Microbe Interactions within the Skin Microbiome
Abstract
1. Introduction
2. Bioactive Molecules Produced during Microbial Interactions in the Human Skin
3. Bacterial Interactions in the Skin That Affect the Regulation of Quorum Sensing Systems
4. Biotechnological Applications of Products Obtained from Skin Interactions
5. Future Directions for the Field
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Romanovsky, A.A. Skin temperature: Its role in thermoregulation. Acta Physiol. 2014, 210, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Yousef, H.; Alhajj, M.; Sharma, S. Anatomy, Skin (Integument), Epidermis. In StatPearls; [Updated 2022 Nov 14]; StatPearls Publishing: Treasure Island, FL, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK470464/ (accessed on 1 December 2023).
- Chen, Y.E.; Fischbach, M.A.; Belkaid, Y. Skin microbiota-host interactions. Nature 2018, 553, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Boxberger, M.; Cenizo, V.; Cassir, N.; La Scola, B. Challenges in exploring and manipulating the human skin microbiome. Microbiome 2021, 9, 125. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Staphylococci in the human microbiome: The role of host and interbacterial interactions. Curr. Opin. Microbiol. 2020, 53, 71–77. [Google Scholar] [CrossRef]
- Scholz, C.F.P.; Kilian, M. The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 4422–4432. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; NISC Comparative Sequencing Program; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; et al. Topographical and temporal diversity of the human skin microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef]
- Sfriso, R.; Egert, M.; Gempeler, M.; Voegeli, R.; Campiche, R. Revealing the secret life of skin-with the microbiome you never walk alone. Int. J. Cosmet. Sci. 2020, 42, 116–126. [Google Scholar] [CrossRef]
- Swaney, M.H.; Kalan, L.R. Living in Your Skin: Microbes, Molecules, and Mechanisms. Infect. Immun. 2021, 89, e00695-20. [Google Scholar] [CrossRef]
- Bäumler, A.J.; Sperandio, V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 2016, 535, 85–93. [Google Scholar] [CrossRef]
- Mirzaei, M.K.; Maurice, C.F. Ménage à trois in the human gut: Interactions between host, bacteria and phages. Nat. Rev. Microbiol. 2017, 15, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Y.; Du, Z.; Zhang, L.; Chen, J.; Shen, Z.; Liu, Q.; Qin, J.; Lv, H.; Wang, H.; et al. Skin microbiota analysis-inspired development of novel anti-infectives. Microbiome 2020, 8, 85. [Google Scholar] [CrossRef] [PubMed]
- Christensen, G.J.; Brüggemann, H. Bacterial skin commensals and their role as host guardians. Benef. Microbes 2014, 5, 201–215. [Google Scholar] [CrossRef] [PubMed]
- Cogen, A.L.; Yamasaki, K.; Muto, J.; Sanchez, K.M.; Crotty Alexander, L.; Tanios, J.; Lai, Y.; Kim, J.E.; Nizet, V.; Gallo, R.L. Staphylococcus epidermidis antimicrobial delta-toxin (phenol-soluble modulin-gamma) cooperates with host antimicrobial peptides to kill group A Streptococcus. PLoS ONE 2010, 5, e8557. [Google Scholar] [CrossRef]
- Wang, Y.; Kuo, S.; Shu, M.; Yu, J.; Huang, S.; Dai, A.; Two, A.; Gallo, R.L.; Huang, C.M. Staphylococcus epidermidis in the human skin microbiome mediates fermentation to inhibit the growth of Propionibacterium acnes: Implications of probiotics in acne vulgaris. Appl. Microbiol. Biotechnol. 2014, 98, 411–424. [Google Scholar] [CrossRef]
