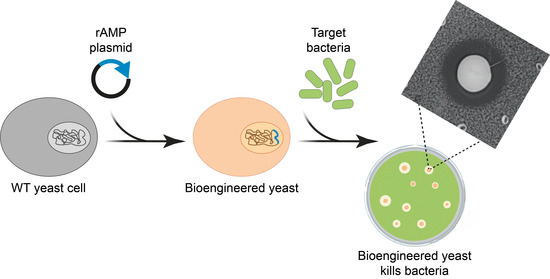
Bioengineering the Antimicrobial Activity of Yeast by Recombinant Thanatin Production
Abstract
:1. Introduction
2. Results
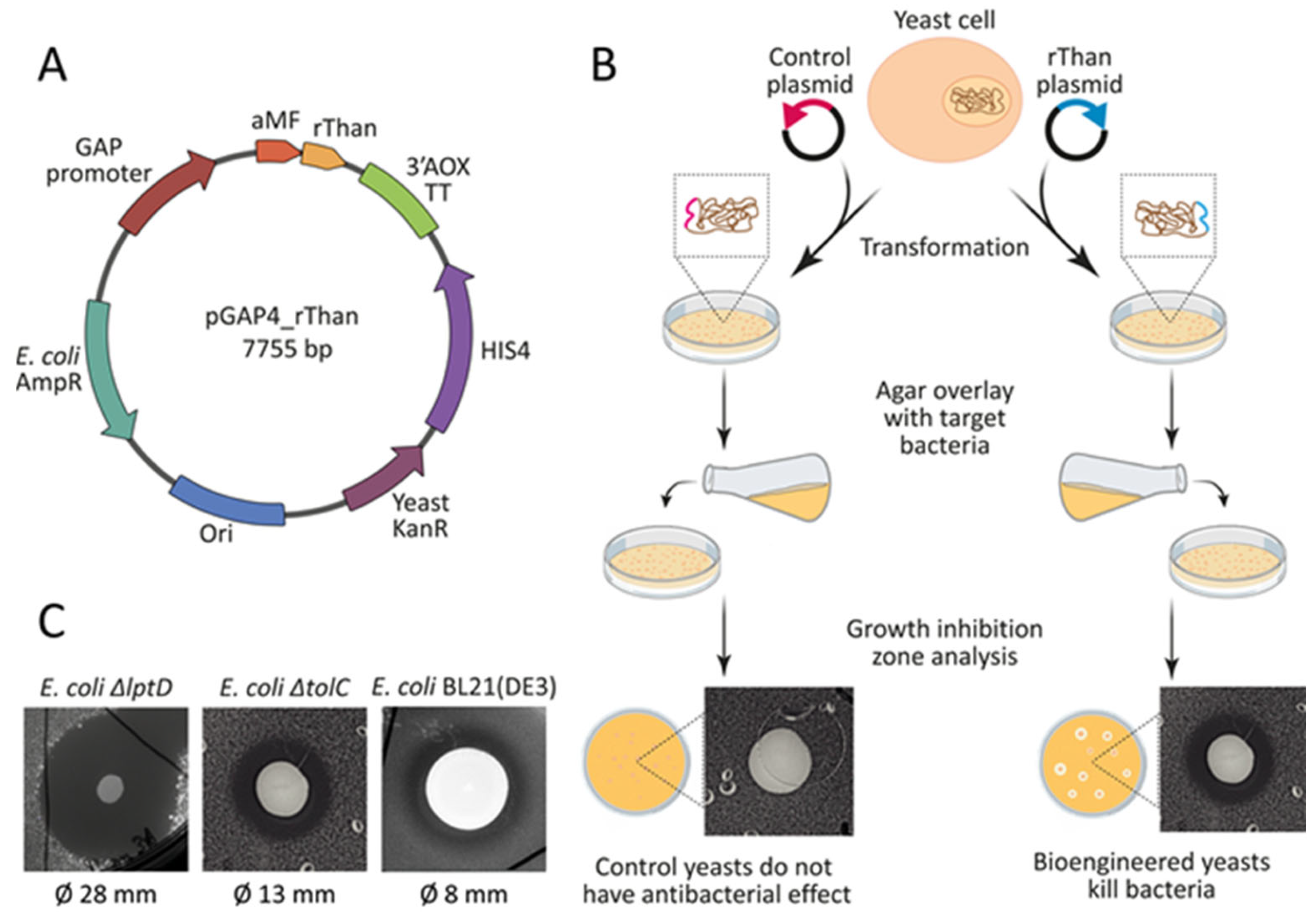
2.1. Bioengineering rThan—Producing Yeast
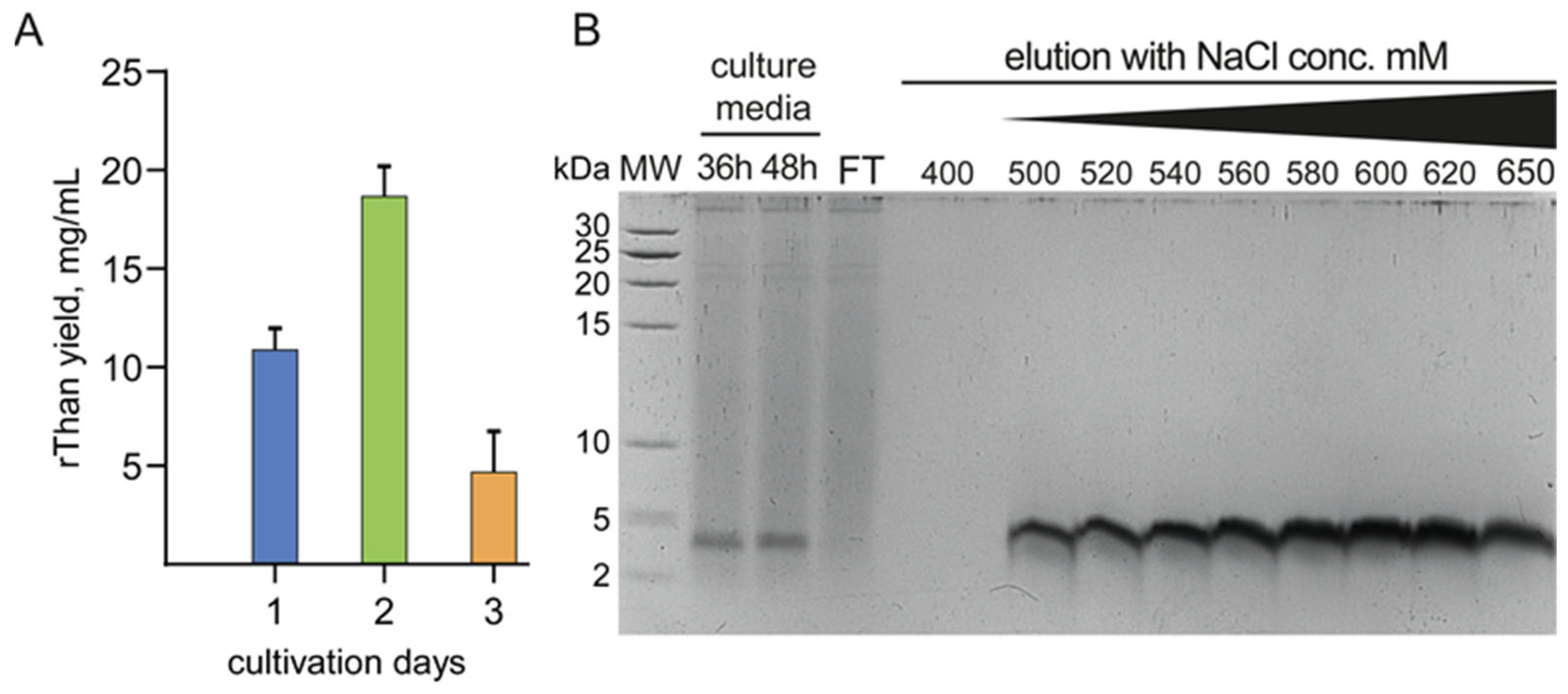
2.2. Constitutive Production and Purification of rThan
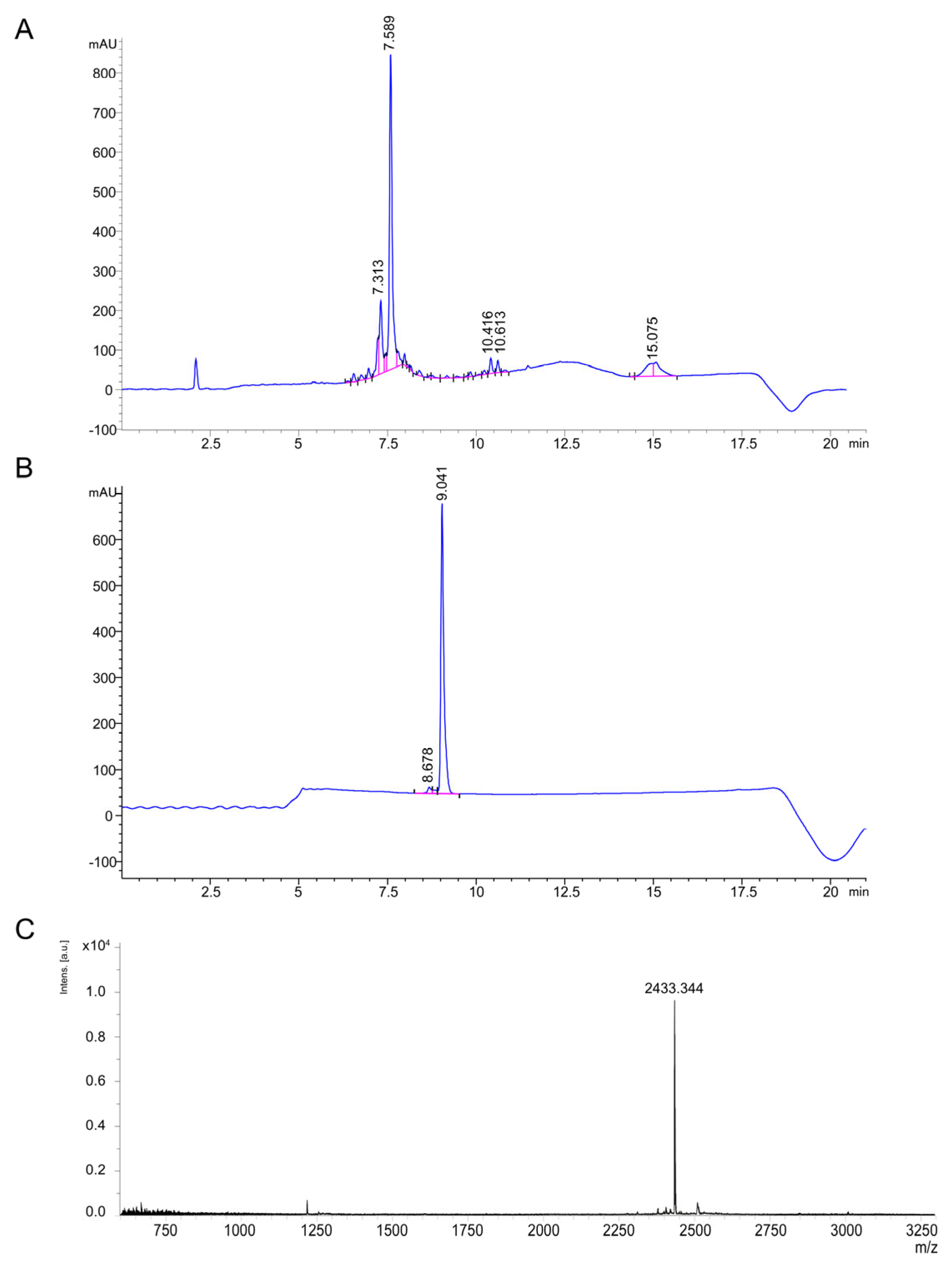
2.3. Chemical Synthesis of Thanatin
2.4. Antimicrobial Activity Spectra of rThan
2.5. Cocultivation Experiments Reveal Biocontrol Potential of rThan-Producing Yeasts
3. Discussion
4. Materials and Methods
4.1. Bacterial and Yeast Strains
4.2. Expression Vector Design and Construction
4.3. Yeast Transformation, rThan Production and Purification
4.4. Agar Overlay Assay
4.5. Cocultivation with Target Bacteria
4.6. Peptide Chemical Synthesis
4.6.1. Materials
4.6.2. Peptide Synthesis
4.6.3. Disulfide Bond Formation in Thanatin
4.6.4. Peptide Purification
4.7. Evaluation of Antimicrobial Activity
4.8. Evaluation of Cytotoxicity
4.9. Liquid Chromatography and Mass Spectrometry
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nicolaou, K.C.; Rigol, S. A Brief History of Antibiotics and Select Advances in Their Synthesis. J. Antibiot. 2017, 71, 153–184. [Google Scholar] [CrossRef] [PubMed]
- Xie, J. Grand Challenge of Antibiotics Resistance: A Global, Multidisciplinary Effort Is Needed. Front. Antibiot. 2022, 1, 984076. [Google Scholar] [CrossRef]
- de Breij, A.; Riool, M.; Cordfunke, R.A.; Malanovic, N.; de Boer, L.; Koning, R.I.; Ravensbergen, E.; Franken, M.; van der Heijde, T.; Boekema, B.K.; et al. The Antimicrobial Peptide SAAP-148 Combats Drug-Resistant Bacteria and Biofilms. Sci. Transl. Med. 2018, 10, eaan4044. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Kizhakkedathu, J.; Straus, S. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Laslop, N.; Mankin, A.S. How Macrolide Antibiotics Work. Trends Biochem. Sci. 2018, 43, 668–684. [Google Scholar] [CrossRef] [PubMed]
