Assessing the Effect of Oxytetracycline on the Selection of Resistant Escherichia coli in Treated and Untreated Broiler Chickens
Abstract
1. Introduction
2. Results
2.1. Isolation and Confirmation of E. coli
2.2. Determination of Antimicrobial Susceptibility
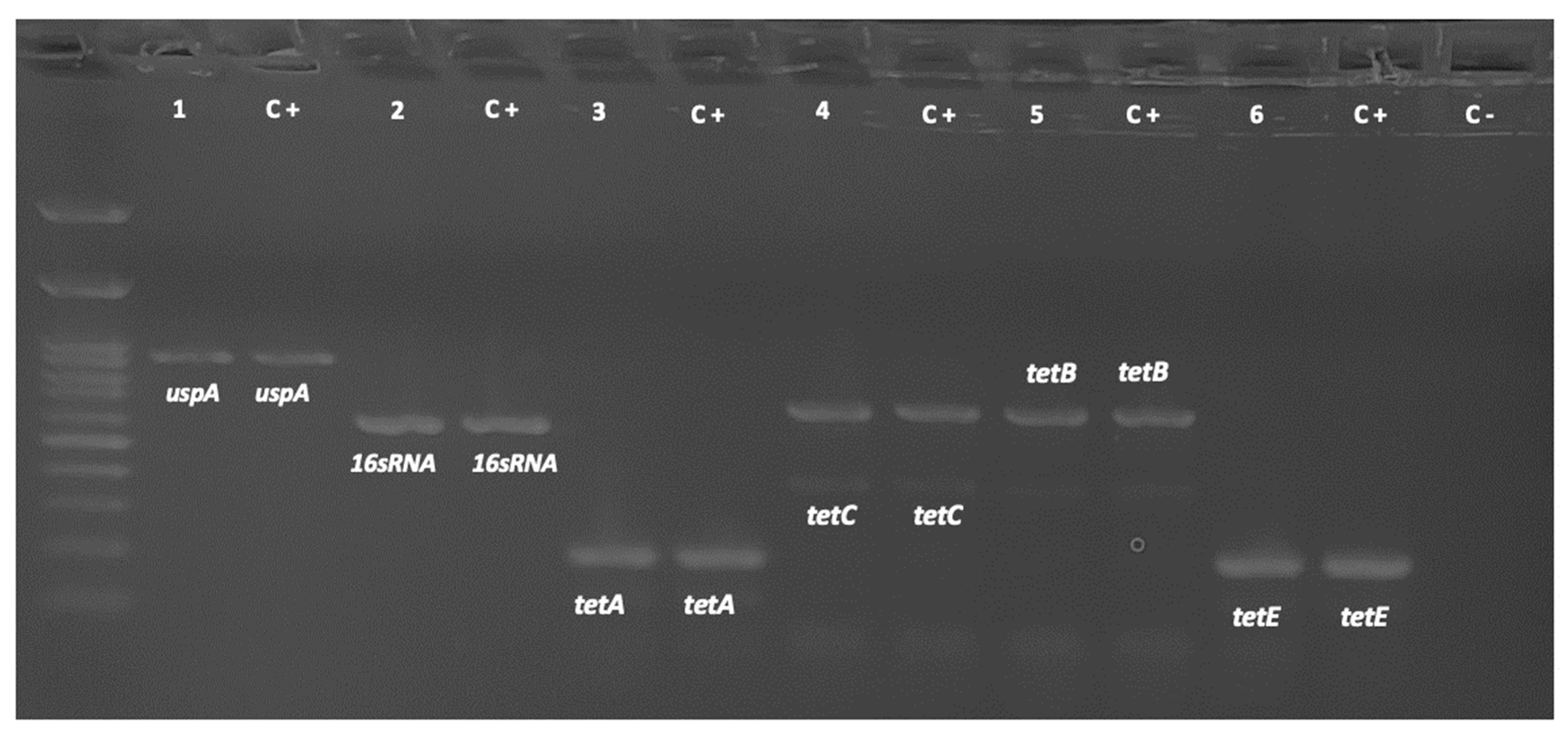
2.3. Detection of Resistance Genes
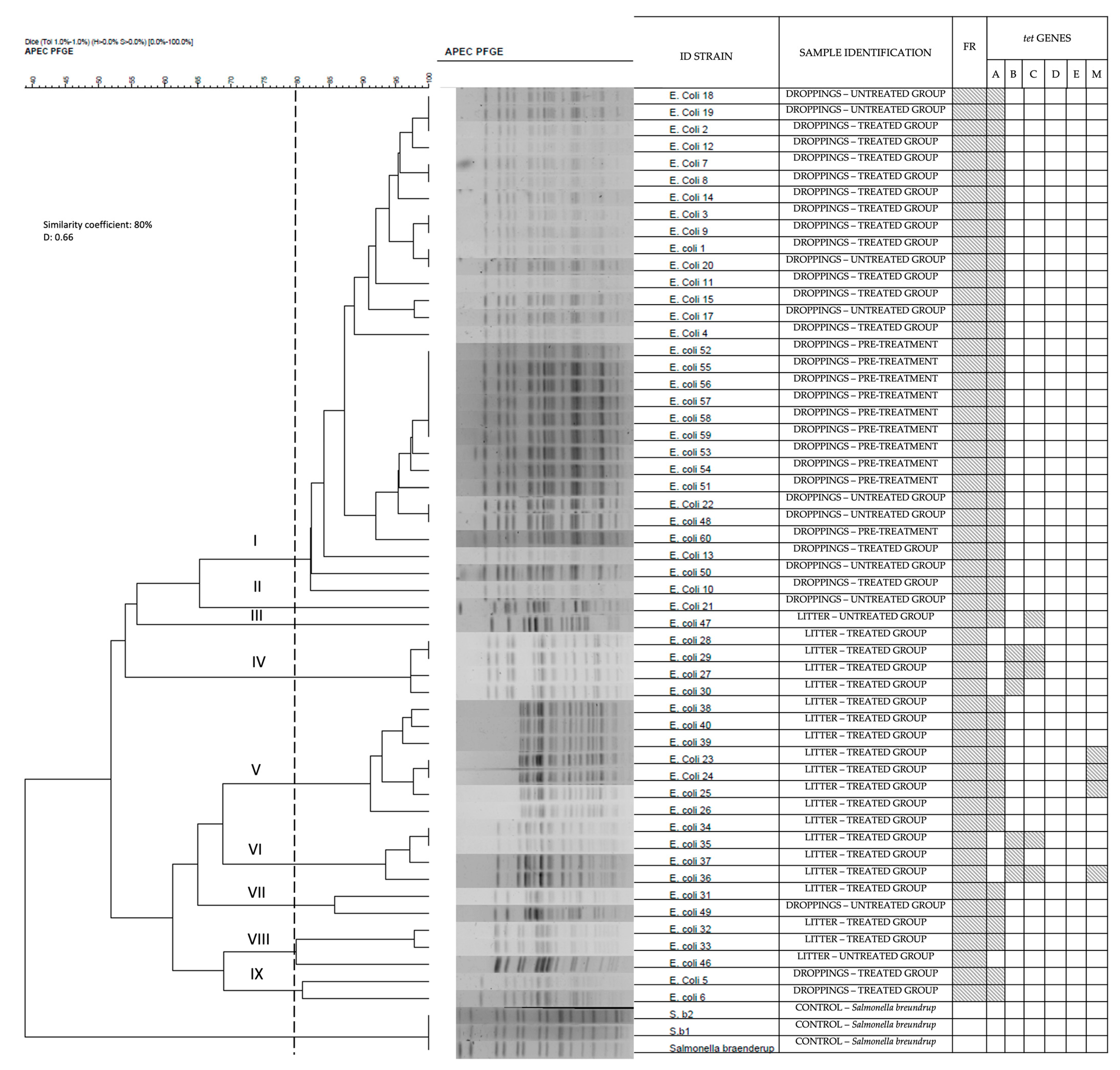
2.4. Isolate Subtyping
3. Discussion
4. Materials and Methods
4.1. Experimental Animals
4.2. Sampling Collection
4.3. E. coli Identification and Isolation
4.4. Antimicrobial Susceptibility Testing
4.5. PCR Detection of Tet Genes
4.6. Pulsed-Field Gel Electrophoresis
4.7. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sumpradit, N.; Wongkongkathep, S.; Malathum, K.; Janejai, N.; Paveenkittiporn, W.; Yingyong, T.; Chuxnum, T.; Vijitleela, A.; Boonyarit, P.; Akaleephan, C.; et al. Thailand’s national strategic plan on antimicrobial resistance: Progress and challenges. Bull. World Health Organ. 2021, 99, 661. [Google Scholar] [CrossRef]
- Chopra, I.; Roberts, M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C. Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 2005, 245, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Thaker, M.; Spanogiannopoulos, P.; Wright, G.D. The tetracycline resistome. Cell. Mol. Life Sci. 2010, 67, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Bryskier, A. Antimicrobial Agents: Antibacterials and Antifungals; Bryskier, A., Ed.; ASM Press: Washington, DC, USA, 2005. [Google Scholar]
- Pruden, A.; Pei, R.; Storteboom, H.; Carlson, K.H. Antibiotic Resistance Genes as Emerging Contaminants: Studies in Northern Colorado. Environ. Sci. Technol. 2006, 40, 7445–7450. [Google Scholar] [CrossRef] [PubMed]
- Zalewska, M.; Błażejewska, A.; Czapko, A.; Popowska, M. Antibiotics and Antibiotic Resistance Genes in Animal Manure—Consequences of Its Application in Agriculture. Front. Microbiol. 2021, 12, 640. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Sun, Y.; Zeng, Z. Antimicrobial use and antimicrobial resistance in food animals. Environ. Sci. Pollut. Res. 2018, 25, 18377–18384. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, J.; Zhu, L.; Ge, W.; Wang, J. Environmental analysis of typical antibiotic-resistant bacteria and ARGs in farmland soil chronically fertilized with chicken manure. Sci. Total Environ. 2017, 593, 10–17. [Google Scholar] [CrossRef]
