Shotgun Metagenomics-Guided Prediction Reveals the Metal Tolerance and Antibiotic Resistance of Microbes in Poly-Extreme Environments in the Danakil Depression, Afar Region
Abstract
:1. Introduction
2. Results
2.1. Physicochemical Analysis
2.2. Metagenomic Data Analysis
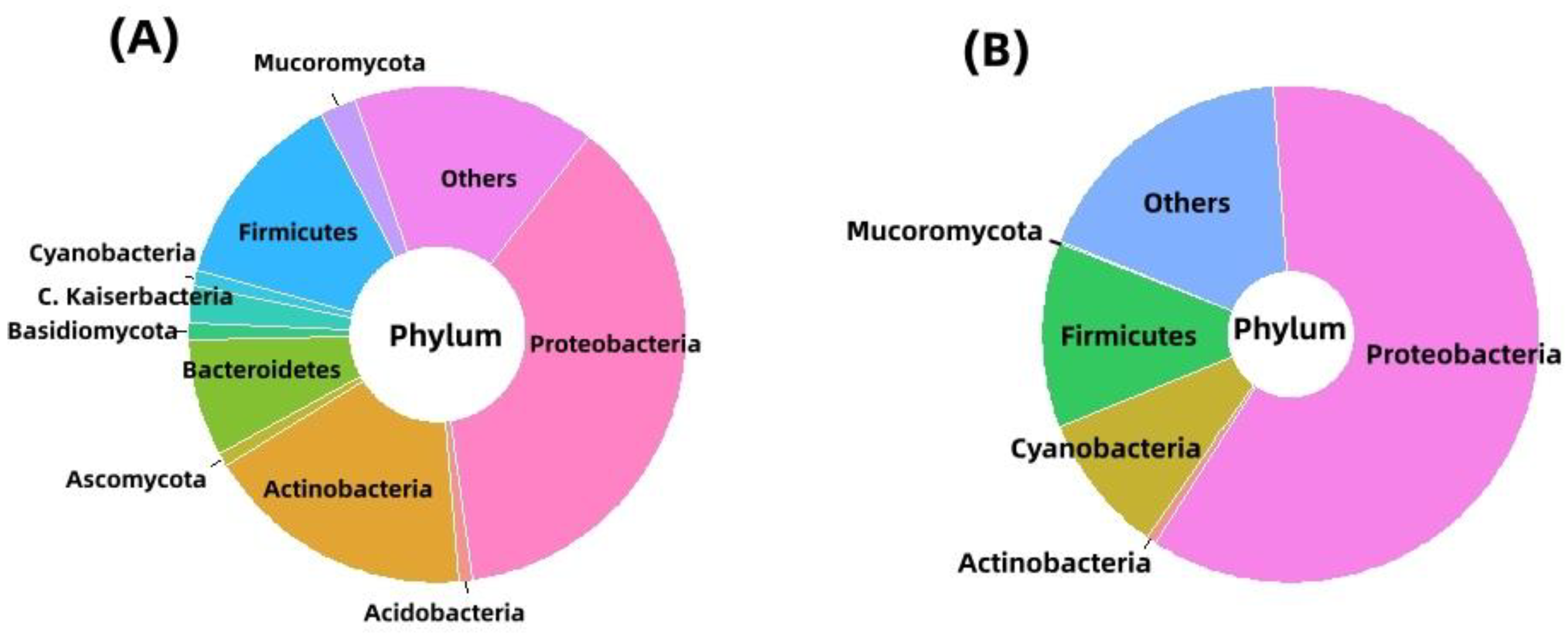
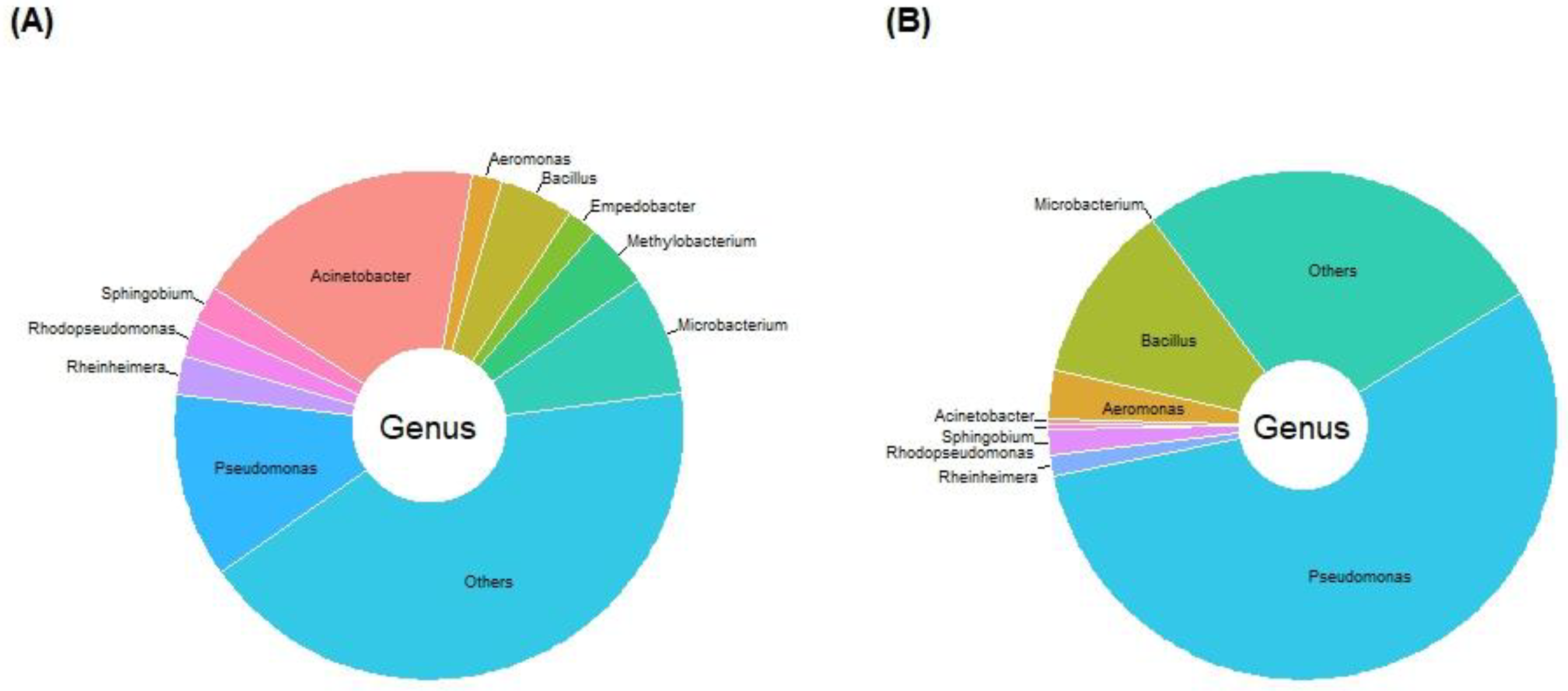
2.3. Bacterial Community Profile
2.4. Metagenomic Studies of ARGs
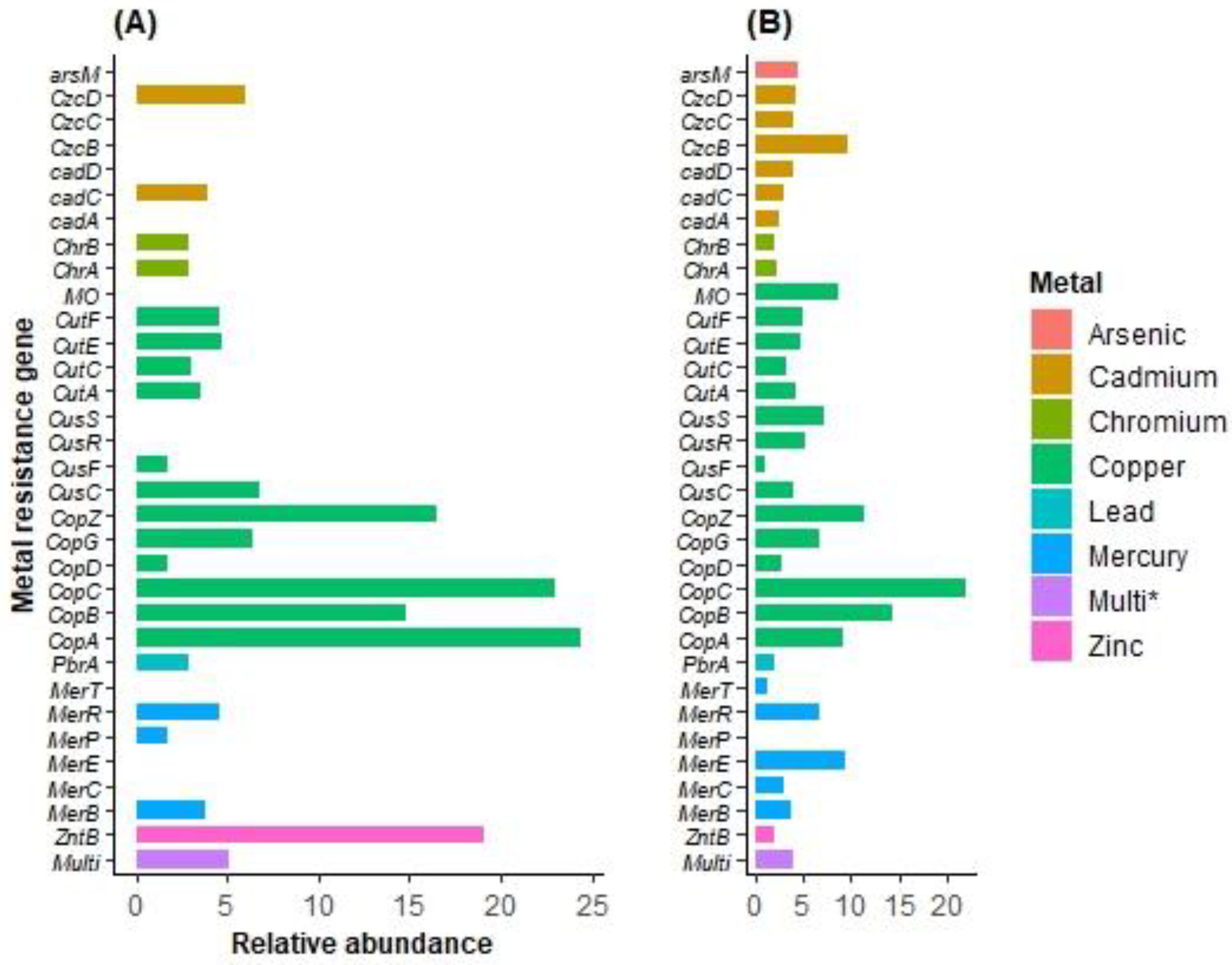
2.5. Metagenomic Studies of MRGs
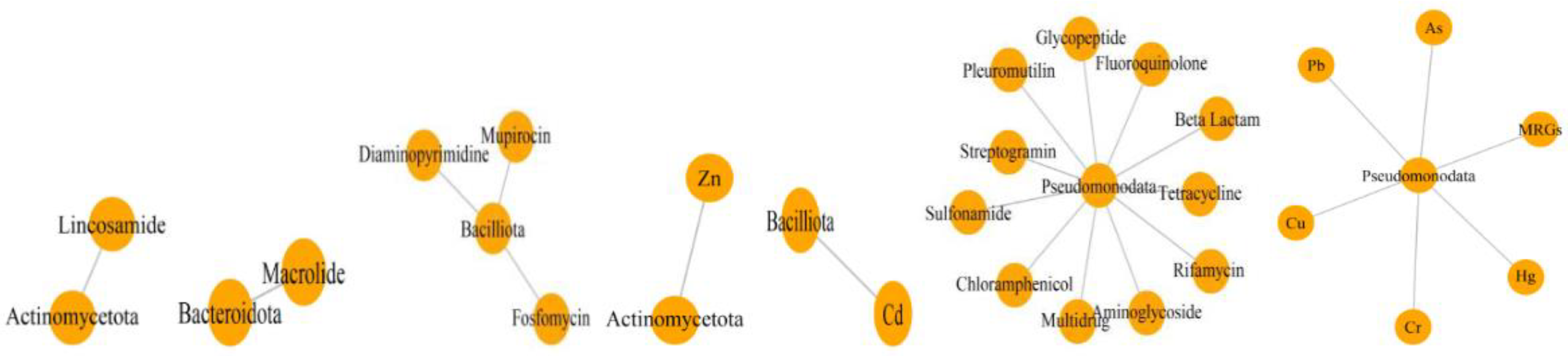
2.6. Co-Occurrence of Bacterial Hosts and Resistance Genes
2.7. Correlations between MRGs and ARGs
3. Discussion
4. Materials and Methods
4.1. Study Area and Sampling Sites
4.2. Determining Metal Compositions
4.3. Metagenomic DNA Extractions and Shotgun Sequencing
4.4. Assembly, Binning, and Annotation of Reads
4.5. Taxonomic Assignment of Contigs
4.6. Extracting Antibiotic and Metal Resistance Genes
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dafale, N.A.; Semwal, U.P.; Rajput, R.K.; Singh, G. Selection of appropriate analytical tools to determine the potency and bioactivity of antibiotics and antibiotic resistance. J. Pharm. Anal. 2016, 6, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Roca, I.; Akova, M.; Baquero, F.; Carlet, J.; Cavaleri, M.; Coenen, S.; Cohen, J.; Findlay, D.; Gyssens, I.; Heure, O.E.; et al. The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect. 2015, 6, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Abraham, E.P. The Antibiotics. In Comprehensive Biochemistry; Elsevier: Amsterdam, The Netherlands, 1963; Volume 11. [Google Scholar]
- World health organization (WHO). Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Cadernos de Pesquisa. 2013, Volume 43. Available online: http://www.cdc.gov/drugresistance/threat-report-2013/ (accessed on 24 September 2023).
- Miller, J.H.; Novak, J.T.; Knocke, W.R.; Pruden, A. Survival of antibiotic resistant bacteria and horizontal gene transfer control an-tibiotic resistance gene content in anaerobic digesters. Front. Microbiol. 2016, 7, 263. [Google Scholar] [CrossRef] [PubMed]
- Shankar, P.R. Book review: Tackling drug-resistant infections globally. Arch. Pharm. Pract. 2016, 7, 110–111. [Google Scholar] [CrossRef]
- WHO. Antimicrobial Resistance: An Emerging Water, Sanitation and Hygiene Issue. Briefing Note. 2014. Available online: www.who.int/about/licensing/copyright_form/en/index.html (accessed on 25 September 2023).
- Wuijts, S.; van den Berg, H.H.; Miller, J.; Abebe, L.; Sobsey, M.; Andremont, A.; de Roda Husman, A.M. Towards a research agenda for water, sani-tation and antimicrobial resistance. J. Water Health 2017, 15, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Chapman, B.; Pintar, K.; Smith, B.A. Multi-Exposure Pathway Model to Compare Escherichia coli O157 Risks and Interventions. Risk Anal. 2017, 38, 392–409. [Google Scholar] [CrossRef]