- Kotra, L.P.; Haddad, J.; Mobashery, S. Aminoglycosides: Perspectives on Mechanisms of Action and Resistance and Strategies to Counter Resistance. Antimicrob. Agents Chemother. 2000, 44, 3249–3256. [Google Scholar] [CrossRef] [PubMed]
- Yocum, R.R.; Rasmussen, J.R.; Strominger, J.L. The Mechanism of Action of Penicillin. Penicillin Acylates the Active Site of Bacillus Stearothermophilus D-Alanine Carboxypeptidase. J. Biol. Chem. 1980, 255, 3977–3986. [Google Scholar] [CrossRef]
- Fàbrega, A.; Madurga, S.; Giralt, E.; Vila, J. Mechanism of Action of and Resistance to Quinolones. Microb. Biotechnol. 2009, 2, 40–61. [Google Scholar] [CrossRef]
- Lee, T.-H.; Hall, K.N.; Aguilar, M.-I. Antimicrobial Peptide Structure and Mechanism of Action: A Focus on the Role of Membrane Structure. Curr. Top. Med. Chem. 2016, 16, 25–39. [Google Scholar] [CrossRef]
- Wimley, W.C. Describing the Mechanism of Antimicrobial Peptide Action with the Interfacial Activity Model. ACS Chem. Biol. 2010, 5, 905–917. [Google Scholar] [CrossRef]
- Sitaram, N.; Nagaraj, R. Interaction of Antimicrobial Peptides with Biological and Model Membranes: Structural and Charge Requirements for Activity. Biochim. Biophys. Acta 1999, 1462, 29–54. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial Peptides: Pore Formers or Metabolic Inhibitors in Bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Vetterli, S.U.; Zerbe, K.; Müller, M.; Urfer, M.; Mondal, M.; Wang, S.-Y.; Moehle, K.; Zerbe, O.; Vitale, A.; Pessi, G.; et al. Thanatin Targets the Intermembrane Protein Complex Required for Lipopolysaccharide Transport in Escherichia coli. Sci. Adv. 2018, 4, eaau2634. [Google Scholar] [CrossRef]
- Dijksteel, G.S.; Ulrich, M.M.W.; Middelkoop, E.; Boekema, B.K.H.L. Review: Lessons Learned from Clinical Trials Using Antimicrobial Peptides (AMPs). Front. Microbiol. 2021, 12, 616979. [Google Scholar] [CrossRef] [PubMed]
