. Foodborne Pathog. Dis. 2009, 6, 959–964. [Google Scholar] [CrossRef] [PubMed]
- Trobos, M.; Lester, C.H.; Olsen, J.E.; Frimodt-Møller, N.; Hammerum, A.M. Natural transfer of sulphonamide and ampicillin resistance between Escherichia coli residing in the human intestine. J. Antimicrob. Chemoth. 2009, 63, 80–86. [Google Scholar] [CrossRef]
- FAOSTAT Crops and Livestocks Products. Available online: https://www.fao.org/faostat/en/#data/TCL (accessed on 2 October 2023).
- Mulchandani, R.; Wang, Y.; Gilbert, M.; Boeckel, T.P.V. Global trends in antimicrobial use in food-producing animals: 2020 to 2030. PLoS Glob. Public Health 2023, 3, e0001305. [Google Scholar] [CrossRef] [PubMed]
- Ludden, C.; Raven, K.E.; Jamrozy, D.; Gouliouris, T.; Blane, B.; Coll, F.; de Goffau, M.; Naydenova, P.; Horner, C.; Hernandez-Garcia, J.; et al. One Health Genomic Surveillance of Escherichia coli Demonstrates Distinct Lineages and Mobile Genetic Elements in Isolates from Humans versus Livestock. mBio 2019, 10, e02693-18. [Google Scholar] [CrossRef]
- Dorado-García, A.; Smid, J.H.; van Pelt, W.; Bonten, M.J.M.J.; Fluit, A.C.; van den Bunt, G.; Wagenaar, J.A.; Hordijk, J.; Dierikx, C.M.; Veldman, K.T.; et al. Molecular relatedness of ESBL/AmpC-producing Escherichia coli from humans, animals, food and the environment: A pooled analysis. J. Antimicrob. Chemother. 2018, 73, 339–347. [Google Scholar] [CrossRef]
- Day, M.J.; Hopkins, K.L.; Wareham, D.W.; Toleman, M.A.; Elviss, N.; Randall, L.; Teale, C.; Cleary, P.; Wiuff, C.; Doumith, M.; et al. Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and food chain-derived samples from England, Wales, and Scotland: An epidemiological surveillance and typing study. Lancet Infect. Dis. 2019, 19, 1325–1335. [Google Scholar] [CrossRef]
- Mughini-Gras, L.; Dorado-García, A.; van Duijkeren, E.; van den Bunt, G.; Dierikx, C.M.; Bonten, M.J.; Bootsma, M.C.; Schmitt, H.; Hald, T.; Evers, E.G.; et al. Attributable sources of community-acquired carriage of Escherichia coli containing β-lactam antibiotic resistance genes: A population-based modelling study. Lancet Planet Health 2019, 3, e357–e369. [Google Scholar] [CrossRef]
- Akwar, T.H.; Poppe, C.; Wilson, J.; Reid-Smith, R.J.; Dyck, M.; Waddington, J.; Shang, D.; Dassie, N.; McEwen, S.A. Risk factors for antimicrobial resistance among fecal Escherichia coli from residents on forty-three swine farms. Microb. Drug Resist. 2007, 13, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Dohmen, W.; Bonten, M.J.M.; Bos, M.E.H.; van Marm, S.; Scharringa, J.; Wagenaar, J.A.; Heederik, D.J.J. Carriage of extended-spectrum β-lactamases in pig farmers is associated with occurrence in pigs. Clin. Microbiol. Infect. 2015, 21, 917–923. [Google Scholar] [CrossRef] [PubMed]
- Dohmen, W.; Gompel, L.V.; Schmitt, H.; Liakopoulos, A.; Heres, L.; Urlings, B.A.; Mevius, D.; Bonten, M.J.M.; Heederik, D.J.J. ESBL carriage in pig slaughterhouse workers is associated with occupational exposure. Epidemiol. Infect. 2017, 145, 2003–2010. [Google Scholar] [CrossRef]
- Dohmen, W.; Liakopoulos, A.; Bonten, M.J.M.; Mevius, D.J.; Heederik, D.J.J. Longitudinal Study of Dynamic Epidemiology of Extended-Spectrum Beta-Lactamase-Producing Escherichia coli in Pigs and Humans Living and/or Working on Pig Farms. Microbiol. Spectr. 2023, 11, e02947-22. [Google Scholar] [CrossRef]
- UK-VARSS. UK Veterinary Antibiotic Resistance and Sales Surveillance Report (UK-VARSS 2021). Veterinary Medicines Directorate, Addlestone. 2022. Available online: https://www.gov.uk/government/publications/veterinary-antimicrobial-resistance-and-sales-surveillance-2021 (accessed on 6 January 2023).
- DANMAP (Danish Integrated Antimicrobial Resistance Monitoring and Research Programme). DANMAP 2021. Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Food Animals, Food and Humans in Denmark. 2022. Available online: https://www.danmap.org/reports/2021 (accessed on 1 June 2023).
- European Food Safety Authority (EFSA) and European Centre for Disease Prevention and Control (ECDC). The European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2020/2021. EFSA J. 2023, 21, 7867. [Google Scholar] [CrossRef]
- Luppi, A. Swine enteric colibacillosis: Diagnosis, therapy and antimicrobial resistance. Porc. Health Manag. 2017, 3, 16. [Google Scholar] [CrossRef]
- Hayer, S.; Rovira, A.; Olsen, K.; Johnson, T.J.; Vannucci, F.; Rendahl, A.; Perez, A.; Alvarez, J. Prevalence and trend analysis of antimicrobial resistance in clinical Escherichia coli isolates collected from diseased pigs in the USA between 2006 and 2016. Transbound. Emerg. Dis. 2020, 67, 1930–1941. [Google Scholar] [CrossRef]
- Burow, E.; Simoneit, C.; Tenhagen, B.A.; Käsbohrer, A. Oral antimicrobials increase antimicrobial resistance in porcine E. coli—A systematic review. Prev. Vet. Med. 2014, 113, 364–375. [Google Scholar] [CrossRef]
- Hayer, S.S.; Casanova-Higes, A.; Paladino, E.; Elnekave, E.; Nault, A.; Johnson, T.; Bender, J.; Perez, A.; Alvarez, J. Global Distribution of Extended Spectrum Cephalosporin and Carbapenem Resistance and Associated Resistance Markers in Escherichia coli of Swine Origin—A Systematic Review and Meta-Analysis. Front. Microbiol. 2022, 13, 853810. [Google Scholar] [CrossRef]
- Hayer, S.S.; Casanova-Higes, A.; Paladino, E.; Elnekave, E.; Nault, A.; Johnson, T.; Bender, J.; Perez, A.; Alvarez, J. Global Distribution of Fluoroquinolone and Colistin Resistance and Associated Resistance Markers in Escherichia coli of Swine Origin—A Systematic Review and Meta-Analysis. Front. Microbiol. 2022, 13, 834793. [Google Scholar]
- Li, M.; Li, Z.; Zhong, Q.; Liu, J.; Han, G.; Li, Y.; Li, C. Antibiotic resistance of fecal carriage of Escherichia coli from pig farms in China: A meta-analysis. Environ. Sci. Pollut. Res. 2022, 29, 22989–23000. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2023. [Google Scholar]
- Kahlmeter, G.; Turnidge, J. How to: ECOFFs—The why, the how, and the don’ts of EUCAST epidemiological cutoff values. Clin. Microbiol. Infect. 2022, 28, 952–954. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Data from the EUCAST MIC Distribution Website. 2023. Available online: http://www.eucast.org (accessed on 21 July 2023).
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 13.1. 2023. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_13.1_Breakpoint_Tables.pdf (accessed on 4 August 2023).
- Kahlmeter, G.; Giske, C.G.; Kirn, T.J.; Sharp, S.E. Point-Counterpoint: Differences between the European Committee on Antimicrobial Susceptibility Testing and Clinical and Laboratory Standards Institute Recommendations for Reporting Antimicrobial Susceptibility Results. J. Clin. Microbiol. 2019, 57, 10–128. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance. 2017. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_170711.pdf (accessed on 3 November 2023).
- Smith, H.W.; Crabb, W.E. The effect of continuous administration of diets containing low levels of tetracyclines on the incidence of drug-resistant Bacterium coli in the faeces of pigs and chickens: The sensitivity of the Bact. coli to other chemotherapeutic agents. Vet. Rec. 1957, 69, 24–30. [Google Scholar]
- Smith, W.H. Antibiotic-resistant Escherichia coli in market pigs in 1956–1979: The emergence of organisms with plasmid-borne trimethoprim resistance. Epidemiol. Infect. 1980, 84, 467–477. [Google Scholar]
- Aalbæ, B.; Rasmussen, J.; Nielsen, B.; Olsen, J.E. Prevalence of antibiotic-resistant Escherichia coli in Danish pigs and cattle. APMIS 1991, 99, 1103–1110. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Integrated Surveillance of Antimicrobial Resistance in Foodborne Bacteria: Application of a One Health Approach. Geneva, Switzerland, 2017; Licence: CC BY-NC-SA 3.0 IGO. Available online: https://apps.who.int/iris/handle/10665/255747 (accessed on 8 July 2021).
- Food and Drug Administration. National Antimicrobial Resistance Monitoring System. Available online: https://www.fda.gov/animal-veterinary/antimicrobial-resistance/national-antimicrobial-resistance-monitoring-system (accessed on 12 August 2023).
- Government of Canada. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS). Available online: https://www.canada.ca/en/public-health/services/surveillance/canadian-integrated-program-antimicrobial-resistance-surveillance-cipars.html (accessed on 12 August 2023).
- Ministry of Agriculture, Forestry and Fisheries. Available online: https://www.maff.go.jp/nval/english/AMR/Monitoring/index.html (accessed on 30 September 2023).
- de Jong, A.; Thomas, V.; Klein, U.; Marion, H.; Moyaert, H.; Simjee, S.; Vallé, M. Pan-European resistance monitoring programmes encompassing food-borne bacteria and target pathogens of food-producing and companion animals. Int. J. Antimicrob. Agents 2013, 41, 403–409. [Google Scholar] [CrossRef]
- DANMAP (Danish Integrated Antimicrobial Resistance Monitoring and Research Programme). Available online: https://www.danmap.org/ (accessed on 30 September 2023).
- Wageningen University and Research. Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals in the Netherlands. Available online: https://www.wur.nl/en/Research-Results/Research-Institutes/Bioveterinary-Research/In-the-spotlight/Antibiotic-resistance/MARAN-reports.htm (accessed on 30 September 2023).