- Michael, I.; Rizzo, L.; McArdell, C.S.; Manaia, C.M.; Merlin, C.; Schwartz, T.; Dagot, C.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: A review. Water Res. 2013, 47, 957–995. [Google Scholar] [CrossRef]
- Rabkin, C.S.; Galaid, E.I.; Hollis, D.G.; Weaver, R.E.; Dees, S.B.; Kai, A.; Moss, C.W.; Sandhu, K.K.; Broome, C.V. Thermophilic bacteria: A new cause of human disease. J. Clin. Microbiol. 1985, 21, 553–557. [Google Scholar] [CrossRef]
- Jardine, J.L.; Abia AL, K.; Mavumengwana, V.; Ubomba-Jaswa, E. Phylogenetic Analysis and Antimicrobial Profiles. Int. J. Environ. Res. Public Health 2017, 14, 1070. [Google Scholar] [CrossRef]
- Iyer, A.; Mody, K.; Jha, B. Biosorption of heavy metals by a marine bacterium. Mar. Pollut. Bull. 2005, 50, 340–343. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhou, T.; Feng, F.; Huang, S.; Sun, Y. The response of copper resistance genes, antibiotic resistance genes, and intl1/2 to copper addition during anaerobic digestion in laboratory. Ecotoxicol. Environ. Saf. 2021, 210, 111822. [Google Scholar] [CrossRef] [PubMed]
- Summers, A.O.; Wireman, J.; Vimy, M.J.; Lorscheider, F.L.; Marshall, B.; Levy, S.B.; Billard, L. Mercury Released from Dental “Silver” Fillings Provokes an Increase in Mercury-and Antibiotic-Resistant Bacteria in Oral and Intestinal Floras of Primates. Antimicrob. Agents Chemother. 1993, 37, 825–834. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Sanchez, P.; Martínez, J.L. Environmental selection of antibiotic resistance genes. Environ. Microbiol. 2001, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Tang, J.; Wang, X.; Li, Y.; Yao, X.; Sun, H.; Wu, Y. Fate of antibiotic resistance genes and metal resistance genes during the thermophilic fermentation of solid and liquid swine manures in an ectopic fermentation system. Ecotoxicol. Environ. Saf. 2021, 213, 111981. [Google Scholar] [CrossRef] [PubMed]
- Summers, A.O. Generally Overlooked Fundamentals of Bacterial Genetics and Ecology. Clin. Infect. Dis. 2002, 34, S85–S92. Available online: http://cid.oxfordjournals.org/ (accessed on 24 September 2023). [CrossRef] [PubMed]
- Di Cesare, A.; Eckert, E.; Corno, G. Co-selection of antibiotic and heavy metal resistance in freshwater bacteria. J. Limnol. 2016, 75, 59–66. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Y.; Xie, X.; Zhu, W.; Meng, X. Fate of adsorbed Pb(II) on graphene oxide under variable redox potential controlled by electrochemical method. J. Hazard. Mater. 2019, 367, 152–159. [Google Scholar] [CrossRef]
- Seiler, C.; Berendonk, T.U. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agri-culture and aquaculture. Front. Microbiol. 2012, 3, 399. [Google Scholar] [CrossRef]
- Yuan, L.; Li, Z.-H.; Zhang, M.-Q.; Shao, W.; Fan, Y.-Y.; Sheng, G.-P. Mercury/silver resistance genes and their association with antibiotic resistance genes and microbial community in a municipal wastewater treatment plant. Sci. Total. Environ. 2019, 657, 1014–1022. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, Y.; Song, H.; Lu, J.; Yuan, Z.; Guo, J. Copper nanoparticles and copper ions promote horizontal transfer of plas-mid-mediated multi-antibiotic resistance genes across bacterial genera. Environ. Int. 2019, 129, 478–487. [Google Scholar] [CrossRef]