- Spohn, R.; Daruka, L.; Lázár, V.; Martins, A.; Vidovics, F.; Grézal, G.; Méhi, O.; Kintses, B.; Számel, M.; Jangir, P.K.; et al. Integrated Evolutionary Analysis Reveals Antimicrobial Peptides with Limited Resistance. Nat. Commun. 2019, 10, 4538. [Google Scholar] [CrossRef] [PubMed]
- Jahangiri, A.; Neshani, A.; Mirhosseini, S.A.; Ghazvini, K.; Zare, H.; Sedighian, H. Synergistic Effect of Two Antimicrobial Peptides, Nisin and P10 with Conventional Antibiotics against Extensively Drug-Resistant Acinetobacter Baumannii and Colistin-Resistant Pseudomonas Aeruginosa Isolates. Microb. Pathog. 2021, 150, 104700. [Google Scholar] [CrossRef]
- Lázár, V.; Martins, A.; Spohn, R.; Daruka, L.; Grézal, G.; Fekete, G.; Számel, M.; Jangir, P.K.; Kintses, B.; Csörgő, B.; et al. Antibiotic-Resistant Bacteria Show Widespread Collateral Sensitivity to Antimicrobial Peptides. Nat. Microbiol. 2018, 3, 718–731. [Google Scholar] [CrossRef]
- Ruden, S.; Rieder, A.; Chis Ster, I.; Schwartz, T.; Mikut, R.; Hilpert, K. Synergy Pattern of Short Cationic Antimicrobial Peptides against Multidrug-Resistant Pseudomonas Aeruginosa. Front. Microbiol. 2019, 10, 2740. [Google Scholar] [CrossRef]
- Fehlbaum, P.; Bulet, P.; Chernysh, S.; Briand, J.P.; Roussel, J.P.; Letellier, L.; Hetru, C.; Hoffmann, J.A. Structure-Activity Analysis of Thanatin, a 21-Residue Inducible Insect Defense Peptide with Sequence Homology to Frog Skin Antimicrobial Peptides. Proc. Natl. Acad. Sci. USA 1996, 93, 1221–1225. [Google Scholar] [CrossRef]
- Pagès, J.-M.; Dimarcq, J.-L.; Quenin, S.; Hetru, C. Thanatin Activity on Multidrug Resistant Clinical Isolates of Enterobacter Aerogenes and Klebsiella Pneumoniae. Int. J. Antimicrob. Agents 2003, 22, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Fang, C.; Lu, L.; Wang, M.; Xue, X.; Zhou, Y.; Li, M.; Hu, Y.; Luo, X.; Hou, Z. The Antimicrobial Peptide Thanatin Disrupts the Bacterial Outer Membrane and Inactivates the NDM-1 Metallo-β-Lactamase. Nat. Commun. 2019, 10, 3517. [Google Scholar] [CrossRef]