- Torres, A.G. (Ed.) Escherichia coli in the Americas; Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Agyare, C.; Boamah, V.E.; Zumbi, C.N.; Osei, F.B. Antibiotic Use in Poultry Production and Its Effects on Bacterial Resistance. In Antimicrobial Resistance: A Global Threat; Kumar, Y., Ed.; IntechOpen: London, UK, 2019; pp. 1–20. [Google Scholar] [CrossRef]
- Moreno, M.; Dominguez, L.; Teshager, T.; Herrero, I.A.; Porrero, M.C. Antibiotic resistance monitoring: The Spanish programme. Int. J. Antimicrob. Agents 2000, 14, 285–290. [Google Scholar] [CrossRef]
- Nguyen, M.C.; Woerther, P.L.; Bouvet, M.; Andremont, A.; Leclercq, R.; Canu, A. Escherichia coli as Reservoir for Macrolide Resistance Genes. Emerg. Infect. Dis. 2009, 15, 1648–1650. [Google Scholar] [CrossRef]
- Burow, E.; Simoneit, C.; Tenhagen, B.A.; Käsbohrer, A. Oral antimicrobials increase antimicrobial resistance in porcine E. coli—A systematic review. Prev. Vet. Med. 2014, 113, 364–375. [Google Scholar] [CrossRef]
- Chantziaras, I.; Boyen, F.; Callens, B.; Dewulf, J. Correlation between veterinary antimicrobial use and antimicrobial resistance in food-producing animals: A report on seven countries. J. Antimicrob. Chemother. 2014, 69, 827–834. [Google Scholar] [CrossRef]
- Da Costa, P.M.; Bica, A.; Vaz-Pires, P.; Bernardo, F. Effects of Antimicrobial Treatment on Selection of Resistant Escherichia coli in Broiler Fecal Flora. Microb. Drug Resist. 2008, 14, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Herrero-Fresno, A.; Zachariasen, C.; Hansen, M.H.; Nielsen, A.; Hendriksen, R.S.; Nielsen, S.S.; Olsen, J.E. Apramycin treatment affects selection and spread of a multidrug-resistant Escherichia coli strain able to colonize the human gut in the intestinal microbiota of pigs. Vet. Res. 2016, 47, 12. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Dhar, P.K.; Dutta, A.; Jalal, M.S.; Ghosh, P.; Das, T.; Barua, H.; Biswas, P.K. Circulation of oxytetracycline- and ciprofloxacin-resistant commensal Escherichia coli strains in broiler chickens and farm environments, Bangladesh. Vet. World 2020, 13, 2395–2400. [Google Scholar] [CrossRef]
- Shah, S.H.; Sheikh, I.S.; Kakar, N.; Sumaira; Afzal, S.; Mehmood, K.; Rehman, H.U. In vivo analysis the effect of antibiotic growth promoters (AGPs), Oxytetracycline di-hydrate and Tylosin phosphate on the intestinal microflora in broiler chicken. Braz. J. Biol. 2022, 84, e258114. [Google Scholar] [CrossRef] [PubMed]