- Costa, M.; Cardo, M.; d’Anjo, M.C.; Leite, A. Assessing antimicrobial resistance occurrence in the Portuguese food system: Poultry, pigs and derived food, 2014–2018. Zoonoses Public Health 2022, 69, 312–324. [Google Scholar] [CrossRef]
- Duarte, A.S.R.; Marques, A.R.; Andersen, V.D.; Korsgaard, H.B.; Mordhorst, H.; Møller, F.D.; Petersen, T.N.; Vigre, H.; Hald, T.; Aarestrup, F.M. Antimicrobial resistance monitoring in the Danish swine production by phenotypic methods and metagenomics from 1999 to 2018. Eurosurveillance 2023, 28, 2200678. [Google Scholar] [CrossRef]
- Sodagari, H.R.; Varga, C. Evaluating Antimicrobial Resistance Trends in Commensal Escherichia coli Isolated from Cecal Samples of Swine at Slaughter in the United States, 2013–2019. Microorganisms 2023, 11, 1033. [Google Scholar] [CrossRef]
- Schrijver, R.; Stijntjes, M.; Rodríguez-Baño, J.; Tacconelli, E.; Rajendran, B.N.; Voss, A. Review of antimicrobial resistance surveillance pro-grammes in livestock and meat in EU with focus on humans. Clin. Microbiol. Infect. 2018, 24, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Criscuolo, N.G.; Pires, J.; Zhao, C.; Boeckel, T.P.V. resistancebank.org, an open-access repository for surveys of antimicrobial resistance in animals. Sci. Data 2021, 8, 189. [Google Scholar] [CrossRef] [PubMed]
- van Boeckel, T.P.; Pires, J.; Silvester, R.; Zhao, C.; Song, J.; Criscuolo, N.G.; Gilbert, M.; Bonhoeffer, S.; Laxminarayan, R. Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 2019, 365, eaaw1944. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Wang, Y.; Tiseo, K.; Pires, J.; Criscuolo, N.G.; van Boeckel, T.P. Geographically targeted surveillance of livestock could help prioritize intervention against antimicrobial resistance in China. Nat. Food 2021, 2, 596–602. [Google Scholar] [CrossRef]
- Ramos, S.; Silva, N.; Caniça, M.; Capelo-Martinez, J.; Brito, F.; Igrejas, G.; Poeta, P. High prevalence of antimicrobial-resistant Escherichia coli from animals at slaughter: A food safety risk. J. Sci. Food Agric. 2013, 93, 517–526. [Google Scholar] [CrossRef]
- Wasyl, D.; Hoszowski, A.; Zając, M.; Szulowski, K. Antimicrobial resistance in commensal Escherichia coli isolated from animals at slaughter. Front. Microbiol. 2013, 4, 221. [Google Scholar] [CrossRef]
- Gibbons, J.; Boland, F.; Egan, J.; Fanning, S.; Markey, B.; Leonard, F. Antimicrobial Resistance of Faecal Escherichia coli Isolates from Pig Farms with Different Durations of In-feed Antimicrobial Use. Zoonoses Public Health 2016, 63, 241–250. [Google Scholar] [CrossRef]
- Österberg, J.; Wingstrand, A.; Jensen, A.; Kerouanton, A.; Cibin, V.; Barco, L.; Denis, M.; Aabo, S.; Bengtsson, B. Antibiotic Resistance in Escherichia coli from Pigs in Organic and Conventional Farming in Four European Countries. PLoS ONE 2016, 11, e0157049. [Google Scholar] [CrossRef]
- Aasmäe, B.; Häkkinen, L.; Kaart, T.; Kalmus, P. Antimicrobial resistance of Escherichia coli and Enterococcus spp. isolated from Estonian cattle and swine from 2010 to 2015. Acta Vet. Scand. 2019, 61, 5. [Google Scholar] [CrossRef] [PubMed]
- Food Drug Administration (FDA) NARMSNow Rockville MD. U.S. Department of Health and Human Services. 2023. Available online: https://www.fda.gov/animal-veterinary/national-antimicrobial-resistance-monitoring-system/narms-now-integrated-data (accessed on 30 September 2023).
- Adenipekun, E.O.; Jackson, C.R.; Oluwadun, A.; Iwalokun, B.A.; Frye, J.G.; Barrett, J.B.; Hiott, L.M.; Woodley, T.A. Prevalence and Antimicrobial Resistance in Escherichia coli from Food Animals in Lagos, Nigeria. Microb. Drug Resist. 2015, 21, 358–365. [Google Scholar] [CrossRef]
- Ikwap, K.; Gertzell, E.; Hansson, I.; Dahlin, L.; Selling, K.; Magnusson, U.; Dione, M.; Jacobson, M. The presence of antibiotic-resistant Staphylococcus spp. and Escherichia coli in smallholder pig farms in Uganda. BMC Vet. Res. 2021, 17, 31. [Google Scholar] [CrossRef]
- Manishimwe, R.; Moncada, P.M.; Musanayire, V.; Shyaka, A.; Scott, M.H.; Loneragan, G.H. Antibiotic-Resistant Escherichia coli and Salmonella from the Feces of Food Animals in the East Province of Rwanda. Animals 2021, 11, 1013. [Google Scholar] [CrossRef] [PubMed]
- Katakweba, A.; Muhairwa, A.P.; Lupindu, A.M.; Damborg, P.; Rosenkrantz, J.T.; Minga, U.M.; Mtambo, M.; Olsen, J.E. First Report on a Randomized Investigation of Antimicrobial Resistance in Fecal Indicator Bacteria from Livestock, Poultry, and Humans in Tanzania. Microb. Drug Resist. 2018, 24, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Kimera, Z.I.; Mgaya, F.X.; Misinzo, G.; Mshana, S.E.; Moremi, N.; Matee, M.I. Multidrug-Resistant, Including Extended-Spectrum Beta Lactamase-Producing and Quinolone-Resistant, Escherichia coli Isolated from Poultry and Domestic Pigs in Dar es Salaam, Tanzania. Antibiotics 2021, 10, 406. [Google Scholar] [CrossRef]
- Fang, J.; Shen, Y.; Qu, D.; Han, J. Antimicrobial resistance profiles and characteristics of integrons in Escherichia coli strains isolated from a large-scale centralized swine slaughterhouse and its downstream markets in Zhejiang, China. Food Control 2019, 95, 215–222. [Google Scholar] [CrossRef]
- Zhang, X.; Li, X.; Wang, W.; Qi, J.; Wang, D.; Xu, L.; Liu, Y.; Zhang, Y.; Guo, K. Diverse Gene Cassette Arrays Prevail in Commensal Escherichia coli From Intensive Farming Swine in Four Provinces of China. Front. Microbiol. 2020, 11, 565349. [Google Scholar] [CrossRef]
- Lei, T.; Tian, W.; He, L.; Huang, X.H.; Sun, Y.X.; Deng, Y.T.; Sun, Y.; Lv, D.H.; Wu, C.M.; Huang, L.Z.; et al. Antimicrobial resistance in Escherichia coli isolates from food animals, animal food products and companion animals in China. Vet. Microbiol. 2010, 146, 85–89. [Google Scholar] [CrossRef]
- Jiang, H.X.; Lü, D.H.; Chen, Z.L.; Wang, X.M.; Chen, J.R.; Liu, Y.H.; Liao, X.P.; Liu, J.H.; Zeng, Z.L. High prevalence and widespread distribution of multi-resistant Escherichia coli isolates in pigs and poultry in China. Vet. J. 2011, 187, 99–103. [Google Scholar] [CrossRef]
- Zhang, P.; Shen, Z.; Zhang, C.; Song, L.; Wang, B.; Shang, J.; Yue, X.; Qu, Z.; Li, X.; Wu, L.; et al. Surveillance of antimicrobial resistance among Escherichia coli from chicken and swine, China, 2008–2015. Vet. Microbiol. 2017, 203, 49–55. [Google Scholar] [CrossRef]
- Lv, C.; Shang, J.; Zhang, W.; Sun, B.; Li, M.; Guo, C.; Zhou, N.; Guo, X.; Huang, S.; Zhu, Y. Dynamic antimicrobial resistant patterns of Escherichia coli from healthy poultry and swine over 10 years in Chongming Island, Shanghai. Infect. Dis. Poverty 2022, 11, 98. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zhou, W.; Wu, J.; Liu, X.; Lin, J.; Ji, X.; Lin, H.; Wang, J.; Jiang, H.; Zhou, Q.; et al. Large-Scale Studies on Antimicrobial Resistance and Molecular Characterization of Escherichia coli from Food Animals in Developed Areas of Eastern China. Microbiol. Spectr. 2022, 10, e02015-22. [Google Scholar] [CrossRef]
- Peng, Z.; Hu, Z.; Li, Z.; Zhang, X.; Jia, C.; Li, T.; Dai, M.; Tan, C.; Xu, Z.; Wu, B.; et al. Antimicrobial resistance and population genomics of multidrug-resistant Escherichia coli in pig farms in mainland China. Nat. Commun. 2022, 13, 1116. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Yang, Y.; Cao, S.; Liu, H.; Li, X.; Sun, J.; Li, F.; Ishfaq, M.; Zhang, X. Prevalence and Characteristic of Swine-Origin mcr-1-Positive Escherichia coli in Northeastern China. Front. Microbiol. 2021, 12, 712707. [Google Scholar] [CrossRef]
- Lugsomya, K.; Chatsuwan, T.; Niyomtham, W.; Tummaruk, P.; Hampson, D.J.; Prapasarakul, N. Routine Prophylactic Antimicrobial Use Is Associated with Increased Phenotypic and Genotypic Resistance in Commensal Escherichia coli Isolates Recovered from Healthy Fattening Pigs on Farms in Thailand. Microb. Drug Resist. 2018, 24, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Pholwat, S.; Pongpan, T.; Chinli, R.; McQuade, E.T.; Thaipisuttikul, I.; Ratanakorn, P.; Liu, J.; Taniuchi, M.; Houpt, E.R.; Foongladda, S. Antimicrobial Resistance in Swine Fecal Specimens Across Different Farm Management Systems. Front. Microbiol. 2020, 11, 1238. [Google Scholar] [CrossRef]
- Trongjit, S.; Angkittitrakul, S.; Chuanchuen, R. Occurrence and molecular characteristics of antimicrobial resistance of Escherichia coli from broilers, pigs and meat products in Thailand and Cambodia provinces. Microbiol. Immunol. 2016, 60, 575–585. [Google Scholar] [CrossRef]
- Tuat, C.V.; Hue, P.T.; Loan, N.T.P.; Thuy, N.T.; Hue, L.T.; Giang, V.N.; Erickson, V.I.; Padungtod, P. Antimicrobial Resistance Pilot Surveillance of Pigs and Chickens in Vietnam, 2017–2019. Front. Vet. Sci. 2021, 8, 618497. [Google Scholar] [CrossRef]
- Government of Canada. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) 2019: Figures and Tables. Public Health Agency of Canada, Guelph. 2020. Available online: https://publications.gc.ca/collections/collection_2022/aspc-phac/HP2-4-2019-eng-5.pdf (accessed on 4 November 2023).