- Moura, E.C.C.M.; Baeta, T.; Romanelli, A.; Laguri, C.; Martorana, A.M.; Erba, E.; Simorre, J.-P.; Sperandeo, P.; Polissi, A. Thanatin Impairs Lipopolysaccharide Transport Complex Assembly by Targeting LptC-LptA Interaction and Decreasing LptA Stability. Front. Microbiol. 2020, 11, 909. [Google Scholar] [CrossRef]
- Wu, G.; Ding, J.; Li, H.; Li, L.; Zhao, R.; Shen, Z.; Fan, X.; Xi, T. Effects of Cations and PH on Antimicrobial Activity of Thanatin and S-Thanatin Against Escherichia coli ATCC25922 and B. Subtilis ATCC 21332. Curr. Microbiol. 2008, 57, 552–557. [Google Scholar] [CrossRef] [PubMed]
- Mochnáčová, E.; Petroušková, P.; Danišová, O.; Hudecová, P.; Bhide, K.; Kulkarni, A.; Bhide, M. Simple and Rapid Pipeline for the Production of Cyclic and Linear Small-Sized Peptides in E. Coli. Protein Expr. Purif. 2022, 191, 106026. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Li, Y.; Zhang, L.; Ding, Z.; Shi, G. Microbial Production of Small Peptide: Pathway Engineering and Synthetic Biology. Microb. Biotechnol. 2021, 14, 2257–2278. [Google Scholar] [CrossRef] [PubMed]
- Li, Y. Recombinant Production of Antimicrobial Peptides in Escherichia coli: A Review. Protein Expr. Purif. 2011, 80, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Lamer, T.; Vederas, J.C. Simplified Cloning and Isolation of Peptides from “Sandwiched” SUMO-Peptide-Intein Fusion Proteins. BMC Biotechnol. 2023, 23, 11. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; de la Fuente-Nunez, C.; Ou, R.W.; Torres, M.D.T.; Pande, S.G.; Sinskey, A.J.; Lu, T.K. Yeast-Based Synthetic Biology Platform for Antimicrobial Peptide Production. ACS Synth. Biol. 2018, 7, 896–902. [Google Scholar] [CrossRef]
- Meng, D.-M.; Dai, H.-X.; Gao, X.-F.; Zhao, J.-F.; Guo, Y.-J.; Ling, X.; Dong, B.; Zhang, Z.-Q.; Fan, Z.-C. Expression, Purification and Initial Characterization of a Novel Recombinant Antimicrobial Peptide Mytichitin-A in Pichia pastoris. Protein Expr. Purif. 2016, 127, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Kuddus, M.R.; Rumi, F.; Tsutsumi, M.; Takahashi, R.; Yamano, M.; Kamiya, M.; Kikukawa, T.; Demura, M.; Aizawa, T. Expression, Purification and Characterization of the Recombinant Cysteine-Rich Antimicrobial Peptide Snakin-1 in Pichia pastoris. Protein Expr. Purif. 2016, 122, 15–22. [Google Scholar] [CrossRef]