- Berg, J.; Thorsen, M.K.; Holm, P.E.; Jensen, J.; Nybroe, O.; Brandt, K.K. Cu Exposure under Field Conditions Coselects for Antibiotic Resistance as Determined by a Novel Cultivation-Independent Bacterial Community Tolerance Assay. Environ. Sci. Technol. 2010, 44, 8724–8728. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.-W.; Wang, J.-T.; Li, J.; Shi, X.-Z.; Ma, Y.-B.; Chen, D.; He, J.-Z. Long-Term Nickel Contamination Increases the Occurrence of Antibiotic Resistance Genes in Agricultural Soils. Environ. Sci. Technol. 2017, 51, 790–800. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, H.; Hu, J.; Zheng, T.; Zhang, R.; Zhong, H.; Gao, Q.; Sun, W.; Chen, Q.; Ni, J. Unveil the role of dissolved and sedimentary metal(loid)s on bacterial communities and metal resistance genes (MRGs) in an urban river of the Qinghai-Tibet Plateau. Water Res. 2022, 211, 118050. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Xu, Z.; Fan, L. Response of heavy metal and antibiotic resistance genes and related microorganisms to different heavy metals in activated sludge. J. Environ. Manag. 2021, 300, 113754. [Google Scholar] [CrossRef] [PubMed]
- Selgelid, M.J. Ethics and infectious DISEASE1. Bioethics 2005, 19, 272–289. [Google Scholar] [CrossRef] [PubMed]
- Clemente, J.C.; Pehrsson, E.C.; Blaser, M.J.; Sandhu, K.; Gao, Z.; Wang, B.; Magris, M.; Hidalgo, G.; Contreras, M.; Noya-Alarcón, Ó.; et al. The microbiome of uncontacted Amerindians. Sci. Adv. 2015, 1, e1500183. [Google Scholar] [CrossRef] [PubMed]
- Pawlowski, A.C.; Wang, W.; Koteva, K.; Barton, H.A.; McArthur, A.G.; Wright, G.D. A diverse intrinsic antibiotic resistome from a cave bacterium. Nat. Commun. 2016, 7, 13803. [Google Scholar] [CrossRef]
- Van Goethem, M.W.; Pierneef, R.; Bezuidt, O.K.I.; Van De Peer, Y.; Cowan, D.A.; Makhalanyane, T.P. A reservoir of “historical” anti-biotic resistance genes in remote pristine Antarctic soils. Microbiome 2018, 6, 1–12. [Google Scholar] [CrossRef]
- Fillinger, L.; Hug, K.; Griebler, C. Selection imposed by local environmental conditions drives differences in microbial com-munity composition across geographically distinct groundwater aquifers. FEMS Microbiol Ecol. 2019, 95, 1–12. [Google Scholar] [CrossRef]
- Gómez, F.; Cavalazzi, B.; Rodríguez, N.; Amils, R.; Ori, G.G.; Olsson-Francis, K.; Escudero, C.; Martínez, J.M.; Miruts, H. Ultra-small microorganisms in the polyextreme conditions of the Dallol volcano, Northern Afar, Ethiopia. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Cavalazzi, B.; Barbieri, R.; Gómez, F.; Capaccioni, B.; Olsson-Francis, K.; Pondrelli, M.; Hagos, M. The Dallol Geothermal Area, Northern Afar (Ethiopia)—An Exceptional Planetary Field Analog on Earth. Astrobiology 2019, 19, 553–578. [Google Scholar] [CrossRef] [PubMed]