- Ministry of Agriculture, Forestry and Fisheries. Report on the Japanese Veterinary Antimicrobial Resistance Monitoring System 2016–2017. 2020. Available online: https://www.maff.go.jp/nval/yakuzai/pdf/200731_JVARMReport_2016–2017.pdf (accessed on 20 July 2021).
- Teshager, T.; Herrero, I.A.; Porrero, M.; Garde, J.; Moreno, M.A.; Domínguez, L. Surveillance of antimicrobial resistance in Escherichia coli strains isolated from pigs at Spanish slaughterhouses. Int. J. Antimicrob. Agents 2000, 15, 137–142. [Google Scholar] [CrossRef]
- Sáenz, Y.; Zarazaga, M.; Briñas, L.; Lantero, M.; Ruiz-Larrea, F.; Torres, C. Antibiotic resistance in Escherichia coli isolates obtained from animals, foods and humans in Spain. Int. J. Antimicrob. Agents 2001, 18, 353–358. [Google Scholar] [CrossRef] [PubMed]
- de Jong, A.; Thomas, V.; Simjee, S.; Godinho, K.; Schiessl, B.; Klein, U.; Butty, P.; Vallé, M.; Marion, H.; Shryock, T.R. Pan-European monitoring of susceptibility to human-use antimicrobial agents in enteric bacteria isolated from healthy food-producing animals. J. Antimicrob. Chemoth. 2012, 67, 638–651. [Google Scholar] [CrossRef] [PubMed]
- de Jong, A.; Garch, F.E.; Hocquet, D.; Prenger-Berninghoff, E.; Dewulf, J.; Migura-Garcia, L.; Perrin-Guyomard, A.; Veldman, K.T.; Janosi, S.; Skarzynska, M.; et al. European-wide antimicrobial resistance monitoring in commensal Escherichia coli isolated from healthy food animals between 2004 and 2018. J. Antimicrob. Chemother. 2022, 77, 3301–3311. [Google Scholar] [CrossRef] [PubMed]
- Dunlop, R.H.; McEwen, S.A.; Meek, A.H.; Black, W.D.; Friendship, R.M.; Clarke, R.C. Prevalences of resistance to seven antimicrobials among fecal Escherichia coli of swine on thirty-four farrow-to-finish farms in Ontario, Canada. Prev. Vet. Med. 1998, 34, 265–282. [Google Scholar] [CrossRef] [PubMed]
- Varga, C.; Rajić, A.; McFall, M.E.; Avery, B.P.; Reid-Smith, R.J.; Deckert, A.; Checkley, S.L.; McEwen, S.A. Antimicrobial resistance in generic Escherichia coli isolated from swine fecal samples in 90 Alberta finishing farms. Can. J. Vet. Res. 2008, 72, 175–180. [Google Scholar] [PubMed]
- Rosengren, L.B.; Waldner, C.L.; Reid-Smith, R.J.; Checkley, S.L.; McFall, M.E.; Rajić, A. Antimicrobial resistance of fecal Escherichia coli isolated from grow-finish pigs in 20 herds in Alberta and Saskatchewan. Can. J. Vet. Res. 2008, 72, 160–167. [Google Scholar]
- Akwar, H.; Poppe, C.; Wilson, J.; Reid-Smith, R.J.; Dyck, M.; Waddington, J.; Shang, D.; McEwen, S.A. Prevalence and patterns of antimicrobial resistance of fecal Escherichia coli among pigs on 47 farrow-to-finish farms with different in-feed medication policies. Can. J. Vet. Res. 2008, 72, 195–201. [Google Scholar]
- Bunner, C.A.; Norby, B.; Bartlett, P.C.; Erskine, R.J.; Downes, F.P.; Kaneene, J.B. Prevalence and pattern of antimicrobial susceptibility in Escherichia coli isolated from pigs reared under antimicrobial-free and conventional production methods. J. Am. Vet. Med. Assoc. 2007, 231, 275–283. [Google Scholar] [CrossRef]
- Smith, M.; Jordan, D.; Gibson, J.; Cobbold Chapman, T.; Abraham, S.; Trott, D. Phenotypic and genotypic profiling of antimicrobial resistance in enteric Escherichia coli communities isolated from finisher pigs in Australia. Aust. Vet. J. 2016, 94, 371–376. [Google Scholar] [CrossRef]
- Kidsley, A.K.; Abraham, S.; Bell, J.M.; O’Dea, M.; Laird, T.J.; Jordan, D.; Mitchell, P.; McDevitt, C.A.; Trott, D.J. Antimicrobial Susceptibility of Escherichia coli and Salmonella spp. Isolates From Healthy Pigs in Australia: Results of a Pilot National Survey. Front. Microbiol. 2018, 9, 1207. [Google Scholar] [CrossRef]
- Lim, S.K.; Lee, H.S.; Nam, H.M.; Cho, Y.S.; Kim, J.M.; Song, S.W.; Park, Y.H.; Jung, S.C. Antimicrobial resistance observed in Escherichia coli strains isolated from fecal samples of cattle and pigs in Korea during 2003–2004. Int. J. Food Microbiol. 2007, 116, 283–286. [Google Scholar] [CrossRef]
- Lee, M.; Shin, E.; Lee, Y. Antimicrobial Resistance and Integron Profiles in Multidrug-Resistant Escherichia coli Isolates from Pigs. Foodborne Pathog. Dis. 2014, 11, 988–997. [Google Scholar] [CrossRef]
- Song, H.J.; Kim, S.J.; Moon, D.C.; Mechesso, A.F.; Choi, J.H.; Kang, H.Y.; Boby, N.; Yoon, S.S.; Lim, S.K. Antimicrobial Resistance in Escherichia coli Isolates from Healthy Food Animals in South Korea, 2010–2020. Microorganisms 2022, 10, 524. [Google Scholar] [CrossRef]
- Agga, G.; Scott, H.; Amachawadi, R.; Nagaraja, T.; Vinasco, J.; Bai, J.; Norby, B.; Renter, D.; Dritz, S.; Nelssen, J.; et al. Effects of chlortetracycline and copper supplementation on antimicrobial resistance of fecal Escherichia coli from weaned pigs. Prev. Vet. Med. 2014, 114, 231–246. [Google Scholar] [CrossRef]
- Bibbal, D.; Dupouy, V.; Ferré, J.; Toutain, P.; Fayet, O.; Prère, M.; Bousquet-Mélou, A. Impact of Three Ampicillin Dosage Regimens on Selection of Ampicillin Resistance in Enterobacteriaceae and Excretion of blaTEM Genes in Swine Feces. Appl. Environ. Microbiol. 2007, 73, 4785–4790. [Google Scholar] [CrossRef]
- Gaire, T.N.; Scott, H.M.; Sellers, L.; Nagaraja, T.G.; Volkova, V.V. Age Dependence of Antimicrobial Resistance Among Fecal Bacteria in Animals: A Scoping Review. Front. Vet. Sci. 2021, 7, 622495. [Google Scholar] [CrossRef]
- Lekagul, A.; Tangcharoensathien, V.; Yeung, S. Patterns of antibiotic use in global pig production: A systematic review. Vet. Anim. Sci. 2019, 7, 00058. [Google Scholar] [CrossRef] [PubMed]
- Sarrazin, S.; Joosten, P.; Van Gompel, L.; Luiken, R.E.; Mevius, D.J.; Wagenaar, J.A.; Heederik, D.J.; Dewulf, J. Quantitative and qualitative analysis of antimicrobial usage patterns in 180 selected farrow-to-finish pig farms from nine European countries based on single batch and purchase data. J. Antimicrob. Chemother. 2019, 74, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Alali, W.Q.; Scott, H.M.; Harvey, R.B.; Norby, B.; Lawhorn, D.B.; Pillai, S.D. Longitudinal Study of Antimicrobial Resistance among Escherichia coli Isolates from Integrated Multisite Cohorts of Humans and Swine. Appl. Environ. Microbiol. 2008, 74, 3672–3681. [Google Scholar] [CrossRef] [PubMed]
- Marchant, M.; Moreno, M.A. Dynamics and Diversity of Escherichia coli in Animals and System Management of the Manure on a Commercial Farrow-to-Finish Pig Farm. Appl. Environ. Microbiol. 2013, 79, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Pissetti, C.; Kich, J.; Allen, H.K.; Navarrete, C.; de Costa, E.; Morés, N.; Cardoso, M. Antimicrobial resistance in commensal Escherichia coli and Enterococcus spp. isolated from pigs subjected to different antimicrobial administration protocols. Res. Vet. Sci. 2021, 137, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Langlois, B.; Dawson, K.; Leak, I.; Aaron, D. Effect of age and housing location on antibiotic resistance of fecal coliforms from pigs in a non-antibiotic-exposed herd. Appl. Environ. Microbiol. 1988, 54, 1341–1344. [Google Scholar] [CrossRef]
- Yun, J.; Muurinen, J.; Nykäsenoja, S.; Seppä-Lassila, L.; Sali, V.; Suomi, J.; Tuominen, P.; Joutsen, S.; Hämäläinen, M.; Olkkola, S.; et al. Antimicrobial use, biosecurity, herd characteristics, and antimicrobial resistance in indicator Escherichia coli in ten Finnish pig farms. Prev. Vet. Med. 2021, 193, 105408. [Google Scholar] [CrossRef] [PubMed]
- Burow, E.; Rostalski, A.; Harlizius, J.; Gangl, A.; Simoneit, C.; Grobbel, M.; Kollas, C.; Tenhagen, B.A.A.; Käsbohrer, A. Antibiotic resistance in Escherichia coli from pigs from birth to slaughter and its association with antibiotic treatment. Prev. Vet. Med. 2019, 165, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Mathew, A.; Beckmann, M.; Saxton, A.M. A comparison of antibiotic resistance in bacteria isolated from swine herds in which antibiotics were used or excluded. J. Swine Health Prod. 2001, 9, 125–129. [Google Scholar]
- Græsbøll, K.; Damborg, P.; Mellerup, A.; Herrero-Fresno, A.; Larsen, I.; Holm, A.; Nielsen, J.P.; Christiansen, L.E.; Angen, Ø.; Ahmed, S.; et al. Effect of Tetracycline Dose and Treatment Mode on Selection of Resistant Coliform Bacteria in Nursery Pigs. Appl. Environ. Microbiol. 2017, 83, e00538-17. [Google Scholar] [CrossRef] [PubMed]