- Traisaeng, S.; Herr, D.R.; Kao, H.J.; Chuang, T.H.; Huang, C.M. A Derivative of Butyric Acid, the Fermentation Metabolite of Staphylococcus epidermidis, Inhibits the Growth of a Staphylococcus aureus Strain Isolated from Atopic Dermatitis Patients. Toxins 2019, 11, 311. [Google Scholar] [CrossRef]
- Iwase, T.; Uehara, Y.; Shinji, H.; Tajima, A.; Seo, H.; Takada, K.; Agata, T.; Mizunoe, Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 2010, 465, 346–349. [Google Scholar] [CrossRef]
- Sugimoto, S.; Iwamoto, T.; Takada, K.; Okuda, K.; Tajima, A.; Iwase, T.; Mizunoe, Y. Staphylococcus epidermidis Esp degrades specific proteins associated with Staphylococcus aureus biofilm formation and host-pathogen interaction. J. Bacteriol. 2013, 195, 1645–1655. [Google Scholar] [CrossRef]
- Glatthardt, T.; Campos, J.C.M.; Chamon, R.C.; de Sá Coimbra, T.F.; Rocha, G.A.; de Melo, M.A.F.; Parente, T.E.; Lobo, L.A.; Antunes, L.C.M.; Dos Santos, K.R.N.; et al. Small Molecules Produced by Commensal Staphylococcus epidermidis Disrupt Formation of Biofilms by Staphylococcus aureus. Appl. Environ. Microbiol. 2020, 86, e02539-19. [Google Scholar] [CrossRef]
- Lynch, D.; O’Connor, P.M.; Cotter, P.D.; Hill, C.; Field, D.; Begley, M. Identification and characterisation of capidermicin, a novel bacteriocin produced by Staphylococcus capitis. PLoS ONE 2019, 14, e0223541. [Google Scholar] [CrossRef]
- O’Sullivan, J.N.; O’Connor, P.M.; Rea, M.C.; O’Sullivan, O.; Walsh, C.J.; Healy, B.; Mathur, H.; Field, D.; Hill, C.; Ross, R.P. Nisin J, a Novel Natural Nisin Variant, Is Produced by Staphylococcus capitis Sourced from the Human Skin Microbiota. J. Bacteriol. 2020, 202, e00639-19. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, A.M.; Nakatsuji, T.; Hayachi, A.; Williams, M.R.; Mills, R.H.; Gonzalez, D.J.; Gallo, R.L. Identification of a Human Skin Commensal Bacterium that Selectively Kills Cutibacterium acnes. J. Investig. Dermatol. 2020, 140, 1619–1628.e2. [Google Scholar] [CrossRef] [PubMed]
- Zipperer, A.; Konnerth, M.C.; Laux, C.; Berscheid, A.; Janek, D.; Weidenmaier, C.; Burian, M.; Schilling, N.A.; Slavetinsky, C.; Marschal, M.; et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 2016, 535, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Claesen, J.; Spagnolo, J.B.; Ramos, S.F.; Kurita, K.L.; Byrd, A.L.; Aksenov, A.A.; Melnik, A.V.; Wong, W.R.; Wang, S.; Hernandez, R.D.; et al. A Cutibacterium acnes antibiotic modulates human skin microbiota composition in hair follicles. Sci. Transl. Med. 2020, 12, eaay5445. [Google Scholar] [CrossRef] [PubMed]
- Shu, M.; Wang, Y.; Yu, J.; Kuo, S.; Coda, A.; Jiang, Y.; Gallo, R.L.; Huang, C.M. Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PLoS ONE 2013, 8, e55380. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; O’Neill, A.M.; Williams, M.R.; Cau, L.; Nakatsuji, T.; Horswill, A.R.; Gallo, R.L. Short chain fatty acids produced by Cutibacterium acnes inhibit biofilm formation by Staphylococcus epidermidis. Sci. Rep. 2020, 10, 21237. [Google Scholar] [CrossRef]
- Lima, R.D.; Dos Reis, G.A.; da Silva Reviello, J.; Glatthardt, T.; da Silva Coimbra, L.; Lima, C.O.G.X.; Antunes, L.C.M.; Ferreira, R.B.R. Antibiofilm activity of Cutibacterium acnes cell-free conditioned media against Staphylococcus spp. Braz. J. Microbiol. 2021, 52, 2373–2383. [Google Scholar] [CrossRef]
- Lebeer, S.; Oerlemans, E.F.M.; Claes, I.; Henkens, T.; Delanghe, L.; Wuyts, S.; Spacova, I.; van den Broek, M.F.L.; Tuyaerts, I.; Wittouck, S.; et al. Selective targeting of skin pathobionts and inflammation with topically applied lactobacilli. Cell Rep. Med. 2022, 3, 100521. [Google Scholar] [CrossRef]