- Liu, Z.; Zhu, M.; Chen, X.; Yang, G.; Yang, T.; Yu, L.; Hui, L.; Wang, X. Expression and Antibacterial Activity of Hybrid Antimicrobial Peptide cecropinA-Thanatin in Pichia pastoris. Front. Lab. Med. 2018, 2, 23–29. [Google Scholar] [CrossRef]
- Çelik, E.; Çalık, P. Production of Recombinant Proteins by Yeast Cells. Biotechnol. Adv. 2012, 30, 1108–1118. [Google Scholar] [CrossRef] [PubMed]
- Moridi, K.; Hemmaty, M.; Akbari Eidgahi, M.R.; Fathi Najafi, M.; Zare, H.; Ghazvini, K.; Neshani, A. Construction, Cloning, and Expression of Melittin Antimicrobial Peptide Using Pichia pastoris Expression System. Gene Rep. 2020, 21, 100900. [Google Scholar] [CrossRef]
- Pipiya, S.O.; Mirzoeva, N.Z.; Baranova, M.N.; Eliseev, I.E.; Mokrushina, Y.A.; Shamova, O.V.; Gabibov, A.G.; Smirnov, I.V.; Terekhov, S.S. Creation of Recombinant Biocontrol Agents by Genetic Programming of Yeast. Acta Naturae 2023, 15, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Pipiya, S.O.; Mokrushina, Y.A.; Gabibov, A.G.; Smirnov, I.V.; Terekhov, S.S. Selective Eradication of Staphylococcus aureus by the Designer Genetically Programmed Yeast Biocontrol Agent. Antibiotics 2020, 9, 527. [Google Scholar] [CrossRef] [PubMed]
- Shariati, A.; Arshadi, M.; Khosrojerdi, M.A.; Abedinzadeh, M.; Ganjalishahi, M.; Maleki, A.; Heidary, M.; Khoshnood, S. The Resistance Mechanisms of Bacteria against Ciprofloxacin and New Approaches for Enhancing the Efficacy of This Antibiotic. Front. Public. Health 2022, 10, 1025633. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Santajit, S.; Indrawattana, N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. BioMed Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef]