- Pokrant, E.; Yévenes, K.; Trincado, L.; Terraza, G.; Galarce, N.; Maddaleno, A.; Martín, B.S.; Lapierre, L.; Cornejo, J. Evaluation of antibiotic dissemination into the environment and untreated animals, by analysis of oxytetracycline in poultry droppings and litter. Animals 2021, 11, 853. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.L. Effect of antibiotics on bacterial populations: A multi-hierarchical selection process. F1000Research 2017, 6, 51. [Google Scholar] [CrossRef] [PubMed]
- Berge, A.C.B.; Epperson, W.B.; Pritchard, R.H. Assessing the effect of a single dose florfenicol treatment in feedlot cattle on the antimicrobial resistance patterns in faecal Escherichia coli. Vet. Res. 2005, 36, 723–734. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fairchild, A.S.; Smith, J.L.; Idris, U.; Lu, J.; Sanchez, S.; Purvis, L.B.; Hofacre, C.; Lee, M.D. Effects of Orally Administered Tetracycline on the Intestinal Community Structure of Chickens and on tet Determinant Carriage by Commensal Bacteria and Campylobacter jejuni. Appl. Environ. Microbiol. 2005, 71, 5865–5872. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Microbiological effects of sublethal levels of Antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Gullberg, E.; Albrecht, L.M.; Karlsson, C.; Sandegren, L.; Andersson, D.I. Selection of a Multidrug Resistance Plasmid by Sublethal Levels of Antibiotics and Heavy Metals. MBio 2014, 5, e01918-14. [Google Scholar] [CrossRef] [PubMed]
- Wistrand-Yuen, E.; Knopp, M.; Hjort, K.; Koskiniemi, S.; Berg, O.G.; Andersson, D.I. Evolution of high-level resistance during low-level antibiotic exposure. Nat. Commun. 2018, 9, 1599. [Google Scholar] [CrossRef] [PubMed]
- Roth, N.; Käsbohrer, A.; Mayrhofer, S.; Zitz, U.; Hofacre, C.; Domig, K.J. The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: A global overview. Poult. Sci. 2019, 98, 1791–1804. [Google Scholar] [CrossRef] [PubMed]
- Rapún Mas, L.; Angulo López, I.; Cecilio Irazola, Á.; Betrán Escartín, A.I. Discrepancy in the genotypic versus phenotypic testing for resistance to rifampicin in Mycobacterium tuberculosis. A case report. Enf. Infecc. Microbiol. Clin. (Engl. ed.) 2019, 37, 212–213. [Google Scholar] [CrossRef]
