Whole-Genome Sequencing Analysis of Non-Typhoidal Salmonella Isolated from Breeder Poultry Farm Sources in China, 2020–2021
Abstract
:1. Introduction
2. Results
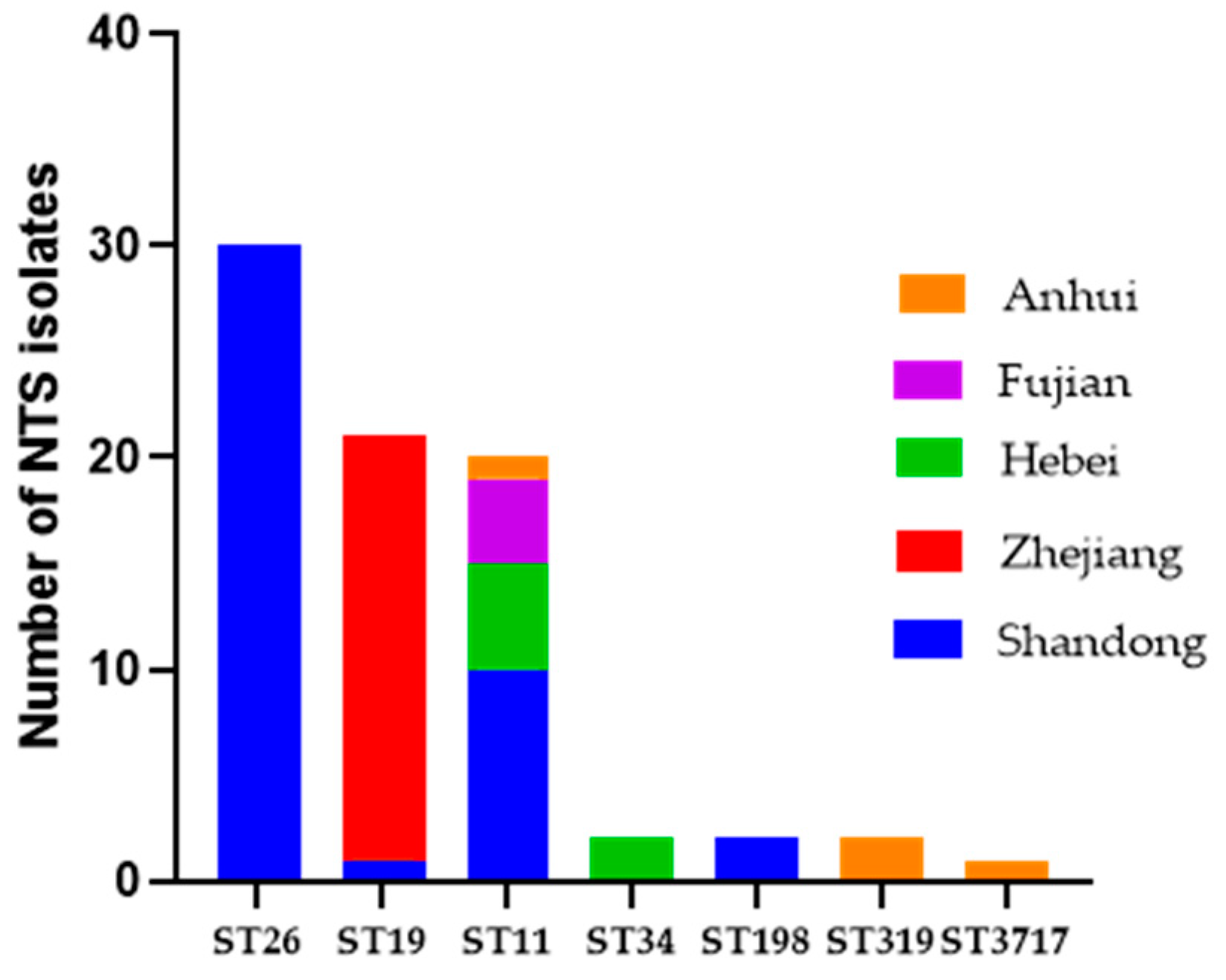
2.1. Distribution of Serotypes and Multi-Locus Sequence Typing Profiles
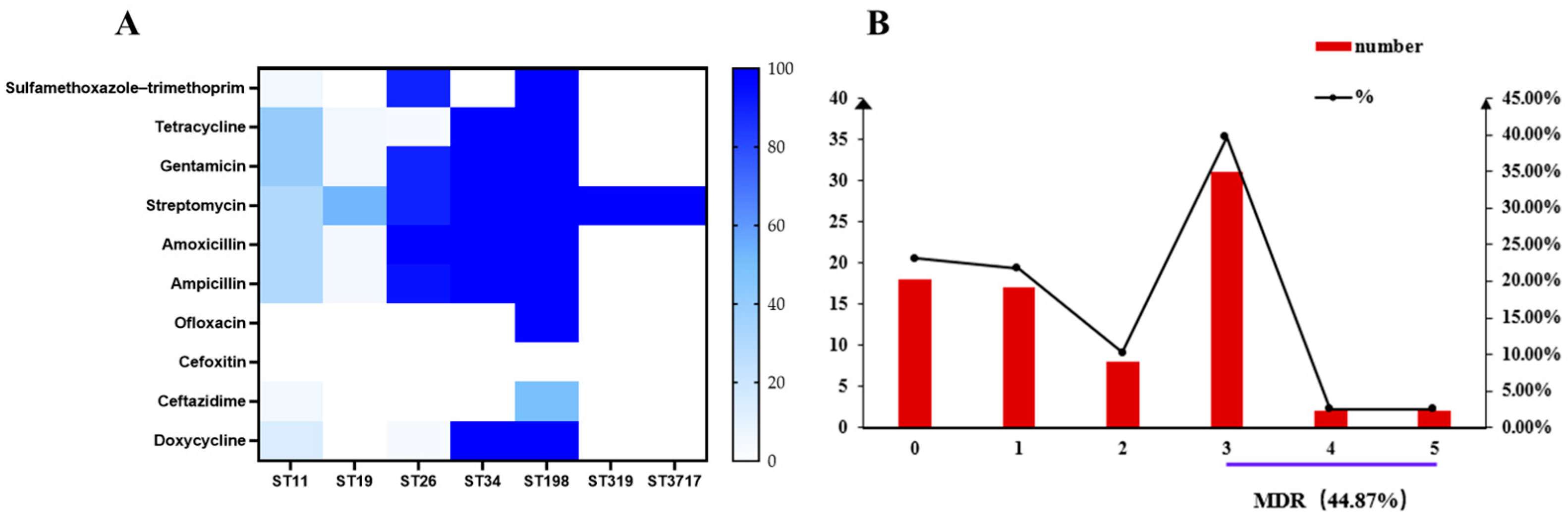
2.2. Phenotypic Antimicrobial Resistance
2.3. Genotypic Antimicrobial Resistance Profiles
2.4. Prediction of Virulence Genes
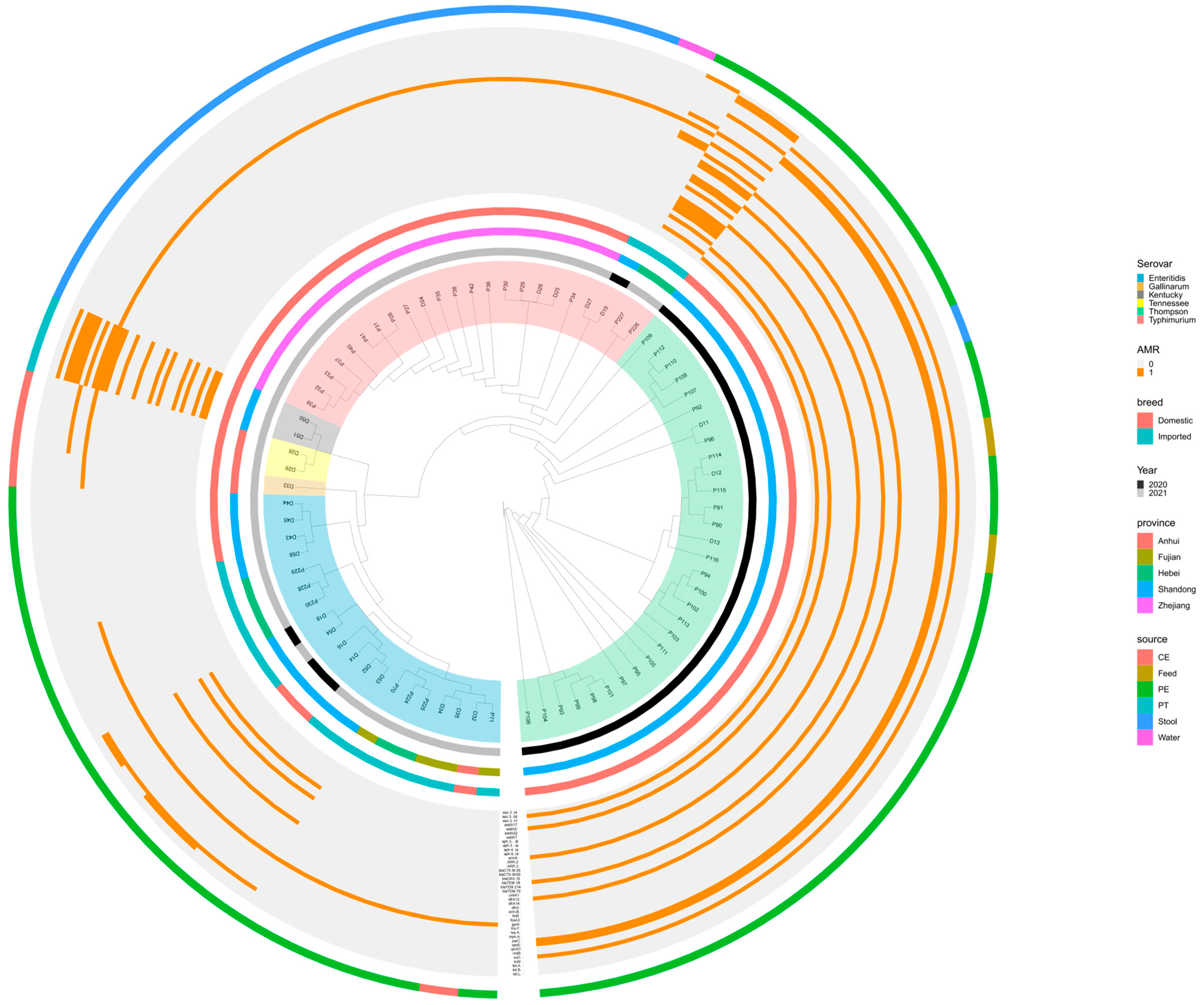
2.5. Whole-Genome SNP Analysis of Salmonella Strains
3. Discussion
4. Materials and Methods
4.1. Salmonella Strains
4.2. Antimicrobial Resistance Tests
4.3. Whole-Genome Sequencing
4.4. Gene Annotation and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tay, M.Y.; Pathirage, S.; Chandrasekaran, L.; Wickramasuriya, U.; Sadeepanie, N.; Waidyarathna, K.D.; Liyanage, L.D.; Seow, K.L.; Hendriksen, R.S.; Takeuchi, M.T.; et al. Whole-Genome Sequencing Analysis of Nontyphoidal Salmonella enterica of Chicken Meat and Human Origin Under Surveillance in Sri Lanka. Foodborne Pathog. Dis. 2019, 16, 531–537. [Google Scholar] [CrossRef]
- Takaichi, M.; Osawa, K.; Nomoto, R.; Nakanishi, N.; Kameoka, M.; Miura, M.; Shigemura, K.; Kinoshita, S.; Kitagawa, K.; Uda, A.; et al. Antibiotic Resistance in Non-Typhoidal Salmonella enterica Strains Isolated from Chicken Meat in Indonesia. Pathogens 2022, 11, 543. [Google Scholar] [CrossRef] [PubMed]
- Rabsch, W.; Tschäpe, H.; Bäumler, A.J. Non-Typhoidal Salmonellosis: Emerging Problems. Microbes Infect. 2001, 3, 237–247. [Google Scholar] [CrossRef]
- Sedrakyan, A.; Ktsoyan, Z.; Arakelova, K.; Gevorgyan, Z.; Zakharyan, M.; Hakobyan, S.; Hovhannisyan, A.; Arakelyan, A.; Aminov, R. Molecular Epidemiology and Virulence of Non-Typhoidal Salmonella in Armenia. Int. J. Mol. Sci. 2022, 23, 9330. [Google Scholar] [CrossRef] [PubMed]