- Pazgier, M.; Lubkowski, J. Expression and Purification of Recombinant Human α-Defensins in Escherichia coli. Protein Expr. Purif. 2006, 49, 1–8. [Google Scholar] [CrossRef]
- Tanhaeian, A.; Azghandi, M.; Mousavi, Z.; Javadmanesh, A. Expression of Thanatin in HEK293 Cells and Investigation of Its Antibacterial Effects on Some Human Pathogens. Protein Pept. Lett. 2020, 27, 41–47. [Google Scholar] [CrossRef]
- Huynh, E.; Akhtar, N.; Li, J. Efficient Production of Recombinant Protegrin-1 From, and Its Antimicrobial and Cell Migration Activity. Front. Microbiol. 2018, 9, 2300. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Lu, J.; Fang, C.; Zhou, Y.; Bai, H.; Zhang, X.; Xue, X.; Chen, Y.; Luo, X. Underlying Mechanism of in vivo and in vitro Activity of C-Terminal-Amidated Thanatin against Clinical Isolates of Extended-Spectrum Beta-Lactamase-Producing Escherichia coli. J. Infect. Dis. 2011, 203, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Edwards, I.A.; Elliott, A.G.; Kavanagh, A.M.; Zuegg, J.; Blaskovich, M.A.T.; Cooper, M.A. Contribution of Amphipathicity and Hydrophobicity to the Antimicrobial Activity and Cytotoxicity of β-Hairpin Peptides. ACS Infect. Dis. 2016, 2, 442–450. [Google Scholar] [CrossRef]
- Huang, Y.; Gao, L.; Lin, M.; Yu, T. Recombinant Expression of Antimicrobial Peptides in Pichia pastoris: A Strategy to Inhibit the Penicillium Expansum in Pears. Postharvest Biol. Technol. 2021, 171, 111298. [Google Scholar] [CrossRef]
- de los Santos, D.G.; de los Santos, J.R.G.; Gil-Turnes, C.; Gaboardi, G.; Silva, L.F.; França, R.; Fernandes, C.G.; Conceição, F.R. Probiotic Effect of Pichia pastoris X-33 Produced in Parboiled Rice Effluent and YPD Medium on Broiler Chickens. PLoS ONE 2018, 13, e0192904. [Google Scholar] [CrossRef]
- Becerril-García, M.Á.; Flores-Maldonado, O.E.; González, G.M.; García-González, G.; Hernández-Bello, R.; Palma-Nicolás, J.P. Safety Profile of Intravenous Administration of Live Pichia pastoris Cells in Mice. FEMS Yeast Res. 2022, 22, foac023. [Google Scholar] [CrossRef]