- Myles, I.A.; Williams, K.W.; Reckhow, J.D.; Jammeh, M.L.; Pincus, N.B.; Sastalla, I.; Saleem, D.; Stone, K.D.; Datta, S.K. Transplantation of human skin microbiota in models of atopic dermatitis. JCI Insight 2016, 1, e86955. [Google Scholar] [CrossRef]
- Myles, I.A.; Earland, N.J.; Anderson, E.D.; Moore, I.N.; Kieh, M.D.; Williams, K.W.; Saleem, A.; Fontecilla, N.M.; Welch, P.A.; Darnell, D.A.; et al. First-in-human topical microbiome transplantation with Roseomonas mucosa for atopic dermatitis. JCI Insight 2018, 3, e120608. [Google Scholar] [CrossRef]
- Myles, I.A.; Castillo, C.R.; Barbian, K.D.; Kanakabandi, K.; Virtaneva, K.; Fitzmeyer, E.; Paneru, M.; Otaizo-Carrasquero, F.; Myers, T.G.; Markowitz, T.E.; et al. Therapeutic responses to Roseomonas mucosa in atopic dermatitis may involve lipid-mediated TNF-related epithelial repair. Sci. Transl. Med. 2020, 12, eaaz8631. [Google Scholar] [CrossRef] [PubMed]
- Carothers, K.E.; Liang, Z.; Mayfield, J.; Donahue, D.L.; Lee, M.; Boggess, B.; Ploplis, V.A.; Castellino, F.J.; Lee, S.W. The Streptococcal Protease SpeB Antagonizes the Biofilms of the Human Pathogen Staphylococcus aureus USA300 through Cleavage of the Staphylococcal SdrC Protein. J. Bacteriol. 2020, 202, e00008-20. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yuan, C.; Xing, L.; Humbert, P. Topographical diversity of common skin microflora and its association with skin environment type: An observational study in Chinese women. Sci. Rep. 2017, 7, 18046. [Google Scholar] [CrossRef] [PubMed]
- Kumariya, R.; Garsa, A.K.; Rajput, Y.S.; Sood, S.K.; Akhtar, N.; Patel, S. Bacteriocins: Classification, synthesis, mechanism of action and resistance development in food spoilage causing bacteria. Microb. Pathog. 2019, 128, 171–177. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, J.N.; Rea, M.C.; O’Connor, P.M.; Hill, C.; Ross, R.P. Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol. Ecol. 2019, 95, fiy241. [Google Scholar] [CrossRef]
- Fernández-Fernández, R.; Lozano, C.; Eguizábal, P.; Ruiz-Ripa, L.; Martínez-Álvarez, S.; Abdullahi, I.N.; Zarazaga, M.; Torres, C. Bacteriocin-Like Inhibitory Substances in Staphylococci of Different Origins and Species with Activity against Relevant Pathogens. Front. Microbiol. 2022, 13, 870510. [Google Scholar] [CrossRef]
- Janek, D.; Zipperer, A.; Kulik, A.; Krismer, B.; Peschel, A. High Frequency and Diversity of Antimicrobial Activities Produced by Nasal Staphylococcus Strains against Bacterial Competitors. PLoS Pathog. 2016, 12, e1005812. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Chen, T.H.; Narala, S.; Chun, K.A.; Two, A.M.; Yun, T.; Shafiq, F.; Kotol, P.F.; Bouslimani, A.; Melnik, A.V.; et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 2017, 9, eaah4680. [Google Scholar] [CrossRef]
- Bitschar, K.; Sauer, B.; Focken, J.; Dehmer, H.; Moos, S.; Konnerth, M.; Schilling, N.A.; Grond, S.; Kalbacher, H.; Kurschus, F.C.; et al. Lugdunin amplifies innate immune responses in the skin in synergy with host- and microbiota-derived factors. Nat. Commun. 2019, 10, 2730. [Google Scholar] [CrossRef]
- Christensen, G.J.; Scholz, C.F.; Enghild, J.; Rohde, H.; Kilian, M.; Thürmer, A.; Brzuszkiewicz, E.; Lomholt, H.B.; Brüggemann, H. Antagonism between Staphylococcus epidermidis and Propionibacterium acnes and its genomic basis. BMC Genom. 2016, 17, 152. [Google Scholar] [CrossRef]