- Katouli, M.; Lund, A.; Wallgren, P.; Kühn, I.; Söderlind, O.; Möllby, R. Phenotypic characterization of intestinal Escherichia coli of pigs during suckling, postweaning, and fattening periods. Appl. Environ. Microbiol. 1995, 61, 778–783. [Google Scholar] [CrossRef]
- Schierack, P.; Kadlec, K.; Guenther, S.; Filter, M.; Schwarz, S.; Ewers, C.; Wieler, L.H. Antimicrobial resistances do not affect colonization parameters of intestinal E. coli in a small piglet group. Gut Pathog. 2009, 1, 18. [Google Scholar] [CrossRef]
- Ahmed, S.; Olsen, J.E.; Herrero-Fresno, A. The genetic diversity of commensal Escherichia coli strains isolated from non-antimicrobial treated pigs varies according to age group. PLoS ONE 2017, 12, e0178623. [Google Scholar] [CrossRef]
- Hansen, K.; Damborg, P.; Andreasen, M.; Nielsen, S.; Guardabassi, L. Carriage and Fecal Counts of Cefotaxime M-Producing Escherichia coli in Pigs: A Longitudinal Study. Appl. Environ. Microbiol. 2013, 79, 794–798. [Google Scholar] [CrossRef]
- Amsler, M.; Zurfluh, K.; Hartnack, S.; Sidler, X.; Stephan, R.; Kümmerlen, D. Occurrence of Escherichia coli non-susceptible to quinolones in faecal samples from fluoroquinolone-treated, contact and control pigs of different ages from 24 Swiss pig farms. Porc. Health Manag. 2021, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Mathew, A.G.; Garner, K.N.; Ebner, P.D.; Saxton, A.M.; Clift, R.E.; Liamthong, S. Effects of Antibiotic Use in Sows on Resistance of E. coli and Salmonella enterica Typhimurium in Their Offspring. Foodborne Pathog. Dis. 2005, 2, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Callens, B.; Faes, C.; Maes, D.; Catry, B.; Boyen, F.; Francoys, D.; de Jong, E.; Haesebrouck, F.; Dewulf, J. Presence of Antimicrobial Resistance and Antimicrobial Use in Sows Are Risk Factors for Antimicrobial Resistance in Their Offspring. Microb. Drug Resist. 2015, 21, 50–58. [Google Scholar] [CrossRef]
- Cameron-Veas, K.; Solà-Ginés, M.; Moreno, M.A.; Fraile, L.; Migura-Garcia, L. Impact of the Use of β-Lactam Antimicrobials on the Emergence of Escherichia coli Isolates Resistant to Cephalosporins under Standard Pig-Rearing Conditions. Appl. Environ. Microbiol. 2015, 81, 1782–1787. [Google Scholar] [CrossRef]
- Cameron-Veas, K.; Moreno, M.A.; Fraile, L.; Migura-Garcia, L. Shedding of cephalosporin resistant Escherichia coli in pigs from conventional farms after early treatment with antimicrobials. Vet. J. 2016, 211, 21–25. [Google Scholar] [CrossRef]
- Stannarius, C.; Bürgi, E.; Regula, G.; Zychowska, M.; Zweifel, C.; Stephan, R. Antimicrobial resistance in Escherichia coli strains isolated from Swiss weaned pigs and sows. Schweiz. Arch. Für Tierheilkd. 2009, 151, 119–125. [Google Scholar] [CrossRef][Green Version]
- Hallenberg, G.; Jiwakanon, J.; Angkititrakul, S.; Kangair, S.; Osbjer, K.; Lunha, K.; Sunde, M.; Järhult, J.D.; Boeckel, T.P.; Rich, K.M.; et al. Antibiotic use in pig farms at different levels of intensification—Farmers’ practices in northeastern Thailand. PLoS ONE 2020, 15, e0243099. [Google Scholar] [CrossRef] [PubMed]
- Lunha, K.; Leangapichart, T.; Jiwakanon, J.; Angkititrakul, S.; Sunde, M.; Järhult, J.D.; Hallenberg, G.; Hickman, R.A.; Boeckel, T.; Magnusson, U. Antimicrobial Resistance in Fecal Escherichia coli from Humans and Pigs at Farms at Different Levels of Intensification. Antibiotics 2020, 9, 662. [Google Scholar] [CrossRef]
- Kittitat, L.; Jitrapa, Y.; Waree, N.; Chanwit, T.; Padet, T.; Hampson, D.J.; Nuvee, P. Antimicrobial Resistance in Commensal Escherichia coli Isolated from Pigs and Pork Derived from Farms Either Routinely Using or Not Using in-Feed Antimicrobials. Microb. Drug Resist. 2018, 24, 1054–1066. [Google Scholar]
- Sodagari, H.R.; Agrawal, I.; Yudhanto, S.; Varga, C. Longitudinal analysis of differences and similarities in antimicrobial resistance among commensal Escherichia coli isolated from market swine and sows at slaughter in the United States of America, 2013–2019. Int. J. Food Microbiol. 2023, 407, 110388. [Google Scholar] [CrossRef]
- Leekitcharoenphon, P.; Johansson, M.; Munk, P.; Malorny, B.; Skarżyńska, M.; Wadepohl, K.; Moyano, G.; Hesp, A.; Veldman, K.T.; Bossers, A.; et al. Genomic evolution of antimicrobial resistance in Escherichia coli. Sci. Rep. 2021, 11, 15108. [Google Scholar] [CrossRef] [PubMed]
- Stubberfield, E.; AbuOun, M.; Sayers, E.; O’Connor, H.M.; Card, R.M.; Anjum, M.F. Use of whole genome sequencing of commensal Escherichia coli in pigs for antimicrobial resistance surveillance, United Kingdom, 2018. Eurosurveillance 2019, 24, 1900136. [Google Scholar] [CrossRef]
- AbuOun, M.; O’Connor, H.M.; Stubberfield, E.J.; Nunez-Garcia, J.; Sayers, E.; Crook, D.W.; Smith, R.P.; Anjum, M.F. Characterizing Antimicrobial Resistant Escherichia coli and Associated Risk Factors in a Cross-Sectional Study of Pig Farms in Great Britain. Front. Microbiol. 2020, 11, 861. [Google Scholar] [CrossRef]
- Mencía-Ares, O.; Borowiak, M.; Argüello, H.; Cobo-Díaz, J.F.; Malorny, B.; Álvarez-Ordóñez, A.; Carvajal, A.; Deneke, C. Genomic Insights into the Mobilome and Resistome of Sentinel Microorganisms Originating from Farms of Two Different Swine Production Systems. Microbiol. Spectr. 2022, 10, e02896-22. [Google Scholar] [CrossRef] [PubMed]
- Reid, C.J.; Wyrsch, E.R.; Chowdhury, P.R.; Zingali, T.; Liu, M.; Darling, A.E.; Chapman, T.A.; Djordjevic, S.P. Porcine commensal Escherichia coli: A reservoir for class 1 integrons associated with IS26. Microb. Genom. 2017, 3, e000143. [Google Scholar] [CrossRef]
- Zingali, T.; Reid, C.J.; Chapman, T.A.; Gaio, D.; Liu, M.; Darling, A.E.; Djordjevic, S.P. Whole Genome Sequencing Analysis of Porcine Faecal Commensal Escherichia coli Carrying Class 1 Integrons from Sows and Their Offspring. Microorganisms 2020, 8, 843. [Google Scholar] [CrossRef] [PubMed]
- Pires, J.; Huisman, J.S.; Bonhoeffer, S.; Boeckel, T.P.V. Increase in antimicrobial resistance in Escherichia coli in food animals between 1980 and 2018 assessed using genomes from public databases. J. Antimicrob. Chemother. 2021, 77, 646–655. [Google Scholar] [CrossRef]
- Poirel, L.; Madec, J.Y.Y.; Lupo, A.; Schink, A.K.K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia coli. Microbiol. Spectr. 2018, 6, 6-4. [Google Scholar] [CrossRef]
- Jurado-Rabadán, S.; de la Fuente, R.; Ruiz-Santa-Quiteria, J.A.; Orden, J.A.; de Vries, L.E.; Agersø, Y. Detection and linkage to mobile genetic elements of tetracycline resistance gene tet(M) in Escherichia coli isolates from pigs. BMC Vet. Res. 2014, 10, 155. [Google Scholar] [CrossRef]
- Enne, V.I.; Cassar, C.; Sprigings, K.; Woodward, M.J.; Bennett, P.M. A high prevalence of antimicrobial resistant Escherichia coli isolated from pigs and a low prevalence of antimicrobial resistant E. coli from cattle and sheep in Great Britain at slaughter. FEMS Microbiol. Lett. 2008, 278, 193–199. [Google Scholar] [CrossRef]
- Schwarz, S.; Kehrenberg, C.; Doublet, B.; Cloeckaert, A. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Lett. 2004, 28, 519–542. [Google Scholar] [CrossRef]
- Du, Z.; Wang, M.; Cui, G.; Zu, X.; Zhao, Z.; Xue, Y. The prevalence of amphenicol resistance in Escherichia coli isolated from pigs in main-land China from 2000 to 2018: A systematic review and meta-analysis. PLoS ONE 2020, 15, e0228388. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.A.; Blok, H.E.; Donders, R.A.; Paauw, A.; Fluit, A.C.; Verhoef, J. Multidrug Resistance among Enterobacteriaceae Is Strongly Associated with the Presence of Integrons and Is Independent of Species or Isolate Origin. J. Infect. Dis. 2003, 187, 251–259. [Google Scholar]
- Hall, R.M.; Collis, C.M. Mobile gene cassettes and integrons: Capture and spread of genes by site-specific recombination. Mol. Microbiol. 1995, 15, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Domingues, S.; da Silva, G.J.; Nielsen, K.M. Integrons: Vehicles and pathways for horizontal dissemination in bacteria. Mobile genetic elements. Mob. Genet. Elem. 2012, 2, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, K.M.; White, D.G.; Hume, M.E.; Poole, T.L.; Nisbet, D.J. The chloramphenicol resistance gene cmlA is disseminated on transferable plasmids that confer multiple-drug resistance in swine Escherichia coli. FEMS Microbiol. Lett. 2005, 243, 285–291. [Google Scholar] [CrossRef]