- Lee, M.E.; DeLoache, W.C.; Cervantes, B.; Dueber, J.E. A Highly Characterized Yeast Toolkit for Modular, Multipart Assembly. ACS Synth. Biol. 2015, 4, 975–986. [Google Scholar] [CrossRef]
- Obst, U.; Lu, T.K.; Sieber, V. A Modular Toolkit for Generating Pichia pastoris Secretion Libraries. ACS Synth. Biol. 2017, 6, 1016–1025. [Google Scholar] [CrossRef]
- Tian, S.; Yesselman, J.D.; Cordero, P.; Das, R. Primerize: Automated Primer Assembly for Transcribing Non-Coding RNA Domains. Nucleic Acids Res. 2015, 43, W522–W526. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Letchworth, G.J. High Efficiency Transformation by Electroporation of Pichia pastoris Pretreated with Lithium Acetate and Dithiothreitol. Biotechniques 2004, 36, 152–154. [Google Scholar] [CrossRef]
- Baranova, M.N.; Babikova, P.A.; Kudzhaev, A.M.; Mokrushina, Y.A.; Belozerova, O.A.; Yunin, M.A.; Kovalchuk, S.; Gabibov, A.G.; Smirnov, I.V.; Terekhov, S.S. Live Biosensors for Ultrahigh-Throughput Screening of Antimicrobial Activity against Gram-Negative Bacteria. Antibiotics 2021, 10, 1161. [Google Scholar] [CrossRef] [PubMed]
- Barlos, K.; Chatzi, O.; Gatos, D.; Stavropoulos, G. 2-Chlorotrityl Chloride Resin. Studies on Anchoring of Fmoc-Amino Acids and Peptide Cleavage. Int. J. Pept. Protein Res. 1991, 37, 513–520. [Google Scholar] [PubMed]
- Carpino, L.A.; Han, G.Y. 9-Fluorenylmethoxycarbonyl Function, a New Base-Sensitive Amino-Protecting Group. J. Am. Chem. Soc. 1970, 92, 5748–5749. [Google Scholar] [CrossRef]
- Knorr, R.; Trzeciak, A.; Bannwarth, W.; Gillessen, D. New Coupling Reagents in Peptide Chemistry. Tetrahedron Lett. 1989, 30, 1927–1930. [Google Scholar] [CrossRef]
- Kumar, A.; Sharma, A.; de la Torre, B.G.; Albericio, F. In Situ Fmoc Removal—A Sustainable Solid-Phase Peptide Synthesis Approach. Green. Chem. 2022, 24, 4887–4896. [Google Scholar] [CrossRef]