- Agak, G.W.; Kao, S.; Ouyang, K.; Qin, M.; Moon, D.; Butt, A.; Kim, J. Phenotype and Antimicrobial Activity of Th17 Cells Induced by Propionibacterium acnes Strains Associated with Healthy and Acne Skin. J. Investig. Dermatol. 2018, 138, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Tsuru, A.; Hamazaki, Y.; Tomida, S.; Ali, M.S.; Komura, T.; Nishikawa, Y.; Kage-Nakadai, E. Nonpathogenic Cutibacterium acnes Confers Host Resistance against Staphylococcus aureus. Microbiol. Spectr. 2021, 9, e0056221. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Phenol-soluble modulins. Int. J. Med. Microbiol. 2014, 304, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Cogen, A.L.; Yamasaki, K.; Sanchez, K.M.; Dorschner, R.A.; Lai, Y.; MacLeod, D.T.; Torpey, J.W.; Otto, M.; Nizet, V.; Kim, J.E.; et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J. Investig. Dermatol. 2010, 130, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dai, A.; Huang, S.; Kuo, S.; Shu, M.; Tapia, C.P.; Yu, J.; Two, A.; Zhang, H.; Gallo, R.L.; et al. Propionic acid and its esterified derivative suppress the growth of methicillin-resistant Staphylococcus aureus USA300. Benef Microbes 2014, 5, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Fluhr, J.W.; Darlenski, R.; Surber, C. Glycerol and the skin: Holistic approach to its origin and functions. Br. J. Dermatol. 2008, 159, 23–34. [Google Scholar] [CrossRef]
- Yang, J.J.; Chang, T.W.; Jiang, Y.; Kao, H.J.; Chiou, B.H.; Kao, M.S.; Huang, C.M. Commensal Staphylococcus aureus Provokes Immunity to Protect against Skin Infection of Methicillin-Resistant Staphylococcus aureus. Int. J. Mol. Sci. 2018, 19, 1290. [Google Scholar] [CrossRef]
- Nishijima, S.; Kurokawa, I.; Katoh, N.; Watanabe, K. The bacteriology of acne vulgaris and antimicrobial susceptibility of Propionibacterium acnes and Staphylococcus epidermidis isolated from acne lesions. J. Dermatol. 2000, 27, 318–323. [Google Scholar] [CrossRef]
- Kao, M.S.; Wang, Y.; Marito, S.; Huang, S.; Lin, W.Z.; Gangoiti, J.A.; Barshop, B.A.; Hyun, C.; Lee, W.R.; Sanford, J.A.; et al. The mPEG-PCL Copolymer for Selective Fermentation of Staphylococcus lugdunensis against Candida parapsilosis in the Human Microbiome. J. Microb. Biochem. Technol. 2016, 8, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Kwaszewska, A.; Sobiś-Glinkowska, M.; Szewczyk, E.M. Cohabitation-relationships of corynebacteria and staphylococci on human skin. Folia Microbiol. 2014, 59, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.H.; Oh, J.; Deming, C.; Conlan, S.; Grice, E.A.; Beatson, M.A.; Nomicos, E.; Polley, E.C.; Komarow, H.D.; NISC Comparative Sequence Program; et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012, 22, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Hamady, M.; Lauber, C.L.; Knight, R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 17994–17999. [Google Scholar] [CrossRef] [PubMed]
- Zeeuwen, P.L.; Boekhorst, J.; van den Bogaard, E.H.; de Koning, H.D.; van de Kerkhof, P.M.; Saulnier, D.M.; van Swam, I.I.; van Hijum, S.A.; Kleerebezem, M.; Schalkwijk, J.; et al. Microbiome dynamics of human epidermis following skin barrier disruption. Genome Biol. 2012, 13, R101. [Google Scholar] [CrossRef]
- Chng, K.R.; Tay, A.S.; Li, C.; Ng, A.H.; Wang, J.; Suri, B.K.; Matta, S.A.; McGovern, N.; Janela, B.; Wong, X.F.; et al. Whole metagenome profiling reveals skin microbiome-dependent susceptibility to atopic dermatitis flare. Nat. Microbiol. 2016, 1, 16106. [Google Scholar] [CrossRef] [PubMed]