- Dawes, F.E.; Kuzevski, A.; Bettelheim, K.A.; Hornitzky, M.A.; Djordjevic, S.P.; Walker, M.J. Distribution of Class 1 Integrons with IS26-Mediated Deletions in Their 3′-Conserved Segments in Escherichia coli of Human and Animal Origin. PLoS ONE 2010, 5, e12754. [Google Scholar] [CrossRef]
- Marchant, M.; Vinué, L.; Torres, C.; Moreno, M.A. Change of integrons over time in Escherichia coli isolates recovered from healthy pigs and chickens. Vet. Microbiol. 2013, 163, 124–132. [Google Scholar] [CrossRef]
- Changkaew, K.; Intarapuk, A.; Utrarachkij, F.; Nakajima, C.; Suthienkul, O.; Suzuki, Y. Antimicrobial Resistance, Extended-Spectrum β-Lactamase Productivity, and Class 1 Integrons in Escherichia coli from Healthy Swine. J. Food Protect. 2016, 78, 1442–1450. [Google Scholar] [CrossRef]
- Chaslus-Dancla, E.; Pohl, P.; Meurisse, M.; Marin, M.; Lafont, J. High genetic homology between plasmids of human and animal origins conferring resistance to the aminoglycosides gentamicin and apramycin. Antimicrob. Agents Chemother. 1991, 35, 590–593. [Google Scholar] [CrossRef]
- Jensen, V.F.; Jakobsen, L.; Emborg, H.D.; Seyfarth, A.; Hammerum, A.M. Correlation between apramycin and gentamicin use in pigs and an increasing reservoir of gentamicin-resistant Escherichia coli. J. Antimicrob. Chemother. 2006, 58, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.J.; Lim, S.K.; Nam, H.M.; Kim, A.R.; Jung, S.C.; Kim, M.N. Apramycin and Gentamicin Resistances in Indicator and Clinical Escherichia coli Isolates from Farm Animals in Korea. Foodborne Pathog Dis. 2011, 8, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Pankey, G.A. Tigecycline. J. Antimicrob. Chemother. 2005, 56, 470–480. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Wang, R.; Liu, D.; Walsh, T.R.; Zhang, R.; Lv, Y.; Ke, Y.; Ji, Q.; Wei, R.; Liu, Z.; et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Chantziaras, I.; Boyen, F.; Callens, B.; Dewulf, J. Correlation between veterinary antimicrobial use and antimicrobial resistance in food-producing animals: A report on seven countries. J. Antimicrob. Chemother. 2014, 69, 827–834. [Google Scholar] [CrossRef]
- Callens, B.; Cargnel, M.; Sarrazin, S.; Dewulf, J.; Hoet, B.; Vermeersch, K.; Wattiau, P.; Welby, S. Associations between a decreased veterinary antimicrobial use and resistance in commensal Escherichia coli from Belgian livestock species (2011–2015). Prev. Vet. Med. 2018, 157, 50–58. [Google Scholar] [CrossRef]
- Asai, T.; Kojima, A.; Harada, K.; Ishihara, K.; Takahashi, T.; Tamura, Y. Correlation between the usage volume of veterinary therapeutic antimicrobials and resistance in Escherichia coli isolated from the feces of food-producing animals in Japan. Jpn J. Infect. Dis. 2005, 58, 369–372. [Google Scholar]
- European Centre for Disease Prevention and Control (ECDC); European Food Safety Authority (EFSA); European Medicines Agency (EMA). Third joint inter-agency report on integrated analysis of consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food-producing animals in the EU/EEA. EFSA J. 2021, 19, e06712. [Google Scholar] [CrossRef]
- Ceccarelli, D.; Hesp, A.; van der Goot, J.; Joosten, P.; Sarrazin, S.; Wagenaar, J.A.; Dewulf, J.; Mevius, D.J.; Consortium, O. Antimicrobial resistance prevalence in commensal Escherichia coli from broilers, fattening turkeys, fattening pigs and veal calves in European countries and association with antimicrobial usage at country level. J. Med. Microbiol. 2020, 69, 537–547. [Google Scholar] [CrossRef]
- Dorado-García, A.; Mevius, D.J.; Jacobs, J.J.; Geijlswijk, I.M.; Mouton, J.W.; Wagenaar, J.A.; Heederik, D.J. Quantitative assessment of antimicrobial resistance in livestock during the course of a nationwide antimicrobial use reduction in the Netherlands. J. Antimicrob. Chemother. 2016, 71, 3607–3619. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, C.; Zhang, R.; Chen, Y.; Shen, Y.; Hu, F.; Liu, D.; Lu, J.; Guo, Y.; Xia, X.; et al. Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: An epidemiological comparative study. Lancet Infect. Dis. 2020, 20, 1161–1171. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Zhong, L.L.; Yang, Y.; Doi, Y.; Paterson, D.L.; Stoesser, N.; Ma, F.; Ahmed, M.A.E.G.E.S.; Feng, S.; Huang, S.; et al. Dynamics of mcr-1 prevalence and mcr-1-positive Escherichia coli after the cessation of colistin use as a feed additive for animals in China: A prospective cross-sectional and whole genome sequencing-based molecular epidemiological study. Lancet Microbe 2020, 1, e34–e43. [Google Scholar] [CrossRef] [PubMed]
- Langlois, B.; Cromwell, G.; Hays, V. Influence of Type of Antibiotic and Length of Antibiotic Feeding Period on Performance and Persistence of Antibiotic Resistant Enteric Bacteria in Growing-Finishing Swine. J. Anim. Sci. 1978, 46, 1383–1396. [Google Scholar] [CrossRef]
- Langlois, B.; Dawson, K.; Stahly, T.; Cromwell, G. Antibiotic Resistance of Fecal Coliforms from Swine Fed Subtherapeutic and Therapeutic Levels of Chlortetracycline. J. Anim. Sci. 1984, 58, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Delsol, A.A.; Anjum, M.; Woodward, M.J.; Sunderland, J.; Roe, J.M. The effect of chlortetracycline treatment and its subsequent withdrawal on multi-resistant Salmonella enterica serovar Typhimurium DT104 and commensal Escherichia coli in the pig. J. Appl. Microbiol. 2003, 95, 1226–1234. [Google Scholar] [CrossRef]
- Funk, J.A.; Lejeune, J.T.; Wittum, T.E.; Rajala-Schultz, P.J. The Effect of Subtherapeutic Chlortetracycline on Antimicrobial Resistance in the Fecal Flora of Swine. Microb. Drug Resist. 2006, 12, 210–218. [Google Scholar] [CrossRef]
- Mathew, A.G.; Arnett, D.B.; Cullen, P.; Ebner, P.D. Characterization of resistance patterns and detection of apramycin resistance genes in Escherichia coli isolated from swine exposed to various environmental conditions. Int. J. Food Microbiol. 2003, 89, 11–20. [Google Scholar] [CrossRef]
- Moro, M.; Beran, G.; Hoffman, L.; Griffith, R. Effects of cold stress on the antimicrobial drug resistance of Escherichia coli of the intestinal flora of swine. Lett. Appl. Microbiol. 1998, 27, 251–254. [Google Scholar] [CrossRef]
- Moro, M.H.; Beran, G.W.; Griffith, R.W.; Hoffman, L.J. Effects of heat stress on the antimicrobial drug resistance of Escherichia coli of the intestinal flora of swine. J. Appl. Microbiol. 2000, 88, 836–844. [Google Scholar] [CrossRef]
- Mathew, A.; Jackson, F.; Saxton, A.M. Effects of antibiotic regimens on resistance of Escherichia coli and Salmonella serovar Typhimurium in swine. J. Swine Health Prod. 2002, 10, 7–13. [Google Scholar]
- Herrero-Fresno, A.; Zachariasen, C.; Nørholm, N.; Holm, A.; Christiansen, L.; Olsen, J. Effect of different oral oxytetracycline treatment regimes on selection of antimicrobial resistant coliforms in nursery pigs. Vet. Microbiol. 2017, 208, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Bibbal, D.; Dupouy, V.; Prère, M.; Toutain, P.; Bousquet-Mélou, A. Relatedness of Escherichia coli Strains with Different Susceptibility Phenotypes Isolated from Swine Feces during Ampicillin Treatment. Appl. Environ. Microbiol. 2009, 75, 2999–3006. [Google Scholar] [CrossRef]
- Herrero-Fresno, A.; Zachariasen, C.; Hansen, M.; Nielsen, A.; Hendriksen, R.S.; Nielsen, S.; Olsen, J. Apramycin treatment affects selection and spread of a multidrug-resistant Escherichia coli strain able to colonize the human gut in the intestinal microbiota of pigs. Vet. Res. 2015, 47, 12. [Google Scholar] [CrossRef]
- Römer, A.; Scherz, G.; Reupke, S.; Meißner, J.; Wallmann, J.; Kietzmann, M.; Kaspar, H. Effects of intramuscularly administered enrofloxacin on the susceptibility of commensal intestinal Escherichia coli in pigs (Sus scrofa domestica). BMC Vet. Res. 2017, 13, 378. [Google Scholar] [CrossRef]
- Burow, E.; Grobbel, M.; Tenhagen, B.A.; Simoneit, C.; Ladwig, M.; Szabo, I.; Wendt, D.; Banneke, S.; Kasbohrer, A. Antimicrobial susceptibility in faecal Escherichia coli from pigs after enrofloxacin administration in an experimental environment. Berl. Munch. Tierarztl. Wochenschr. 2018, 131, 170–181. [Google Scholar]