- Ngogo, F.A.; Joachim, A.; Abade, A.M.; Rumisha, S.F.; Mizinduko, M.M.; Majigo, M.V. Factors Associated with Salmonella Infection in Patients with Gastrointestinal Complaints Seeking Health Care at Regional Hospital in Southern Highland of Tanzania. BMC Infect. Dis. 2020, 20, 135. [Google Scholar] [CrossRef]
- Braden, C.R. Salmonella enterica Serotype Enteritidis and Eggs: A National Epidemic in the United States. Clin. Infect. Dis. 2006, 43, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Foley, S.L.; Nayak, R.; Hanning, I.B.; Johnson, T.J.; Han, J.; Ricke, S.C. Population Dynamics of Salmonella Enterica Serotypes in Commercial Egg and Poultry Production. Appl. Environ. Microb. 2011, 77, 4273–4279. [Google Scholar] [CrossRef]
- Cheng, R.A.; Eade, C.R.; Wiedmann, M. Embracing Diversity: Differences in Virulence Mechanisms, Disease Severity, and Host Adaptations Contribute to the Success of Nontyphoidal Salmonella as A Foodborne Pathogen. Front. Microbiol. 2019, 10, 1368. [Google Scholar] [CrossRef] [PubMed]
- Gong, B.; Li, H.; Feng, Y.; Zeng, S.; Zhuo, Z.; Luo, J.; Chen, X.; Li, X. Prevalence, Serotype Distribution and Antimicrobial Resistance of Non-Typhoidal Salmonella in Hospitalized Patients in Conghua District of Guangzhou, China. Front. Cell. Infect. Microbiol. 2022, 12, 54. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-Borne Outbreaks in 2017. EFSA J. 2018, 16, e05077. [Google Scholar]
- Kimura, A.C.; Palumbo, M.S.; Meyers, H.; Abbott, S.; Rodriguez, R.; Werner, S.B. A Multi-State Outbreak of Salmonella Serotype Thompson Infection from Commercially Distributed Bread Contaminated by An Ill Food Handler. Epidemiol. Infect. 2005, 133, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Friesema, I.; de Jong, A.; Hofhuis, A.; Heck, M.; van den Kerkhof, H.; de Jonge, R.; Hameryck, D.; Nagel, K.; van Vilsteren, G.; van Beek, P.; et al. Large Outbreak of Salmonella Thompson Related to Smoked Salmon in the Netherlands, August to December 2012. Euro. Surveill. 2014, 19, 20918. [Google Scholar] [CrossRef] [PubMed]