- Geoghegan, J.A.; Irvine, A.D.; Foster, T.J. Staphylococcus aureus and Atopic Dermatitis: A Complex and Evolving Relationship. Trends Microbiol. 2018, 26, 484–497. [Google Scholar] [CrossRef]
- Bowen, A.C.; Tong, S.Y.; Chatfield, M.D.; Carapetis, J.R. The microbiology of impetigo in indigenous children: Associations between Streptococcus pyogenes, Staphylococcus aureus, scabies, and nasal carriage. BMC Infect. Dis. 2014, 14, 727. [Google Scholar] [CrossRef]
- Abbott, C.; Grout, E.; Morris, T.; Brown, H.L. Cutibacterium acnes biofilm forming clinical isolates modify the formation and structure of Staphylococcus aureus biofilms, increasing their susceptibility to antibiotics. Anaerobe 2022, 76, 102580. [Google Scholar] [CrossRef]
- Borda, L.J.; Perper, M.; Keri, J.E. Treatment of seborrheic dermatitis: A comprehensive review. J. Dermatol. Treat. 2019, 30, 158–169. [Google Scholar] [CrossRef]
- An, Q.; Sun, M.; Qi, R.Q.; Zhang, L.; Zhai, J.L.; Hong, Y.X.; Song, B.; Chen, H.D.; Gao, X.H. High Staphylococcus epidermidis Colonization and Impaired Permeability Barrier in Facial Seborrheic Dermatitis. Chin. Med. J. 2017, 130, 1662–1669. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhang, Y.J.; Wang, H.X.; Sun, Y.Z.; Yang, Y.; Li, Z.X.; Qi, R.Q.; Gao, X.H. Malassezia furfur promoting growth of Staphylococcus epidermidis by increasing pH when cultured in a lipid-free environment. Chin. Med. J. 2019, 132, 873–876. [Google Scholar] [CrossRef] [PubMed]
- Abisado, R.G.; Benomar, S.; Klaus, J.R.; Dandekar, A.A.; Chandler, J.R. Bacterial Quorum Sensing and Microbial Community Interactions. mBio 2018, 9, e02331-17. [Google Scholar] [CrossRef] [PubMed]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Chen, J.; Yang, C.; Yin, Y.; Yao, K. Quorum Sensing: A Prospective Therapeutic Target for Bacterial Diseases. Biomed. Res. Int. 2019, 2019, 2015978. [Google Scholar] [CrossRef] [PubMed]
- Otto, M.; Echner, H.; Voelter, W.; Götz, F. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 2001, 69, 1957–1960. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.R.; Costa, S.K.; Zaramela, L.S.; Khalil, S.; Todd, D.A.; Winter, H.L.; Sanford, J.A.; O’Neill, A.M.; Liggins, M.C.; Nakatsuji, T.; et al. Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci. Transl. Med. 2019, 11, eaat8329. [Google Scholar] [CrossRef]
- Paharik, A.E.; Parlet, C.P.; Chung, N.; Todd, D.A.; Rodriguez, E.I.; Van Dyke, M.J.; Cech, N.B.; Horswill, A.R. Coagulase-Negative Staphylococcal Strain Prevents Staphylococcus aureus Colonization and Skin Infection by Blocking Quorum Sensing. Cell Host Microbe 2017, 22, 746–756.e5. [Google Scholar] [CrossRef]
- Brown, M.M.; Kwiecinski, J.M.; Cruz, L.M.; Shahbandi, A.; Todd, D.A.; Cech, N.B.; Horswill, A.R. Novel Peptide from Commensal Staphylococcus simulans Blocks Methicillin-Resistant Staphylococcus aureus Quorum Sensing and Protects Host Skin from Damage. Antimicrob. Agents Chemother. 2020, 64, e00172-20. [Google Scholar] [CrossRef]
- Severn, M.M.; Williams, M.R.; Shahbandi, A.; Bunch, Z.L.; Lyon, L.M.; Nguyen, A.; Zaramela, L.S.; Todd, D.A.; Zengler, K.; Cech, N.B.; et al. The Ubiquitous Human Skin Commensal Staphylococcus hominis Protects against Opportunistic Pathogens. mBio 2022, 13, e0093022. [Google Scholar] [CrossRef]
- Ramsey, M.M.; Freire, M.O.; Gabrilska, R.A.; Rumbaugh, K.P.; Lemon, K.P. Staphylococcus aureus Shifts toward Commensalism in Response to Corynebacterium Species. Front. Microbiol. 2016, 7, 1230. [Google Scholar] [CrossRef] [PubMed]