- Gellin, G.; Langlois, B.; Dawson, K.; Aaron, D. Antibiotic resistance of gram-negative enteric bacteria from pigs in three herds with different histories of antibiotic exposure. Appl. Environ. Microbiol. 1989, 55, 2287–2292. [Google Scholar] [CrossRef] [PubMed]
- Mathew, A.G.; Upchurch, W.G.; Chattin, S.E. Incidence of antibiotic resistance in fecal Escherichia coli isolated from commercial swine farms. J. Anim. Sci. 1998, 76, 429. [Google Scholar] [CrossRef]
- Mencía-Ares, O.; Argüello, H.; Puente, H.; Gómez-García, M.; Manzanilla, E.G.; Álvarez-Ordóñez, A.; Carvajal, A.; Rubio, P. Antimicrobial resistance in commensal Escherichia coli and Enterococcus spp. is influenced by production system, antimicrobial use, and biosecurity measures on Spanish pig farms. Porc. Health Manag. 2021, 7, 27. [Google Scholar] [CrossRef]
- Hickman, R.A.; Leangapichart, T.; Lunha, K.; Jiwakanon, J.; Angkititrakul, S.; Magnusson, U.; Sunde, M.; Järhult, J.D. Exploring the Antibiotic Resistance Burden in Livestock, Livestock Handlers and Their Non-Livestock Handling Contacts: A One Health Perspective. Front. Microbiol. 2021, 12, 651461. [Google Scholar] [CrossRef]
- Dewulf, J.; Catry, B.; Timmerman, T.; Opsomer, G.; de Kruif, A.; Maes, D. Tetracycline-resistance in lactose-positive enteric coliforms originating from Belgian fattening pigs: Degree of resistance, multiple resistance and risk factors. Prev. Vet. Med. 2007, 78, 339–351. [Google Scholar] [CrossRef]
- Vieira, A.R.; Houe, H.; Wegener, H.C.; Wong, D.M.L.F.; Emborg, H.D.D. Association between tetracycline consumption and tetracycline resistance in Escherichia coli from healthy Danish slaughter pigs. Foodborne Pathog. Dis. 2009, 6, 99–109. [Google Scholar] [CrossRef]
- Dunlop, R.H.; McEwen, S.A.; Meek, A.H.; Clarke, R.C.; Black, W.D.; Friendship, R.M. Associations among antimicrobial drug treatments and antimicrobial resistance of fecal Escherichia coli of swine on 34 farrow-to-finish farms in Ontario, Canada. Prev. Vet. Med. 1998, 34, 283–305. [Google Scholar] [PubMed]
- Rosengren, L.B.; Waldner, C.L.; Reid-Smith, R.J.; Dowling, P.M.; Harding, J.C. Associations Between Feed and Water Antimicrobial Use in Farrow-to-Finish Swine Herds and Antimicrobial Resistance of Fecal Escherichia coli from Grow-Finish Pigs. Microb. Drug Resist. 2007, 13, 261–270. [Google Scholar] [PubMed]
- Harada, K.; Asai, T.; Ozawa, M.; Kojima, A.; Takahashi, T. Farm-Level Impact of Therapeutic Antimicrobial Use on Antimicrobial-Resistant Populations of Escherichia coli Isolates from Pigs. Microb. Drug Resist. 2008, 14, 239–244. [Google Scholar] [PubMed]
- Alali, W.Q.; Scott, H.M.; Christian, K.L.; Fajt, V.R.; Harvey, R.B.; Lawhorn, D.B. Relationship between level of antibiotic use and resistance among Escherichia coli isolates from integrated multi-site cohorts of humans and swine. Prev. Vet. Med. 2009, 90, 160–167. [Google Scholar] [CrossRef]
- Varga, C.; Rajić, A.; McFall, M.E.; Reid-Smith, R.J.; Deckert, A.E.; Checkley, S.L.; McEwen, S.A. Associations between reported on-farm anti-microbial use practices and observed antimicrobial resistance in generic fecal Escherichia coli isolated from Alberta finishing swine farms. Prev. Vet. Med. 2009, 88, 185–192. [Google Scholar] [CrossRef]
- Makita, K.; Goto, M.; Ozawa, M.; Kawanishi, M.; Koike, R.; Asai, T.; Tamura, Y. Multivariable Analysis of the Association Between Antimicrobial Use and Antimicrobial Resistance in Escherichia coli Isolated from Apparently Healthy Pigs in Japan. Microb. Drug Resist. 2016, 22, 28–39. [Google Scholar] [CrossRef]
- Akwar, H.; Poppe, C.; Wilson, J.; Reid-Smith, R.J.; Dyck, M.; Waddington, J.; Shang, D.; McEwen, S.A. Associations of antimicrobial uses with antimicrobial resistance of fecal Escherichia coli from pigs on 47 farrow-to-finish farms in Ontario and British Columbia. Can. J. Vet. Res. 2008, 72, 202–210. [Google Scholar]
- European Medicines Agency (EMA). Categorisation of Antibiotics for Use in Animals for Prudent and Responsible Use. 2019. Available online: https://www.ema.europa.eu/en/documents/report/categorisation-antibiotics-european-union-answer-request-european-commission-updating-scientific_en.pdf (accessed on 6 June 2023).
- Regulation (EU) 2019/6 of the European Parliament and of the Council of 11 December 2018 on Veterinary Medicinal Products and Repealing Directive 2001/82/EC (Text with EEA relevance). Off. J. Eur. Union 2019, L4, 43–167. Available online: http://data.europa.eu/eli/reg/2019/6/oj (accessed on 6 July 2021).
- Lambrecht, E.; Meervenne, E.; Boon, N.; de Wiele, T.; Wattiau, P.; Herman, L.; Heyndrickx, M.; Coillie, E. Characterization of Cefotaxime- and Ciprofloxacin-Resistant Commensal Escherichia coli Originating from Belgian Farm Animals Indicates High Antibiotic Resistance Transfer Rates. Microb. Drug Resist. 2018, 24, 707–717. [Google Scholar] [CrossRef]
- Jacoby, G.A. AmpC β-Lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Jacoby, G.A. Updated Functional Classification of β-Lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. β-Lactams and β-Lactamase Inhibitors: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a025247. [Google Scholar] [CrossRef] [PubMed]
- Livermore, D.M.; Canton, R.; Gniadkowski, M.; Nordmann, P.; Rossolini, G.; Arlet, G.; Ayala, J.; Coque, T.M.; Kern-Zdanowicz, I.; Luzzaro, F.; et al. CTX-M: Changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 2007, 59, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Woerther, P.L.; Burdet, C.; Chachaty, E.; Andremont, A. Trends in Human Fecal Carriage of Extended-Spectrum β-Lactamases in the Community: Toward the Globalization of CTX-M. Clin. Microbiol. Rev. 2013, 26, 744–758. [Google Scholar] [CrossRef] [PubMed]
- Bevan, E.R.; Jones, A.M.; Hawkey, P.M. Global epidemiology of CTX-M β-lactamases: Temporal and geographical shifts in genotype. J. Antimicrob. Chemoth. 2017, 72, 2145–2155. [Google Scholar] [CrossRef]
- Liu, J.H.; Wei, S.Y.; Ma, J.Y.; Zeng, Z.L.; Lü, D.H.; Yang, G.X.; Chen, Z.L. Detection and characterisation of CTX-M and CMY-2 β-lactamases among Escherichia coli isolates from farm animals in Guangdong Province of China. Int. J. Antimicrob. Agents 2007, 29, 576–581. [Google Scholar] [CrossRef]
- Tian, G.B.; Wang, H.N.; Zou, L.K.; Tang, J.N.; Zhao, Y.W.; Ye, M.Y.; Tang, J.Y.; Zhang, Y.; Zhang, A.Y.; Yang, X.; et al. Detection of CTX-M-15, CTX-M-22, and SHV-2 Extended-Spectrum β-Lactamases (ESBLs) in Escherichia coli Fecal-Sample Isolates from Pig Farms in China. Foodborne Pathog. Dis. 2009, 6, 297–304. [Google Scholar] [CrossRef]
- Zheng, H.; Zeng, Z.; Chen, S.; Liu, Y.; Yao, Q.; Deng, Y.; Chen, X.; Lv, L.; Zhuo, C.; Chen, Z.; et al. Prevalence and characterisation of CTX-M β-lactamases amongst Escherichia coli isolates from healthy food animals in China. Int. J. Antimicrob. Agents 2012, 39, 305–310. [Google Scholar] [CrossRef]
- Tamang, M.; Nam, H.M.; Kim, S.R.; Chae, M.; Jang, G.C.; Jung, S.C.; Lim, S.K. Prevalence and Molecular Characterization of CTX-M β-Lactamase-Producing Escherichia coli Isolated from Healthy Swine and Cattle. Foodborne Pathog. Dis. 2013, 10, 13–20. [Google Scholar] [CrossRef]
- Rao, L.; Lv, L.; Zeng, Z.; Chen, S.; He, D.; Chen, X.; Wu, C.; Wang, Y.; Yang, T.; Wu, P.; et al. Increasing prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in food animals and the diversity of CTX-M genotypes during 2003–2012. Vet. Microbiol. 2014, 172, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; An, W.; Wang, H.; Zhang, X. Prevalence and characteristics of extended-spectrum β-lactamase genes in Escherichia coli isolated from piglets with post-weaning diarrhea in Heilongjiang province, China. Front. Microbiol. 2015, 6, 1103. [Google Scholar] [CrossRef]