- Gaulin, C.; Fiset, M.; Duchesne, C.; Ramsay, D.; Savard, N.; Urbanek, A.; Pilon, P.A.; Usongo, V.; Bekal, S. Salmonella Thompson Outbreak Associated with Consumption of Chicken Shawarma and the Usefulness of Genome Sequencing in the Investigation. Can. Commun. Dis. Rep. 2017, 43, 186–192. [Google Scholar] [CrossRef]
- Lee, W.; Kim, E.; Zin, H.; Sung, S.; Woo, J.; Lee, M.J.; Yang, S.M.; Kim, S.H.; Kim, S.H.; Kim, H.Y. Genomic Characteristics and Comparative Genomics Analysis of Salmonella enterica subsp. enterica Serovar Thompson Isolated from An Outbreak in South Korea. Sci. Rep. 2022, 12, 20553. [Google Scholar] [CrossRef]
- Elnekave, E.; Hong, S.L.; Lim, S.; Johnson, T.J.; Perez, A.; Alvarez, J. Comparing Serotyping with Whole-Genome Sequencing for Subtyping of Non-Typhoidal Salmonella enterica: A Large-Scale Analysis of 37 Serotypes with A Public Health Impact in the USA. Microb. Genom. 2020, 6, mgen000425. [Google Scholar] [CrossRef] [PubMed]
- Ben Hassena, A.; Haendiges, J.; Zormati, S.; Guermazi, S.; Gdoura, R.; Gonzalez-Escalona, N.; Siala, M. Virulence and Resistance Genes Profiles and Clonal Relationships of Non-Typhoidal Food-Borne Salmonella Strains Isolated in Tunisia by Whole Genome Sequencing. Int. J. Food Microbiol. 2021, 337, 108941. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Hendriksen, R.S.; Lockett, J.; Gay, K.; Teates, K.; McDermott, P.F.; White, D.G.; Hasman, H.; Sorensen, G.; Bangtrakulnonth, A.; et al. International Spread of Multidrug-Resistant Salmonella Schwarzengrund in Food Products. Emerg. Infect. Dis. 2007, 13, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kang, X.; Ed-Dra, A.; Zhou, X.; Jia, C.; Muller, A.; Liu, Y.; Kehrenberg, C.; Yue, M. Genome-Based Assessment of Antimicrobial Resistance and Virulence Potential of Isolates of Non-Pullorum/Gallinarum Salmonella Serovars Recovered from Dead Poultry in China. Microbiol. Spectr. 2022, 10, e0096522. [Google Scholar] [CrossRef]