- Kotopoulou, E.; Huertas, A.D.; Garcia-Ruiz, J.M.; Dominguez-Vera, J.M.; Lopez-Garcia, J.M.; Guerra-Tschuschke, I.; Rull, F. A Polyextreme Hydrothermal System Controlled by Iron: The Case of Dallol at the Afar Triangle. ACS Earth Space Chem. 2019, 3, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Najar, I.N.; Sherpa, M.T.; Das, S.; Das, S.; Thakur, N. Diversity analysis and metagenomic insights into antibiotic and metal resistance among Himalayan hot spring bacteriobiome insinuating inherent environmental baseline levels of antibiotic and metal tolerance. J. Glob. Antimicrob. Resist. 2020, 21, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Hedlund, B.P.; Cole, J.K.; Williams, A.J.; Hou, W.; Zhou, E.; Li, W.; Dong, H. A review of the microbiology of the Rehai geothermal field in Tengchong, Yunnan Province, China. Geosci. Front. 2012, 3, 273–288. [Google Scholar] [CrossRef]
- Coonrod, J.D.; Leadley, P.J.; Eickhoff, T.C. Antibiotic Susceptibility of Bacillus Species. J. Infect. Dis. 1971, 123, 102–105. [Google Scholar] [CrossRef]
- Jardine, J.; Mavumengwana, V.; Ubomba-Jaswa, E. Antibiotic resistance and heavy metal tolerance in cultured bacteria from hot springs as indicators of environmental intrinsic resistance and tolerance levels. Environ. Pollut. 2019, 249, 696–702. [Google Scholar] [CrossRef]
- Martínez, J.L. Natural antibiotic resistance and contamination by antibiotic resistance determinants: The two ages in the evo-lution of resistance to antimicrobials. Front. Microbiol. 2012, 3, 2010–2012. [Google Scholar] [CrossRef]
- Miller, R.V.; Gammon, K.; Day, M.J. Antibiotic resistance among bacteria isolated from seawater and penguin fecal samples collected near Palmer Station, Antarctica. Can. J. Microbiol. 2009, 55, 37–45. [Google Scholar] [CrossRef]
- Wales, A.D.; Davies, R.H. Co-Selection of Resistance to Antibiotics, Biocides and Heavy Metals, and Its Relevance to Foodborne Pathogens. Antibiotics 2015, 4, 567–604. [Google Scholar] [CrossRef]
- Li, A.-D.; Li, L.-G.; Zhang, T. Exploring antibiotic resistance genes and metal resistance genes in plasmid metagenomes from wastewater treatment plants. Front. Microbiol. 2015, 6, 1025. [Google Scholar] [CrossRef]
- Martins, V.V.; Zanetti, M.O.B.; Pitondo-Silva, A.; Stehling, E.G. Aquatic environments polluted with antibiotics and heavy metals: A human health hazard. Environ. Sci. Pollut. Res. 2014, 21, 5873–5878. [Google Scholar] [CrossRef] [PubMed]
- Nies, D.H. Heavy metal-resistant bacteria as extremophiles: Molecular physiology and biotechnological use of Ralstonia sp. CH34. Extremophiles 2000, 4, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Giachino, A.; Waldron, K.J. Copper tolerance in bacteria requires the activation of multiple accessory pathways. Mol. Microbiol. 2020, 114, 377–390. [Google Scholar] [CrossRef]
- Poli, A.; Salerno, A.; Laezza, G.; di Donato, P.; Dumontet, S.; Nicolaus, B. Heavy metal resistance of some thermophiles: Potential use of α-amylase from Anoxybacillus amylolyticus as a microbial enzymatic bioassay. Res. Microbiol. 2009, 160, 99–106. [Google Scholar] [CrossRef]