- Otto, M.; Süssmuth, R.; Vuong, C.; Jung, G.; Götz, F. Inhibition of virulence factor expression in Staphylococcus aureus by the Staphylococcus epidermidis agr pheromone and derivatives. FEBS Lett. 1999, 450, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Le, K.Y.; Otto, M. Quorum-sensing regulation in staphylococci-an overview. Front. Microbiol. 2015, 6, 1174. [Google Scholar] [CrossRef] [PubMed]
- Jenul, C.; Horswill, A.R. Regulation of Staphylococcus aureus Virulence. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Ji, G.; Beavis, R.; Novick, R.P. Bacterial interference caused by autoinducing peptide variants. Science 1997, 276, 2027–2030. [Google Scholar] [CrossRef]
- Jarraud, S.; Lyon, G.J.; Figueiredo, A.M.; Gerard, L.; Vandenesch, F.; Etienne, J.; Muir, T.W.; Novick, R.P. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J. Bacteriol. 2000, 182, 6517–6522. [Google Scholar] [CrossRef]
- Garbacz, K.; Piechowicz, L.; Barańska-Rybak, W.; Dąbrowska-Szponar, M. Staphylococcus aureus isolated from patients with recurrent furunculosis carrying Panton-Valentine leukocidin genes represent agr specificity group IV. Eur. J. Dermatol. 2011, 21, 43–46. [Google Scholar] [CrossRef]
- Cheung, G.Y.; Wang, R.; Khan, B.A.; Sturdevant, D.E.; Otto, M. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect. Immun. 2011, 79, 1927–1935. [Google Scholar] [CrossRef]
- Kobayashi, S.D.; Malachowa, N.; Whitney, A.R.; Braughton, K.R.; Gardner, D.J.; Long, D.; Bubeck Wardenburg, J.; Schneewind, O.; Otto, M.; Deleo, F.R. Comparative analysis of USA300 virulence determinants in a rabbit model of skin and soft tissue infection. J. Infect. Dis. 2011, 204, 937–941. [Google Scholar] [CrossRef]
- Grundstad, M.L.; Parlet, C.P.; Kwiecinski, J.M.; Kavanaugh, J.S.; Crosby, H.A.; Cho, Y.S.; Heilmann, K.; Diekema, D.J.; Horswill, A.R. Quorum Sensing, Virulence, and Antibiotic Resistance of USA100 Methicillin-Resistant Staphylococcus aureus Isolates. mSphere 2019, 4, e00553-19. [Google Scholar] [CrossRef]
- Lomholt, H.; Andersen, K.E.; Kilian, M. Staphylococcus aureus clonal dynamics and virulence factors in children with atopic dermatitis. J. Investig. Dermatol. 2005, 125, 977–982. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.J.; Jeon, H.S.; Sung, H.; Kim, M.N.; Hong, S.J. Epidemiological characteristics of methicillin-resistant Staphylococcus aureus isolates from children with eczematous atopic dermatitis lesions. J. Clin. Microbiol. 2008, 46, 991–995. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Takahashi, H.; Takaya, A.; Inoue, Y.; Katayama, Y.; Kusuya, Y.; Shoji, T.; Takada, S.; Nakagawa, S.; Oguma, R.; et al. Staphylococcus Agr virulence is critical for epidermal colonization and associates with atopic dermatitis development. Sci. Transl. Med. 2020, 12, eaay4068. [Google Scholar] [CrossRef] [PubMed]
- Olson, M.E.; Todd, D.A.; Schaeffer, C.R.; Paharik, A.E.; Van Dyke, M.J.; Büttner, H.; Dunman, P.M.; Rohde, H.; Cech, N.B.; Fey, P.D.; et al. Staphylococcus epidermidis agr quorum-sensing system: Signal identification, cross talk, and importance in colonization. J. Bacteriol. 2014, 196, 3482–3493. [Google Scholar] [CrossRef] [PubMed]
- Milshteyn, A.; Colosimo, D.A.; Brady, S.F. 2018. Accessing Bioactive Natural Products from the Human Microbiome. Cell Host Microbe 1230, 23, 725–736. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ravichandran, V.; Yin, Y.; Yin, J.; Zhang, Y. Natural Products from Mammalian Gut Microbiota. Trends Biotechnol. 2019, 37, 492–504. [Google Scholar] [CrossRef] [PubMed]