- Ed-Dra, A.; Filali, F.R.; Karraouan, B.; El Allaoui, A.; Aboulkacem, A.; Bouchrif, B. Prevalence, Molecular and Antimicrobial Resistance of Salmonella Isolated from Sausages in Meknes, Morocco. Microb. Pathog. 2017, 105, 340–345. [Google Scholar] [CrossRef]
- Haeusler, G.M.; Curtis, N. Non-Typhoidal Salmonella in Children: Microbiology, Epidemiology and Treatment. Adv. Exp. Med. Biol. 2013, 764, 13–26. [Google Scholar]
- Butaye, P.; Devriese, L.A.; Haesebrouck, F. Antimicrobial Growth Promoters Used in Animal Feed: Effects of Less Well Known Antibiotics on Gram-Positive Bacteria. Clin. Microbiol. Rev. 2003, 16, 175–188. [Google Scholar] [CrossRef]
- Silbergeld, E.K.; Graham, J.; Price, L.B. Industrial Food Animal Production, Antimicrobial Resistance, and Human Health. Annu. Rev. Public Health 2008, 29, 151–169. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Borah, P.; Dutta, R.; Das, L.; Hazarika, G.; Choudhury, M.; Deka, N.K.; Malakar, D.; Hussain, M.I.; Barkalita, L.M. Prevalence, Antimicrobial Resistance and Virulence Genes of Salmonella Serovars Isolated from Humans and Animals. Vet. Res. Commun. 2022, 46, 799–810. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Tyson, G.H.; Hsu, C.H.; Harrison, L.; Strain, E.; Tran, T.T.; Tillman, G.E.; Dessai, U.; McDermott, P.F.; Zhao, S. Long-Read Sequencing Reveals Evolution and Acquisition of Antimicrobial Resistance and Virulence Genes in Salmonella enterica. Front. Microbiol. 2021, 12, 777817. [Google Scholar] [CrossRef] [PubMed]
- McMillan, E.A.; Gupta, S.K.; Williams, L.E.; Jove, T.; Hiott, L.M.; Woodley, T.A.; Barrett, J.B.; Jackson, C.R.; Wasilenko, J.L.; Simmons, M.; et al. Antimicrobial Resistance Genes, Cassettes, and Plasmids Present in Salmonella enterica Associated with United States Food Animals. Front. Microbiol. 2019, 10, 832. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.L.; Low, A.J.; Koziol, A.G.; Thomas, M.C.; Leclair, D.; Tamber, S.; Wong, A.; Blais, B.W.; Carrillo, C.D. Systematic Evaluation of Whole Genome Sequence-Based Predictions of Salmonella Serotype and Antimicrobial Resistance. Front. Microbiol. 2020, 11, 549. [Google Scholar] [CrossRef] [PubMed]