- Knapp, C.W.; McCluskey, S.M.; Singh, B.K.; Campbell, C.D.; Hudson, G.; Graham, D.W. Antibiotic resistance gene abundances corre-late with metal and geochemical conditions in archived Scottish soils. PLoS ONE 2011, 9, 6. [Google Scholar]
- Li, B.; Yang, Y.; Ma, L.; Ju, F.; Guo, F.; Tiedje, J.M.; Zhang, T. Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J. 2015, 9, 2490–2502. [Google Scholar] [CrossRef]
- Pal, C.; Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genom. 2015, 16, 1–14. [Google Scholar] [CrossRef]
- Sandegren, L.; Andersson, D.I. Bacterial gene amplification: Implications for the evolution of antibiotic resistance. Nat. Rev. Microbiol. 2009, 7, 578–588. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar] [CrossRef]
- Ji, X.; Shen, Q.; Liu, F.; Ma, J.; Xu, G.; Wang, Y.; Wu, M. Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai; China. J. Hazard. Mater. 2012, 235–236, 178–185. [Google Scholar] [CrossRef]
- Knapp, C.W.; Callan, A.C.; Aitken, B.; Shearn, R.; Koenders, A.; Hinwood, A. Relationship between antibiotic resistance genes and metals in residential soil samples from Western Australia. Environ. Sci. Pollut. Res. 2017, 24, 2484–2494. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, J.; Zhang, H.; Shi, W.; Liu, Y. Bacterial Heavy-Metal and Antibiotic Resistance Genes in a Copper Tailing Dam Area in Northern China. Front. Microbiol. 2019, 10, 1916. [Google Scholar] [CrossRef] [PubMed]
- Wireman, J.; Liebert, C.A.; Smith, T.; Summers, A.O. Association of mercury resistance with antibiotic resistance in the gram-negative fecal bacteria of primates. Appl. Environ. Microbiol. 1997, 63, 4494–4503. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.-G.; Johnson, T.A.; Su, J.-Q.; Qiao, M.; Guo, G.-X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [PubMed]
- Davis, I.J.; Roberts, A.P.; Ready, D.; Richards, H.; Wilson, M.; Mullany, P. Linkage of a novel mercury resistance operon with strep-tomycin resistance on a conjugative plasmid in Enterococcus faecium. Plasmid 2005, 54, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Li, B.; Li, L.-G.; Zhang, T.; Angelidaki, I. Antibiotic Resistance Genes and Correlations with Microbial Community and Metal Resistance Genes in Full-Scale Biogas Reactors as Revealed by Metagenomic Analysis. Environ. Sci. Technol. 2017, 51, 4069–4080. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.; Tom-Petersen, A.; Nybroe, O. Copper amendment of agricultural soil selects for bacterial antibiotic resistance in the field. Lett. Appl. Microbiol. 2005, 40, 146–151. [Google Scholar] [CrossRef]
- Liu, C.; Li, G.; Qin, X.; Xu, Y.; Wang, J.; Wu, G.; Feng, H.; Ye, J.; Zhu, C.; Li, X.; et al. Profiles of antibiotic- and heavy metal-related resistance genes in animal manure revealed using a metagenomic analysis. Ecotoxicol. Environ. Saf. 2022, 239, 113655. [Google Scholar] [CrossRef] [PubMed]