- Davis, M.A.; Baker, K.N.K.; Orfe, L.H.; Shah, D.H.; Besser, T.E.; Call, D.R. Discovery of a Gene Conferring Multiple-Aminoglycoside Resistance in Escherichia coli. Antimicrob. Agents Chemother. 2010, 54, 2666–2669. [Google Scholar] [CrossRef] [PubMed]
- Al-Bahry, S.; Al-Mashani, B.; Al-Ansari, A.; Elshafie, A.; Mahmoud, L. Escherichia coli tetracycline efflux determinants in relation to tetracycline residues in chicken. Asian Pac. J. Trop. Med. 2013, 6, 718–722. [Google Scholar] [CrossRef]
- Seifi, S.; Khoshbakht, R. Prevalence of tetracycline resistance determinants in broiler isolated Escherichia coli in Iran. Br. Poult. Sci. 2016, 57, 729–733. [Google Scholar] [CrossRef]
- Sreejith, S.; Shajahan, S.; Prathiush, P.R.; Anjana, V.M.; Viswanathan, A.; Chandran, V.; Ajith Kumar, G.S.; Jayachandran, R.; Mathew, J.; Radhakrishnan, E.K. Healthy broilers disseminate antibiotic resistance in response to tetracycline input in feed concentrates. Microb. Pathog. 2020, 149, 104562. [Google Scholar] [CrossRef]
- Skočková, A.; Cupáková, Š.; Karpíšková, R.; Janštová, B. Detection of tetracycline resistance genes in Escherichia coli from raw cow’s milk. J. Microbiol. Biotechnol. Food Sci. 2012, 1, 777–784. [Google Scholar]
- Jurado-Rabadán, S.; De La Fuente, R.; Ruiz-Santa-Quiteria, J.A.; Orden, J.A.; De Vries, L.E.; Agersø, Y. Detection and linkage to mobile genetic elements of tetracycline resistance gene tet(M) in Escherichia coli isolates from pigs. BMC Vet. Res. 2014, 10, 155. [Google Scholar] [CrossRef]
- Titilawo, Y.; Obi, L.; Okoh, A. Occurrence of virulence gene signatures associated with diarrhoeagenic and non-diarrhoeagenic pathovars of Escherichia coli isolates from some selected rivers in South-Western Nigeria. BMC Microbiol. 2015, 15, 204. [Google Scholar] [CrossRef]
- Soufi, L.; Sáenz, Y.; Vinué, L.; Abbassi, M.S.; Ruiz, E.; Zarazaga, M.; Ben Hassen, A.; Hammami, S.; Torres, C. Escherichia coli of poultry food origin as reservoir of sulphonamide resistance genes and integrons. Int. J. Food Microbiol. 2011, 144, 497–502. [Google Scholar] [CrossRef]
- Koo, H.J.; Woo, G.J. Distribution and transferability of tetracycline resistance determinants in Escherichia coli isolated from meat and meat products. Int. J. Food Microbiol. 2011, 145, 407–413. [Google Scholar] [CrossRef]
- European Parliament and of the Council. Directive 2010/63/EU of the of 22 September 2010 on the protection of animals used for scientific purposes. OJEU 2010, L276, 33–79. [Google Scholar]