- Crouse, A.; Schramm, C.; Emond-Rheault, J.G.; Herod, A.; Kerhoas, M.; Rohde, J.; Gruenheid, S.; Kukavica-Ibrulj, I.; Boyle, B.; Greenwood, C.M.T.; et al. Combining Whole-Genome Sequencing and Multimodel Phenotyping to Identify Genetic Predictors of Salmonella Virulence. mSphere 2020, 5, 1128. [Google Scholar] [CrossRef] [PubMed]
- Foley, S.L.; Johnson, T.J.; Ricke, S.C.; Nayak, R.; Danzeisen, J. Salmonella Pathogenicity and Host Adaptation in Chicken-Associated Serovars. Microbiol. Mol. Biol. Rev. 2013, 77, 582–607. [Google Scholar] [CrossRef]
- Gerlach, R.G.; Hensel, M. Salmonella Pathogenicity Islands in Host Specificity, Host Pathogen-Interactions and Antibiotics Resistance of Salmonella enterica. Berl. Munchen. Tierarztl. 2007, 120, 317. [Google Scholar]
- Dos Santos, A.M.; Ferrari, R.G.; Panzenhagen, P.; Rodrigues, G.L.; Conte-Junior, C.A. Virulence Genes Identification and Characterization Revealed the Presence of the Yersinia High Pathogenicity Island (HPI) in Salmonella from Brazil. Gene 2021, 787, 145646. [Google Scholar] [CrossRef]
- Betancor, L.; Yim, L.; Fookes, M.; Martinez, A.; Thomson, N.R.; Ivens, A.; Peters, S.; Bryant, C.; Algorta, G.; Kariuki, S.; et al. Genomic and Phenotypic Variation in Epidemic-Spanning Salmonella enterica Serovar Enteritidis Isolates. BMC Microbiol. 2009, 9, 237. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Hu, M.; Zhang, Q.; Zhao, C.; Zhang, Y.; Li, L.; Qi, J.; Luo, Y.; Zhou, D.; Liu, Y. Characterization of Integrons and Antimicrobial Resistance in Salmonella from Broilers in Shandong, China. Poult. Sci. 2020, 99, 7046–7054. [Google Scholar] [CrossRef]
- Yu, X.; Zhu, H.; Bo, Y.; Li, Y.; Zhang, Y.; Liu, Y.; Zhang, J.; Jiang, L.; Chen, G.; Zhang, X. Prevalence and Antimicrobial Resistance of Salmonella enterica Subspecies enterica Serovar Enteritidis Isolated from Broiler Chickens in Shandong Province, China, 2013–2018. Poult. Sci. 2021, 100, 1016–1023. [Google Scholar] [CrossRef]