- Wardwell, L.H.; Jude, B.A.; Moody, J.P.; Olcerst, A.I.; Gyure, R.A.; Nelson, R.E.; Fekete, F.A. Co-selection of mercury and antibiotic resistance in sphagnum core samples dating back 2000 years. Geomicrobiol. J. 2009, 26, 238–247. [Google Scholar] [CrossRef]
- Wright, M.S.; Peltier, G.L.; Stepanauskas, R.; McArthur, J.V. Bacterial tolerances to metals and antibiotics in metal-contaminated and reference streams. FEMS Microbiol. Ecol. 2006, 58, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Wright, M.S.; Stepanauskas, R.; McArthur, J. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006, 14, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Mata, M.; Baquero, F.; Pã©Rez-Dãaz, J. A multidrug efflux transporter in Listeria monocytogenes. FEMS Microbiol. Lett. 2000, 187, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Stepanauskas, R.; Glenn, T.C.; Jagoe, C.H.; Tuckfield, R.C.; Lindell, A.H.; King, C.J.; McArthur, J.V. Coselection for microbial resistance to metals and antibiotics in freshwater microcosms. Environ. Microbiol. 2006, 8, 1510–1514. [Google Scholar] [CrossRef] [PubMed]
- Binega, Y. Chemical analysis of the Assale (Ethiopia) rock salt deposit. Bull. Chem. Soc. Ethiop. 2006, 20, 319–324. [Google Scholar] [CrossRef]
- Belilla, J.; Moreira, D.; Jardillier, L.; Reboul, G.; Benzerara, K.; López-García, J.M.; Bertolino, P.; López-Archilla, A.I.; López-García, P. Hyperdiverse archaea near life limits at the polyextreme geothermal Dallol area. Nat. Ecol. Evol. 2019, 3, 1552–1561. [Google Scholar] [CrossRef] [PubMed]
- Gebregiorgies, H. Determination of Heavy Metal Concentrations in Water Samples Obtained from Lake Afdera, Upper and Lower Dobi Areas, Afar Regional State, Ethiopia. J. Appl. Chem. 2018, 11, 55–61. [Google Scholar]
- Bekele, A.; Schmerold, R. Characterization of brines and evaporite deposits for their lithium contents in the northern part of the Danakil Depression and in some selected areas of the Main Ethiopian Rift lakes. J. Afr. Earth Sci. 2020, 170, 103904. [Google Scholar] [CrossRef]
- van de Wiel, H.J. Determination of elements by ICP-AES and ICP-MS; National Institute of Public Health and the Environment (RIVM): Bilthoven, The Netherlands, 2003; pp. 1–19. [Google Scholar]
- Zhou, J.; Bruns, M.A.; Tiedje, J.M. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 1996, 62, 316–322. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Babraham Bioinformatics. FastQC A Quality Control tool for High Throughput Sequence Data. 2017. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 21 September 2023).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex meta-genomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Chowdhury, D.; Zhang, Z.; Cheung, W.K.; Lu, A.; Bian, Z.; Zhang, L. A review of computational tools for generating meta-genome-assembled genomes from metagenomic sequencing data. Comput. Struct. Biotechnol. J. 2021, 19, 6301–6314. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.D.; Li, F.; Kirton, E.; Thomas, A.; Egan, R.; An, H.; Wang, Z. MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 2019, 7, e7359. [Google Scholar] [CrossRef] [PubMed]