- King, D.S.; Fields, C.G.; Fields, G.B. A Cleavage Method Which Minimizes Side Reactions Following Fmoc Solid Phase Peptide Synthesis. Int. J. Pept. Protein Res. 1990, 36, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Sabatino, G.; D’Ercole, A.; Pacini, L.; Zini, M.; Ribecai, A.; Paio, A.; Rovero, P.; Papini, A.M. An Optimized Scalable Fully Automated Solid-Phase Microwave-Assisted cGMP-Ready Process for the Preparation of Eptifibatide. Org. Process Res. Dev. 2021, 25, 552–563. [Google Scholar] [CrossRef]
- Riss, T.L.; Moravec, R.A.; Niles, A.L.; Duellman, S.; Benink, H.A.; Worzella, T.J.; Minor, L. Cell Viability Assays. In Assay Guidance Manual; Markossian, S., Grossman, A., Brimacombe, K., Arkin, M., Auld, D., Austin, C., Baell, J., Chung, T.D.Y., Coussens, N.P., Dahlin, J.L., et al., Eds.; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2004. [Google Scholar]
- Kovalchuk, S.I.; Jensen, O.N.; Rogowska-Wrzesinska, A. FlashPack: Fast and Simple Preparation of Ultrahigh-Performance Capillary Columns for LC-MS. Mol. Cell. Proteom. 2019, 18, 383–390. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pipiya, S.O.; Kudzhaev, A.M.; Mirzoeva, N.Z.; Mokrushina, Y.A.; Ziganshin, R.H.; Komlev, A.S.; Petrova, P.E.; Smirnov, I.V.; Gabibov, A.G.; Shamova, O.V.; et al. Bioengineering the Antimicrobial Activity of Yeast by Recombinant Thanatin Production. Antibiotics 2023, 12, 1719. https://doi.org/10.3390/antibiotics12121719
Pipiya SO, Kudzhaev AM, Mirzoeva NZ, Mokrushina YA, Ziganshin RH, Komlev AS, Petrova PE, Smirnov IV, Gabibov AG, Shamova OV, et al. Bioengineering the Antimicrobial Activity of Yeast by Recombinant Thanatin Production. Antibiotics. 2023; 12(12):1719. https://doi.org/10.3390/antibiotics12121719
Chicago/Turabian StylePipiya, Sofiya O., Arsen M. Kudzhaev, Nisso Z. Mirzoeva, Yuliana A. Mokrushina, Rustam H. Ziganshin, Alexey S. Komlev, Polina E. Petrova, Ivan V. Smirnov, Alexander G. Gabibov, Olga V. Shamova, and et al. 2023. "Bioengineering the Antimicrobial Activity of Yeast by Recombinant Thanatin Production" Antibiotics 12, no. 12: 1719. https://doi.org/10.3390/antibiotics12121719