- Gharsallaoui, A.; Oulahal, N.; Joly, C.; Degraeve, P. Nisin as a Food Preservative: Part 1: Physicochemical Properties, Antimicrobial Activity, and Main Uses. Crit. Rev. Food Sci. Nutr. 2016, 56, 1262–1274. [Google Scholar] [CrossRef]
- Brand, A.M.; Smith, C.; Dicks, L.M. The Effects of Continuous In Vivo Administration of Nisin on Staphylococcus aureus Infection and Immune Response in Mice. Probiotics Antimicrob. Proteins 2013, 5, 279–286. [Google Scholar] [CrossRef]
- Mouritzen, M.V.; Andrea, A.; Qvist, K.; Poulsen, S.S.; Jenssen, H. Immunomodulatory potential of Nisin A with application in wound healing. Wound Repair. Regen. 2019, 27, 650–660. [Google Scholar] [CrossRef]
- Wang, Y.; Kao, M.S.; Yu, J.; Huang, S.; Marito, S.; Gallo, R.L.; Huang, C.M. A Precision Microbiome Approach Using Sucrose for Selective Augmentation of Staphylococcus epidermidis Fermentation against Propionibacterium acnes. Int. J. Mol. Sci. 2016, 17, 1870. [Google Scholar] [CrossRef]
- Dickey, S.W.; Cheung, G.Y.C.; Otto, M. Different drugs for bad bugs: Antivirulence strategies in the age of antibiotic resistance. Nat. Rev. Drug Discov. 2017, 16, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Pistone, D.; Meroni, G.; Panelli, S.; D’Auria, E.; Acunzo, M.; Pasala, A.R.; Zuccotti, G.V.; Bandi, C.; Drago, L. A Journey on the Skin Microbiome: Pitfalls and Opportunities. Int. J. Mol. Sci. 2021, 22, 9846. [Google Scholar] [CrossRef] [PubMed]
- Hannigan, G.D.; Meisel, J.S.; Tyldsley, A.S.; Zheng, Q.; Hodkinson, B.P.; SanMiguel, A.J.; Minot, S.; Bushman, F.D.; Grice, E.A. The human skin double-stranded DNA virome: Topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome. mBio 2015, 6, e01578-15. [Google Scholar] [CrossRef] [PubMed]
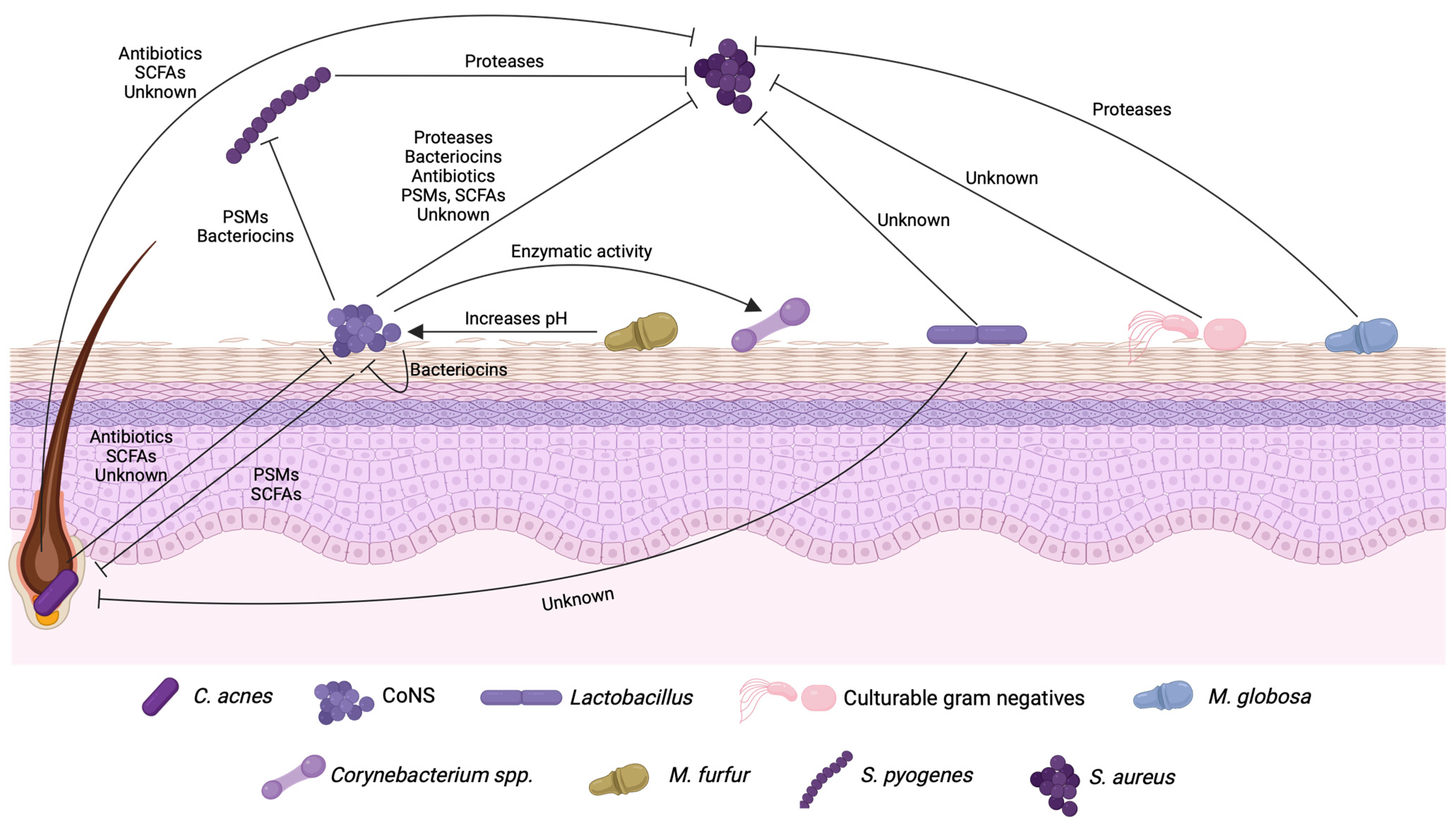
| Microorganism | Type of Molecule | Name of Molecule | Effect | Bacteria Targeted | References |
|---|---|---|---|---|---|
| S. epidermidis | Bacteriocin | Epidermin, Pep5, Epilancin K7 | Growth | MRSA, CoNS | [14] |
| PSM | PSM-γ, PSM-δ | Growth | S. pyogenes, S. aureus | [15] | |
| SCFAs | Acetic acid, butyric acid, lactic acid, succinic acid | Growth | C. acnes, S. aureus | [16,17] | |
| Protease | Esp | Biofilm | S. aureus | [18,19] | |
| Unknown | Unknown | Biofilm | S. aureus | [20] | |
| S. capitis | Bacteriocin | Capidermicin | Growth | L. latis, S. aureus, S. intermedius, S. pseudintermedius, M. luteus | [21] |
| Bacteriocin | Nisin J | Growth | Staphylococcus spp., Streptococcus spp., C. acnes | [22] | |
| PSM | PSMβ | Growth | C. acnes | [23] | |
| S. hominis | Bacteriocin | MP1 | Growth | MRSA strains, penicillin-resistant streptococci, VRE, methicillin-resistant CoNS | [13] |
| S. lugdunensis | Antibiotic | Lugdunin | Growth | S. aureus | [24] |
| C. acnes | Antibiotic | Cutimycin | Growth | MRSA | [25] |
| SCFAs | Acetic acid, lactic acid, propionic acid | Growth | S. aureus | [26] | |
| Acetic acid, propionic acid, isobutyric acid, isovaleric acid | Biofilm | S. epidermidis | [27] | ||
| Unknown | Unknown | Biofilm | S. lugdunensis, S. hominis, S. aureus | [28] | |
| Lactobacillus spp. | Unknown | Unknown | Growth | S. aureus, C. acnes | [29] |
| CGN | Unknown | Unknown | Growth | S. aureus | [30,31,32] |
| S. pyogenes | Protease | SpeB | Biofilm | S. aureus | [33] |
| Malassezia globosa | Protease | MgSAP1 | Biofilm and other virulence factors | S. aureus | [34] |