- Alneberg, J.; Bjarnason, B.S.; de Bruijn, I.; Schirmer, M.; Quick, J.; Ijaz, U.Z.; Lahti, L.; Loman, N.J.; Andersson, A.F.; Quince, C. Binning metagenomic contigs by coverage and composition. Nat. Methods 2014, 11, 1144–1146. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.W.; Simmons, B.A.; Singer, S.W. MaxBin 2.0: An automated binning algorithm to recover genomes from multiple meta-genomic datasets. Bioinformatics 2016, 32, 605–607. [Google Scholar] [CrossRef] [PubMed]
- Sieber, C.M.K.; Probst, A.J.; Sharrar, A.; Thomas, B.C.; Hess, M.; Tringe, S.G.; Banfield, J.F. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 2018, 3, 836–843. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes re-covered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Altschup, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Powell, S.; Forslund, K.; Szklarczyk, D.; Trachana, K.; Roth, A.; Huerta-Cepas, J.; Bork, P. EggNOG v4.0: Nested orthology inference across 3686 organisms. Nucleic Acids Res. 2014, 42, D231–D239. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Natale, D.A. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 1–14. Available online: http://www.biomedcentral.com/1471-2105/4/41 (accessed on 17 August 2023). [CrossRef] [PubMed]
- Huson, D.H.; Mitra, S.; Ruscheweyh, H.-J.; Weber, N.; Schuster, S.C. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011, 21, 1552–1560. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2014, 12, 59–60. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- Forsberg, K.J.; Patel, S.; Gibson, M.K.; Lauber, C.L.; Knight, R.; Fierer, N.; Dantas, G. Bacterial phylogeny structures soil resistomes across habitats. Nature 2014, 509, 612–616. [Google Scholar] [CrossRef]
- Liu, B.; Pop, M. ARDB—Antibiotic resistance genes database. Nucleic Acids Res. 2009, 37, 443–447. [Google Scholar] [CrossRef]
- Martínez, J.L.; Coque, T.M.; Baquero, F. What is a resistance gene? Ranking risk in resistomes. Nat. Rev. Microbiol. 2015, 13, 116–123. [Google Scholar] [CrossRef]
- Pal, C.; Bengtsson-Palme, J.; Rensing, C.; Kristiansson, E.; Larsson, D.G.J. BacMet: Antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 2014, 42, D737–D743. [Google Scholar] [CrossRef]
- Chao, A.; Chazdon, R.L.; Colwell, R.K.; Shen, T. Abundance-Based Similarity Indices and Their Estimation When There Are Unseen Species in Samples. Biometrics 2006, 62, 361–371. [Google Scholar] [CrossRef] [PubMed]
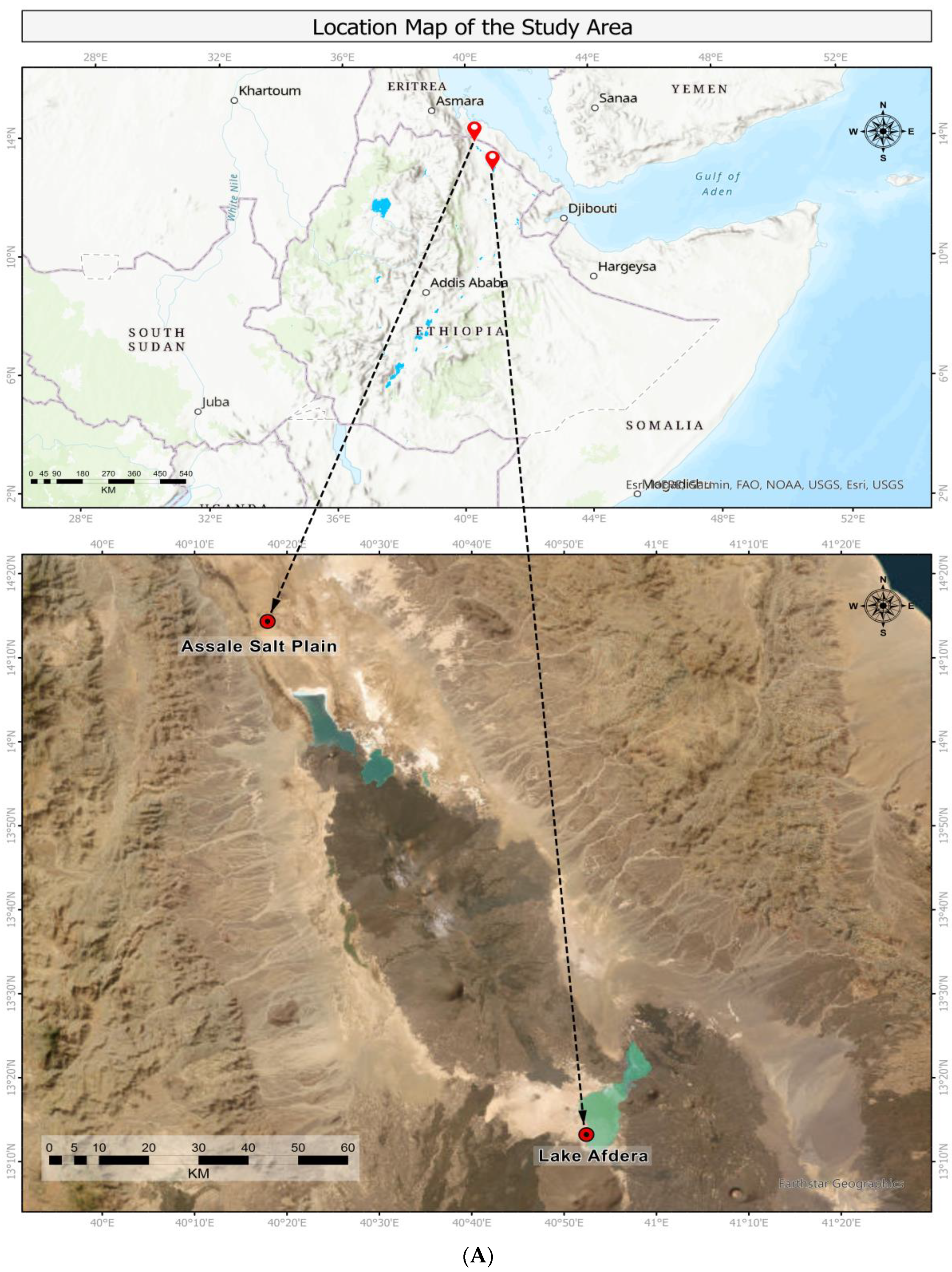



| Measured Parameters | Assale Salt Plain | Lake Afdera |
|---|---|---|
| Coordinates | 0640063 E/1574494 N | 0703016 E/1462263 N |
| pH | 4.61 | 6.29 |
| Temperature (°C) | 39 | 36 |
| Salinity (g/L) | 360 | 137 |
| Elevation (m) | −119 | −111 |
| Na (g/L) | 49 | 38.5 |
| Ca (g/L) | 21.5 | 13.9 |
| Mg (g/L) | 1.98 | 1.1 |
| Cl (g/L) | 1.7 | 1.6 |
| Mn (g/L) | 0.12 | 0.618 |
| Cu (g/L) | 0.012 | 0.008 |
| Pb (g/L) | 0.0001 | 0.0002 |
| Zn (g/L) | 0.007 | 0.0001 |
| Ni (g/L) | 0.002 | 0.001 |