- Song, H.J.; Moon, D.C.; Kim, S.J.; Mechesso, A.F.; Choi, J.H.; Boby, N.; Kang, H.Y.; Na, S.H.; Yoon, S.S.; Lim, S.K. Antimicrobial Resistance Profiles and Molecular Characteristics of Extended-Spectrum β-Lactamase-Producing Escherichia coli Isolated from Healthy Cattle and Pigs in Ko-rea. Foodborne Pathog. Dis. 2023, 20, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; An, J.U.; Guk, J.H.; Song, H.; Yi, S.; Kim, W.H.; Cho, S. Prevalence, Characteristics and Clonal Distribution of Extended-Spectrum β-Lactamase- and AmpC β-Lactamase-Producing Escherichia coli Following the Swine Production Stages, and Potential Risks to Humans. Front. Microbiol. 2021, 12, 710747. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Oh, S.S.; Kim, J.; Park, S.; Shin, J. Clinically Relevant Extended-Spectrum β-Lactamase-Producing Escherichia coli Isolates From Food Animals in South Korea. Front. Microbiol. 2020, 11, 604. [Google Scholar] [CrossRef]
- Seo, K.W.; Do, K.H.; Jung, C.M.; Lee, S.W.; Lee, Y.J.; Lim, S.K.; Lee, W.K. Comparative genetic characterisation of third-generation cephalosporin-resistant Escherichia coli isolated from integrated and conventional pig farm in Korea. J. Glob. Antimicrob. Resist. 2023, 34, 74–82. [Google Scholar] [CrossRef]
- Frye, J.G.; Jackson, C.R. Genetic mechanisms of antimicrobial resistance identified in Salmonella enterica, Escherichia coli, and Enterococcus spp. isolated from U.S. food animals. Front. Microbiol. 2013, 4, 135. [Google Scholar] [CrossRef]
- Jahanbakhsh, S.; Smith, M.G.; Kohan-Ghadr, H.R.; Letellier, A.; Abraham, S.; Trott, D.J.; Fairbrother, J. Dynamics of extended-spectrum cephalosporin resistance in pathogenic Escherichia coli isolated from diseased pigs in Quebec, Canada. Int. J. Antimicrob. Agents 2016, 48, 194–202. [Google Scholar] [CrossRef]
- Hayer, S.; Lim, S.; Hong, S.; Elnekave, E.; Johnson, T.; Rovira, A.; Vannucci, F.; Clayton, J.B.; Perez, A.; Alvarez, J. Genetic Determinants of Resistance to Extended-Spectrum Cephalosporin and Fluoroquinolone in Escherichia coli Isolated from Diseased Pigs in the United States. Msphere 2020, 5, 10–128. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA) and European Centre for Disease Prevention and Control (ECDC), 2021. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA J. 2021, 19, 6490. [Google Scholar] [CrossRef]
- Ewers, C.; de Jong, A.; Prenger-Berninghoff, E.; Garch, F.; Leidner, U.; Tiwari, S.K.; Semmler, T. Genomic Diversity and Virulence Potential of ESBL- and AmpC-β-Lactamase-Producing Escherichia coli Strains From Healthy Food Animals Across Europe. Front. Microbiol. 2021, 12, 626774. [Google Scholar] [CrossRef]
- Endimiani, A.; Rossano, A.; Kunz, D.; Overesch, G.; Perreten, V. First countrywide survey of third-generation cephalosporin-resistant Escherichia coli from broilers, swine, and cattle in Switzerland. Diagn. Micr. Infect. Dis. 2012, 73, 31–38. [Google Scholar] [CrossRef]
- Wu, G.; Day, M.J.; Mafura, M.T.; Nunez-Garcia, J.; Fenner, J.J.; Sharma, M.; van Essen-Zandbergen, A.; Rodríguez, I.; Dierikx, C.; Kadlec, K.; et al. Comparative Analysis of ESBL-Positive Escherichia coli Isolates from Animals and Humans from the UK, The Netherlands and Germany. PLoS ONE 2013, 8, e75392. [Google Scholar] [CrossRef] [PubMed]
- Valentin, L.; Sharp, H.; Hille, K.; Seibt, U.; Fischer, J.; Pfeifer, Y.; Michael, G.B.; Nickel, S.; Schmiedel, J.; Falgenhauer, L.; et al. Subgrouping of ESBL-producing Escherichia coli from animal and human sources: An approach to quantify the distribution of ESBL types between different reservoirs. Int. J. Med. Microbiol. 2014, 304, 805–816. [Google Scholar] [CrossRef] [PubMed]
- García-Cobos, S.; Köck, R.; Mellmann, A.; Frenzel, J.; Friedrich, A.W.; Rossen, J.W. Molecular Typing of Enterobacteriaceae from Pig Holdings in North-Western Germany Reveals Extended-Spectrum and AmpC β-Lactamases Producing but no Carbapenem Resistant Ones. PLoS ONE 2015, 10, e0134533. [Google Scholar] [CrossRef] [PubMed]
- Lalak, A.; Wasyl, D.; Zając, M.; Skarżyńska, M.; Hoszowski, A.; Samcik, I.; Woźniakowski, G.; Szulowski, K. Mechanisms of cephalosporin resistance in indicator Escherichia coli isolated from food animals. Vet. Microbiol. 2016, 194, 69–73. [Google Scholar] [CrossRef]
- Rodrigues, C.; Machado, E.; Peixe, L.; Novais, Â. IncI1/ST3 and IncN/ST1 plasmids drive the spread of blaTEM-52 and blaCTX-M-1/-32 in diverse Escherichia coli clones from different piggeries. J. Antimicrob. Chemoth. 2013, 68, 2245–2248. [Google Scholar] [CrossRef]
- Fournier, C.; Aires-de-Sousa, M.; Nordmann, P.; Poirel, L. Occurrence of CTX-M-15- and MCR-1-producing Enterobacteales in pigs in Portugal: Evidence of direct links with antibiotic selective pressure. Int. J. Antimicrob. Agents 2020, 55, 105802. [Google Scholar] [CrossRef]
- Zelendova, M.; Dolejska, M.; Masarikova, M.; Jamborova, I.; Vasek, J.; Smola, J.; Manga, I.; Cizek, A. CTX-M-producing Escherichia coli in pigs from a Czech farm during production cycle. Lett. Appl. Microbiol. 2020, 71, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K.; Bortolaia, V.; Damborg, P.; Guardabassi, L. Strain Diversity of CTX-M-Producing Enterobacteriaceae in Individual Pigs: In-sights into the Dynamics of Shedding during the Production Cycle. Appl. Environ. Microbiol. 2014, 80, 6620–6626. [Google Scholar] [CrossRef]
- Abraham, S.; Kirkwood, R.N.; Laird, T.; Saputra, S.; Mitchell, T.; Singh, M.; Linn, B.; Abraham, R.J.; Pang, S.; Gordon, D.M.; et al. Dissemination and persistence of extended-spectrum cephalosporin-resistance encoding IncI1-blaCTXM-1 plasmid among Escherichia coli in pigs. ISME J. 2018, 12, 2352–2362. [Google Scholar] [CrossRef]
- Moor, J.; Aebi, S.; Rickli, S.; Mostacci, N.; Overesch, G.; Oppliger, A.; Hilty, M. Dynamics of extended-spectrum cephalosporin-resistant E. coli in pig farms: A longitudinal study. Int. J. Antimicrob. Agents 2021, 58, 106382. [Google Scholar] [CrossRef]
- Dohmen, W.; Dorado-García, A.; Bonten, M.J.M.; Wagenaar, J.A.; Mevius, D.; Heederik, D.J.J. Risk factors for ESBL-producing Escherichia coli on pig farms: A longitudinal study in the context of reduced use of antimicrobials. PLoS ONE 2017, 12, e0174094. [Google Scholar] [CrossRef] [PubMed]
- von Salviati, C.; Friese, A.; Roschanski, N.; Laube, H.; Guerra, B.; Käsbohrer, A.; Kreienbrock, L.; Roesler, U. Extended-spectrum beta-lactamases (ESBL)/AmpC beta-lactamases-producing Escherichia coli in German fattening pig farms: A longitudinal study. Berl. Und Münchener Tierärztliche Wochenschr. 2014, 127, 412–419. [Google Scholar]
- Poulin-Laprade, D.; Brouard, J.S.; Gagnon, N.; Turcotte, A.; Langlois, A.; Matte, J.J.; Carrillo, C.D.; Zaheer, R.; McAllister, T.A.; Topp, E.; et al. Resistance Determinants and Their Genetic Context in Enterobacteria from a Longitudinal Study of Pigs Reared under Various Husbandry Conditions. Appl. Environ. Microbiol. 2021, 87, e02612-20. [Google Scholar] [CrossRef]
- Nguyen, N.T.; Nguyen, H.M.; Nguyen, C.V.; Nguyen, T.V.; Nguyen, M.T.; Thai, H.Q.; Ho, M.H.; Thwaites, G.; Ngo, H.T.; Baker, S.; et al. Use of Colistin and Other Critical Antimicrobials on Pig and Chicken Farms in Southern Vietnam and Its Association with Resistance in Commensal Escherichia coli Bacteria. Appl. Environ. Microbiol. 2016, 82, 3727–3735. [Google Scholar] [CrossRef]
- Jørgensen, C.J.; Cavaco, L.M.; Hasman, H.; Emborg, H.D.; Guardabassi, L. Occurrence of CTX-M-1-producing Escherichia coli in pigs treated with ceftiofur. J. Antimicrob. Chemother. 2007, 59, 1040–1042. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lutz, E.A.; McCarty, M.J.; Mollenkopf, D.F.; Funk, J.A.; Gebreyes, W.A.; Wittum, T.E. Ceftiofur Use in Finishing Swine Barns and the Recovery of Fecal Escherichia coli or Salmonella spp. Resistant to Ceftriaxone. Foodborne Pathog. Dis. 2011, 8, 1229–1234. [Google Scholar] [CrossRef] [PubMed]
- Andersen, V.D.; Jensen, V.F.; Vigre, H.; Andreasen, M.; Agersø, Y. The use of third and fourth generation cephalosporins affects the occurrence of extended-spectrum cephalosporinase-producing Escherichia coli in Danish pig herds. Vet. J. 2015, 204, 345–350. [Google Scholar] [CrossRef]
- Agersø, Y.; Aarestrup, F.M. Voluntary ban on cephalosporin use in Danish pig production has effectively reduced extended-spectrum cephalosporinase-producing Escherichia coli in slaughter pigs. J. Antimicrob. Chemoth. 2013, 68, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, G.A. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 2005, 41, S120–S126. [Google Scholar] [CrossRef]