- American Veterinary Medical Association. AVMA Guidelines for the Euthanasia of Animals: 2020 Edition; AVMA: Schaumburg, IL, USA, 2020. [Google Scholar]
- Comisión Nacional de Investigación Científica y Tecnológica. Manual de Normas de Bioseguridad y Riesgos Asociados: Versión 2018; CONICYT: Santiago, Chile, 2018. [Google Scholar]
- Lupindu, A.M. Isolation and characterization of Escherichia coli from animals, humans, and environment. In Escherichia Coli-Recent Advances on Physiology, Pathogenesis and Biotechnological Applications; Samie, A., Ed.; InTechOpen: Rijeka, Croatia, 2017; pp. 187–206. [Google Scholar] [CrossRef]
- Nkogwe, C.; Raletobana, J.; Stewart-Johnson, A.; Suepaul, S.; Adesiyun, A. Frequency of Detection of Escherichia coli, Salmonella spp., and Campylobacter spp. in the Faeces of Wild Rats (Rattus spp.) in Trinidad and Tobago. Vet. Med. Int. 2011, 2011, 686923. [Google Scholar] [CrossRef]
- Toro, M.; Rivera, D.; Jiménez, M.F.; Díaz, L.; Navarrete, P.; Reyes-Jara, A. Isolation and characterization of non-O157 Shiga toxin-producing Escherichia coli (STEC) isolated from retail ground beef in Santiago, Chile. Food Microbiol. 2018, 75, 4847–4852. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. M100 Performance Standards for Antimicrobial Susceptibility Testing, 29th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- Noll, L.W.; Shridhar, P.B.; Dewsbury, D.M.; Shi, X.; Cernicchiaro, N.; Renter, D.G.; Nagaraja, T.G. A comparison of culture- and PCR- based methods to detect six major non-O157 serogroups of Shiga Toxin-producing Escherichia coli in cattle feces. PLoS ONE 2015, 10, e0135446. [Google Scholar] [CrossRef]
- Ng, L.K.; Martin, I.; Alfa, M.; Mulvey, M. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 2001, 15, 209–215. [Google Scholar] [CrossRef]
- Mamun, M.M.; Parvej, M.S.; Ahamed, S.; Hassan, J.; Nazir, K.H.M.N.H.; Nishikawa, Y.; Rahman, M.T. Prevalence and characterization of shigatoxigenic Escherichia coli in broiler birds in Mymensingh, Bangladesh. J. Vet. Med. 2016, 14, 5–8. [Google Scholar] [CrossRef]
- Standard Operating Procedure for PulseNet PFGE of Escherichia coli O157:H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. Available online: https://www.cdc.gov/pulsenet/pdf/ecoli-shigella-salmonella-pfge-protocol-508c.pdf (accessed on 24 May 2021).