| Bacteria | Molecule | Effect | References |
|---|---|---|---|
| S. epidermidis | AIP (unknown type) | Inhibition of S. aureus Agr types I, II, and III | [67] |
| Agr type I AIP | Inhibition of S. aureus Agr type I | [68] | |
| Unknown | Downregulation of S. aureus Agr | [20] | |
| S. caprae | AIP | Inhibition of all S. aureus Agr types | [69] |
| S. simulans | AIP | Inhibition of all S. aureus Agr types | [70] |
| S. hominis | AIP-II | Inhibition of S. aureus growth | [71] |
| C. striatum | Unknown | Inhibition of S. aureus Agr types I, II, and III | [72] |
| Products | Application | Benefits | Limitations | References |
|---|---|---|---|---|
| Nisin J from S. capitis | Antimicrobial | Inhibitory activity against a wide range of bacterial targets | Not tested in vivo | [36] |
| MP1 from S. hominis | Antimicrobial | Treatment of S. aureus local and systemic infections | No safety tests using probiotic strain | [13] |
| Topical formulation with live S. epidermidis and S. hominis | Atopic dermatitis treatment | Highly potent, selectively killed S. aureus, and synergized with the human AMP LL-37 | A complete catalog of protective bacteria from skin could not be identified | [39] |
| Topical formulation with live R. mucosa | Atopic dermatitis treatment | Enhancement of skin barrier function, innate immune activation, and a reduction in topical steroid requirements without severe adverse events | Small study with children based on historical placebo control data; no data on skin biopsies | [31,32] |
| Topical formulation with live Lactobacillus | Acne treatment | Reduction in inflammatory lesions and microbiome modulation | No information on immunomodulatory mechanisms; viability and activity of lactobacilli | [29] |
| Derivate molecule from butyric acid | Antimicrobial/ Atopic dermatitis treatment | Ameliorate the production of pro-inflammatory interleukin (IL-6) induced by S. aureus, and reduced the colonization of S. aureus in mouse skin | Lack of information about mechanisms of action and possible impacts on microbiome | [17] |
| Derivate molecule from propionic acid | Antimicrobial | Methicillin-resistant S. aureus growth inhibition | Not tested in vivo | [47] |
| mPEG-PCL polymer | Microbiome modulator | Suppression of C. parapsilosis growth and prevention of fungal expansion in human dandruff | Not tested in vivo | [52] |
| Agr interference | Atopic dermatitis treatment | Prevention of skin barrier damage and inflammation | Treatment may promote persistent colonization of S. aureus in the skin; no data on stability of the synthetic autoinducer peptide in the skin | [68] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glatthardt, T.; Lima, R.D.; de Mattos, R.M.; Ferreira, R.B.R. Microbe Interactions within the Skin Microbiome. Antibiotics 2024, 13, 49. https://doi.org/10.3390/antibiotics13010049
Glatthardt T, Lima RD, de Mattos RM, Ferreira RBR. Microbe Interactions within the Skin Microbiome. Antibiotics. 2024; 13(1):49. https://doi.org/10.3390/antibiotics13010049
Chicago/Turabian StyleGlatthardt, Thaís, Rayssa Durães Lima, Raquel Monteiro de Mattos, and Rosana Barreto Rocha Ferreira. 2024. "Microbe Interactions within the Skin Microbiome" Antibiotics 13, no. 1: 49. https://doi.org/10.3390/antibiotics13010049
APA StyleGlatthardt, T., Lima, R. D., de Mattos, R. M., & Ferreira, R. B. R. (2024). Microbe Interactions within the Skin Microbiome. Antibiotics, 13(1), 49. https://doi.org/10.3390/antibiotics13010049