- Strahilevitz, J.; Jacoby, G.A.; Hooper, D.C.; Robicsek, A. Plasmid-Mediated Quinolone Resistance: A Multifaceted Threat. Clin. Microbiol. Rev. 2009, 22, 664–689. [Google Scholar] [CrossRef]
- Taylor, N.M.; Davies, R.H.; Ridley, A.; Clouting, C.; Wales, A.D.; Clifton-Hadley, F.A. A survey of fluoroquinolone resistance in Escherichia coli and thermophilic Campylobacter spp. on poultry and pig farms in Great Britain. J. Appl. Microbiol. 2008, 105, 1421–1431. [Google Scholar] [PubMed]
- Rhouma, M.; Madec, J.Y.; Laxminarayan, R. Colistin: From the shadows to a One Health approach for addressing antimicrobial resistance. Int. J. Antimicrob. Agents 2023, 61, 106713. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Zhang, S.; Abbas, M.; Rehman, M.; Wang, M.; Jia, R.; Chen, S.; Liu, M.; Zhu, D.; Zhao, X.; Gao, Q.; et al. Updates on the global dissemination of colistin-resistant Escherichia coli: An emerging threat to public health. Sci. Total Environ. 2021, 799, 149280. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; An, J.U.; Woo, J.; Song, H.; Yi, S.; Kim, W.H.; Lee, J.H.; Ryu, S.; Cho, S. Prevalence, Characteristics, and Clonal Distribution of Escherichia coli Carrying Mobilized Colistin Resistance Gene mcr-1.1 in Swine Farms and Their Differences According to Swine Production Stages. Front. Microbiol. 2022, 13, 873856. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization (FAO). The FAO Action Plan on Antimicrobial Resistance 2021–2025; FAO: Rome, Italy, 2021. [Google Scholar] [CrossRef]
| Antimicrobial a | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Country | Year b | TET | SUL c | TMP | SXT | AMP | CHL | STR | GEN | AXO d | CTX d | CIP d | CIP HL d | AZM d | COL d | CS | MDR |
| Denmark | 2021 | 29.2% | 41.5% | 30.8% | - | 38.5% | 4.6% | - | 0.0% | - | 1.5% | 0.0% | - | 4.6% | 0.0% | 52.3% | 33.8% |
| France | 2021 | 42.2% | 29.3% | 21.1% | - | 25.4% | 8.6% | - | 0.0% | - | 0.9% | 2.6% | 0.4% | 1.3% | 0.0% | 44.0% | 23.7% |
| Germany | 2021 | 32.1% | 29.5% | 23.7% | - | 28.9% | 6.3% | - | 2.6% | - | 0.0% | 1.6% | 0.5% | 2.6% | 0.0% | 49.5% | 22.6% |
| Ireland | 2021 | 51.8% | 37.1% | 36.5% | - | 28.2% | 9.4% | - | 3.5% | - | 0.0% | 2.4% | 0.0% | 0.6% | 0.0% | 38.2% | 32.9% |
| Netherlands | 2021 | 31.0% | 24.0% | 24.3% | - | 22.3% | 9.3% | - | 0.7% | - | 0.0% | 2.0% | 0.0% | 1.7% | 0.0% | 50.7% | 20.3% |
| Spain | 2021 | 78.8% | 58.8% | 60.0% | - | 83.5% | 41.2% | - | 4.7% | - | 1.2% | 50.6% | 11.8% | 4.7% | 0.0% | 6.5% | 78.8% |
| Sweden | 2021 | 16.8% | 22.5% | 19.7% | - | 24.9% | 7.5% | - | 0.0% | - | 0.6% | 1.7% | 0.0% | 0.6% | 0.0% | 63.6% | 19.7% |
| EU/EEA e | 2021 | 45.9% | 33.8% | 25.9% | - | 32.8% | 11.8% | - | 1.1% | - | 0.9% | 6.4% | 1.2% | 1.6% | 0.0% | 38.3% | 31.2% |
| UK f | 2021 | 52.7% | 40.5% | 37.6% | - | 33.3% | 18.6% | - | 2.1% | - | 1.3% | 4.6% | - | - | 0.0% | - | - |
| USA g | 2021 | 66.5% | 20.3% | - | 9.7% | 25.0% | 7.2% | - | 3.8% | 9.3% | - | 10.2% | 3.0% | 0.4% | - | 27.1% | 16.1% |
| Canada h | 2019 | 55.5% | 35.1% | - | 13.1% | 29.9% | 12.4% | 40.9% | 0.0% | 2.2% | - | - | 0.0% | 0.0% | - | 25.5% | - |
| Japan i | 2017 | 55.4% | - | - | 26.5% | 33.7% | 21.7% | 41.0% | 3.6% | - | 1.2% | - | - | - | 0.0% | - | - |
| Study | Antimicrobial a | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Country | Year b | TET | SUL | TMP | SXT | AMP | STR | GEN | CHL | CTX | TIO | AXO | CIP |
| Europe | |||||||||||||
| Spain [89] | 2000 | 95.6% | 87.8% | 83.4% | - | 72.2% | - | 8.0% | 59.5% | - | - | - | - |
| Spain [90] | 2001 | 68.0% | - | - | 48.0% | 29% | - | 7.0% | 15.0% | 0% | - | - | 3.0% |
| Portugal [63] | 2013 | 93.9% | - | - | 69.7% | 68.2% | 77.3% | 4.5% | 36.4% | 0% | - | - | 1.5% |
| Poland [64] | 2013 | 48.9% | 35.8% | 16.3% | - | 29.5% | 42.6% | 2.6% | 18.9% | 2.6% | - | - | 6.3% |
| Ireland [65] | 2016 | 59.0% | - | - | 27.6% | 18.0% | 33.3% | 5.8% | 9.6% | 0% | 0% | - | 0% |
| Estonia [67] | 2019 | 32.5% | 30.0% | 22.4% | - | 58.7% | 39.2% | 12.5% | 5.8% | 2.5% | - | 0% | 5.8% |
| Denmark c [66] | 2016 | 42.3% | 24.6% | 23.1% | - | 25.0% | 44.2% | 5.8% | 0% | 0% | - | - | 0% |
| France c [66] | 2016 | 74.5% | - | - | 40.4% | 14.9% | 66.0% | 7.5% | 17.0% | 0% | - | - | 4.3% |
| Italy c [66] | 2016 | 74.4% | 61.6% | 50.4% | - | 62.4% | 61.6% | 6.4% | 30.4% | 0% | - | - | 12.0% |
| Sweden c [66] | 2016 | 14.1% | 25.4% | 19.7% | - | 18.3% | 25.4% | 1.4% | 1.4% | 0% | - | - | 1.4% |
| Denmark [91] | 2012 | 36.0% | - | - | 14.7% | 24.0% | - | 0% | 6.7% | 0% | - | - | 0% |
| France [91] | 2012 | 83.2% | - | - | 43.6% | 24.8% | - | 2.0% | 20.8% | 0% | - | - | 0% |
| Germany [91] | 2012 | 64.4% | - | - | 33.7% | 33.7% | - | 0% | 13.5% | 0% | - | - | 1.0% |
| Netherlands [91] | 2012 | 67.9% | - | - | 42.1% | 25.7% | - | 0% | 14.3% | 0% | - | - | 0% |
| Spain [91] | 2012 | 94.0% | - | - | 66.0% | 66.0% | - | 5.0% | 42.0% | 0% | - | - | 0% |
| Europe d [92] | 2022 | 53.3% | - | - | 29.5% | 35.3% | - | 2.2% | 21.3% | 0.8% | - | - | 1.6% |
| France d [92] | 2022 | 61.7% | - | - | 29.0% | 29.4% | - | 0.5% | 13.6% | 0.5% | - | - | 0.9% |
| Germany d [92] | 2022 | 32.9% | - | - | 18.6% | 27.1% | - | 0.5% | 6.2% | 2.9% | - | - | 1.0% |
| Netherlands d [92] | 2022 | 43.1% | - | - | 24.1% | 24.5% | - | 0.5% | 31.0% | 0.0% | - | - | 0.5% |
| Spain d [92] | 2022 | 81.6% | - | - | 50.2% | 66.7% | - | 5.5% | 39.8% | 0.5% | - | - | 6.0% |
| UK d [92] | 2022 | 48.5% | - | - | 26.5% | 30.4% | - | 4.4% | 16.7% | 0.0% | - | - | 0.0% |
| North America | |||||||||||||
| Canada [93] | 1998 | 71.3% | 38.2% | - | - | 29.1% | - | 0.6% | - | - | - | - | - |
| Canada [94] | 2008 | 78.9% | 49.9% | - | 6.4% | 30.6% | 49.6% | 1.1% | 17.6% | - | 0% | 0% | 0% |
| Canada [95] | 2008 | 66.8% | 46.0% | - | 7.4% | 18.6% | 33.4% | 0.8% | 17.3% | - | 0.1% | 0% | 0% |
| Canada [96] | 2008 | 73.0% | 46.6% | - | 2.6% | 25.7% | 27.5% | 0.4% | 15.5% | - | 0% | 0.43% | 0% |
| USA c [97] | 2007 | 90.9% | 31.6% | - | 1.9% | 24.1% | 28.9% | 0.8% | 8.2% | - | 0.3% | 2.0% | 0% |
| Australia | |||||||||||||
| Australia [98] | 2016 | - | - | - | - | 29.4% | - | 17.5% | - | - | 1.8% | - | - |
| Australia [99] | 2018 | 67.7% | - | - | 34.3% | 60.2% | 33.9% | 0% | 22.4% | - | 0% | 0% | 1.0% |
| Asia | |||||||||||||
| Korea [100] | 2007 | 96.3% | - | - | 38.8% | 66.1% | 66.8% | 42.0% | 47.6% | 1.0% | - | - | 7.8% |
| Korea d [101] | 2014 | 89.9% | - | - | - | 71.3% | 61.2% | 23.2% | 68.4% | - | - | - | 8.4% |
| Korea d [102] | 2022 | 73.9% | - | - | 84.8% | 79.4% | 74.5% | 17.6% | 80.0% | - | 5.5% | - | 14.5% |
| China [74] | 2019 | 98.3% | - | - | 71.6% | 90.0% | - | 21.7% | 75.0% | - | - | - | 21.7% |
| China [75] | 2020 | 73.5% | - | - | 71.6% | 58.0% | 53.0% | 21.6% | - | 16.7% | - | - | 23.9% |
| Africa | |||||||||||||
| Tanzania [72] | 2015 | 72.9% | - | - | 60.0% | 38.6% | 50.0% | - | - | 24.3% | - | - | 10% |
| Tanzania [73] | 2021 | 51.3% | - | - | 47.7% | 46.4% | - | 26.0% | 27.3% | 29.5% | - | - | 28.6% |
| Nigeria [69] | 2015 | 50.0% | - | - | 17.9% | 10.7% | - | 0% | - | 3.6% | - | 3.6% | 3.6% |
| Rwanda [71] | 2021 | 26.7% | - | - | - | 12.6% | 13.3% | - | 2.2% | 0.7% | - | 0.7% | 0% |
| Uganda [70] | 2021 | 53.9% | 88.5% | 17.3% | - | 11.5% | - | 3.8% | 5.7% | 7.7% | - | - | 7.6% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Neill, L.; Manzanilla, E.G.; Ekhlas, D.; Leonard, F.C. Antimicrobial Resistance in Commensal Escherichia coli of the Porcine Gastrointestinal Tract. Antibiotics 2023, 12, 1616. https://doi.org/10.3390/antibiotics12111616
O’Neill L, Manzanilla EG, Ekhlas D, Leonard FC. Antimicrobial Resistance in Commensal Escherichia coli of the Porcine Gastrointestinal Tract. Antibiotics. 2023; 12(11):1616. https://doi.org/10.3390/antibiotics12111616
Chicago/Turabian StyleO’Neill, Lorcan, Edgar García Manzanilla, Daniel Ekhlas, and Finola C. Leonard. 2023. "Antimicrobial Resistance in Commensal Escherichia coli of the Porcine Gastrointestinal Tract" Antibiotics 12, no. 11: 1616. https://doi.org/10.3390/antibiotics12111616
APA StyleO’Neill, L., Manzanilla, E. G., Ekhlas, D., & Leonard, F. C. (2023). Antimicrobial Resistance in Commensal Escherichia coli of the Porcine Gastrointestinal Tract. Antibiotics, 12(11), 1616. https://doi.org/10.3390/antibiotics12111616