| Experimental Groups | Strains | Post-Treatment Days | p-Value 1 | ||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 7 | 14 | 21 | |||
| Treated | Total number of isolates | 16 | 20 | 19 | 22 | 23 | |
| Non-susceptible strains (%) | 37.5 | 100 | 84.21 | 63.63 | 34.8 | ||
| Susceptible strains (%) | 62.5 | 0.0 | 15.78 | 36.36 | 65.2 | 3.5 × 10−7 | |
| Untreated | Total number of isolates | 30 | 13 | 17 | 12 | 14 | |
| Non-susceptible strains (%) | 0 | 76.92 | 41.17 | 8.33 | 28.57 | ||
| Susceptible strains (%) | 100 | 23.07 | 58.82 | 91.66 | 71.42 | ||
| Experimental Groups | Strains | Post-Treatment Days | p-Value 1 | |||||
|---|---|---|---|---|---|---|---|---|
| 1 | 7 | 14 | 21 | 29 | 36 | |||
| Treated | Total number of isolates | 29.0 | 12.0 | 18.0 | 17.0 | 22.0 | 25.0 | |
| Non-susceptible strains (%) | 82.8 | 75.0 | 72.2 | 17.6 | 68.2 | 92.0 | ||
| Susceptible strains (%) | 17.2 | 25.0 | 27.8 | 82.4 | 31.8 | 8.0 | 2.2 × 10−16 | |
| Untreated | Total number of isolates | 52 | 15 | 15 | 10 | 10 | 25 | |
| Non-susceptible strains (%) | 3.8 | 0.0 | 0.0 | 30.0 | 30.0 | 8.0 | ||
| Susceptible strains (%) | 96.2 | 100.0 | 100.0 | 70.0 | 70.0 | 92.0 | ||
| Gene | Sequence (5′ > 3′) | Annealing Temperature (°C) | Size | Reference |
|---|---|---|---|---|
| tet(A) | F: GCTACATCCTGCTTGCCTTC | 54 | 210 | [46] |
| R: CATAGATCGCCGTGAAGAGG | ||||
| tet(B) | F: TTGGTTAGGGGCAAGTTTTG | 54 | 659 | [46] |
| R: GTAATGGGCCAATAACACCG | ||||
| tet(C) | F: CTTGAGAGCCTTCAACCCAG | 54 | 418 | [46] |
| R: ATGGTCGTCATCTACCTGCC | ||||
| tet(D) | F: AAACCATTACGGCATTCTGC | 54 | 787 | [46] |
| R: GACCGGATACACCATCCATC | ||||
| tet(E) | F: AAACCACATCCTCCATACGC | 54 | 278 | [46] |
| R: AAATAGGCCACAACCGTCAG | ||||
| tet(M) | F: GTGGACAAAGGTACAACGAG | 54 | 406 | [46] |
| R: CGGTAAAGTTCGTCACACAC | ||||
| 16sRNA | F: GACCTCGGTTTAGTTCACAGA | 54 | 585 | [47] |
| R: CACACGCTGACGCTGACCA | ||||
| E. coli uspA | F: CCGATACGCTGCCAATCAGT | 54 | 884 | [43] |
| R: ACGCAGACCGTAGGCCAGAT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pokrant, E.; Vargas, M.B.; Navarrete, M.J.; Yévenes, K.; Trincado, L.; Cortés, P.; Maddaleno, A.; Lapierre, L.; Cornejo, J. Assessing the Effect of Oxytetracycline on the Selection of Resistant Escherichia coli in Treated and Untreated Broiler Chickens. Antibiotics 2023, 12, 1652. https://doi.org/10.3390/antibiotics12121652
Pokrant E, Vargas MB, Navarrete MJ, Yévenes K, Trincado L, Cortés P, Maddaleno A, Lapierre L, Cornejo J. Assessing the Effect of Oxytetracycline on the Selection of Resistant Escherichia coli in Treated and Untreated Broiler Chickens. Antibiotics. 2023; 12(12):1652. https://doi.org/10.3390/antibiotics12121652
Chicago/Turabian StylePokrant, Ekaterina, María Belén Vargas, María José Navarrete, Karina Yévenes, Lina Trincado, Paula Cortés, Aldo Maddaleno, Lisette Lapierre, and Javiera Cornejo. 2023. "Assessing the Effect of Oxytetracycline on the Selection of Resistant Escherichia coli in Treated and Untreated Broiler Chickens" Antibiotics 12, no. 12: 1652. https://doi.org/10.3390/antibiotics12121652
APA StylePokrant, E., Vargas, M. B., Navarrete, M. J., Yévenes, K., Trincado, L., Cortés, P., Maddaleno, A., Lapierre, L., & Cornejo, J. (2023). Assessing the Effect of Oxytetracycline on the Selection of Resistant Escherichia coli in Treated and Untreated Broiler Chickens. Antibiotics, 12(12), 1652. https://doi.org/10.3390/antibiotics12121652