| Fe (g/L) | 0.049 | 0.002 |
| Cd (g/L) | 0.0001 | 0.00001 |
| Parameters | Assale Salt Plain | Lake Afdera |
|---|---|---|
| Library insert size (bp) | 350 | 350 |
| Length of single read (bp) | 150 | 150 |
| Average raw reads (Mbp) | 6479 | 6762 |
| Total number of cleaned reads (Mbp) | 6459 | 6759 |
| GC content (%) | 62 | 56 |
| No. of contigs | 47,042 | 93,220 |
| Longest contig length (bp) | 16,520 | 176,724 |
| N50 (bp) | 652 | 1212 |
| Sampling Sites | Shannon | Simpson | p-Value |
|---|---|---|---|
| Lake Afdera | 4.20 | 0.89 | 0.01 |
| Assale salt plain | 0.39 | 0.10 | 0.04 |
| Metals | Metal Concentration | Correlation Coefficient | |||
|---|---|---|---|---|---|
| ARG Abundance | p-Value | MRG Abundance | p-Value | ||
| Cu | 11.49 | −0.08 | 0.87 | 0.54 | 0.26 |
| Hg | 0.17 | −0.66 | 0.16 | 0.88 | 0.02 |
| Cd | 0.13 | 0.98 | 0.001 | −0.12 | 0.82 |
| Zn | 7.16 | −0.60 | 0.21 | −0.03 | 0.96 |
| Metals | Metal Concentration | Correlation Coefficient | |||
|---|---|---|---|---|---|
| ARG Abundance | p-Value | MRG Abundance | p-Value | ||
| Cu | 0.88 | −0.54 | 0.26 | 0.31 | 0.54 |
| Hg | 1.57 | −0.31 | 0.54 | 0.83 | 0.04 |
| Cd | 0.02 | 0.08 | 0.87 | −0.37 | 0.47 |
| Zn | 0.07 | 0.93 | 0.001 | −0.32 | 0.53 |
| Samples | ARG Abundance Mean (±SD) | MRG Abundance Mean (±SD) | Correlation Coefficient | p-Value |
|---|---|---|---|---|
| Lake Afdera | 25.02 (11.64) | 4.91 (3.70) | 0.44 | 0.033 |
| Assale salt plain | 2949.94 (1661.09) | 8.76(7.42) | 0.56 | 0.004 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balcha, E.S.; Gómez, F.; Gemeda, M.T.; Bekele, F.B.; Abera, S.; Cavalazzi, B.; Woldesemayat, A.A. Shotgun Metagenomics-Guided Prediction Reveals the Metal Tolerance and Antibiotic Resistance of Microbes in Poly-Extreme Environments in the Danakil Depression, Afar Region. Antibiotics 2023, 12, 1697. https://doi.org/10.3390/antibiotics12121697
Balcha ES, Gómez F, Gemeda MT, Bekele FB, Abera S, Cavalazzi B, Woldesemayat AA. Shotgun Metagenomics-Guided Prediction Reveals the Metal Tolerance and Antibiotic Resistance of Microbes in Poly-Extreme Environments in the Danakil Depression, Afar Region. Antibiotics. 2023; 12(12):1697. https://doi.org/10.3390/antibiotics12121697
Chicago/Turabian StyleBalcha, Ermias Sissay, Felipe Gómez, Mesfin Tafesse Gemeda, Fanuel Belayneh Bekele, Sewunet Abera, Barbara Cavalazzi, and Adugna Abdi Woldesemayat. 2023. "Shotgun Metagenomics-Guided Prediction Reveals the Metal Tolerance and Antibiotic Resistance of Microbes in Poly-Extreme Environments in the Danakil Depression, Afar Region" Antibiotics 12, no. 12: 1697. https://doi.org/10.3390/antibiotics12121697
APA StyleBalcha, E. S., Gómez, F., Gemeda, M. T., Bekele, F. B., Abera, S., Cavalazzi, B., & Woldesemayat, A. A. (2023). Shotgun Metagenomics-Guided Prediction Reveals the Metal Tolerance and Antibiotic Resistance of Microbes in Poly-Extreme Environments in the Danakil Depression, Afar Region. Antibiotics, 12(12), 1697. https://doi.org/10.3390/antibiotics12121697