- Paudyal, N.; Pan, H.; Liao, X.; Zhang, X.; Li, X.; Fang, W.; Yue, M. A Meta-Analysis of Major Foodborne Pathogens in Chinese Food Commodities Between 2006 and 2016. Foodborne Pathog. Dis. 2018, 15, 187–197. [Google Scholar] [CrossRef]
- Chen, J.; Ed-Dra, A.; Zhou, H.; Wu, B.; Zhang, Y.; Yue, M. Antimicrobial Resistance and Genomic Investigation of Non-Typhoidal Salmonella Isolated from Outpatients in Shaoxing city, China. Front. Public Health 2022, 10, 988317. [Google Scholar] [CrossRef]
- Siddique, A.; Ullah, N.; Ali, A.; Patel, A.; Moore, T.; Kenney, S.M.; Ganda, E.; Rahman, A. Draft Genome Sequences of 25 Salmonella enterica Serovar Agona Strains Isolated from Poultry and Associated Food Products Harbouring Multiple Antibiotic Resistance Genes. J. Glob. Antimicrob. Resist. 2022, 29, 131–135. [Google Scholar] [CrossRef]
- Tasmin, R.; Hasan, N.A.; Grim, C.J.; Grant, A.; Choi, S.Y.; Alam, M.S.; Bell, R.; Cavanaugh, C.; Balan, K.V.; Babu, U.S.; et al. Genotypic and Phenotypic Characterization of Multidrug Resistant Salmonella Typhimurium and Salmonella Kentucky Strains Recovered from Chicken Carcasses. PLoS ONE 2017, 12, e0176938. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Y.; Lyu, N.; Li, Z.; Ma, S.; Cao, D.; Pan, Y.; Hu, Y.; Huang, H.; Gao, G.F.; et al. The Temporal Dynamics of Antimicrobial-Resistant Salmonella enterica and Predominant Serovars in China. Natl. Sci. Rev. 2023, 10, nwac269. [Google Scholar] [CrossRef]
- Ao, T.T.; Feasey, N.A.; Gordon, M.A.; Keddy, K.H.; Angulo, F.J.; Crump, J.A. Global Burden of Invasive Nontyphoidal Salmonella Disease, 2010. Emerg. Infect. Dis. 2015, 21, 941–949. [Google Scholar]
- Wołkowicz, T.; Zacharczuk, K.; Gierczyński, R.; Nowakowska, M.; Piekarska, K. Antimicrobial Resistance and Whole-Genome Characterisation of High-Level Ciprofloxacin-Resistant Salmonella enterica Serovar Kentucky ST 198 Strains Isolated from Human in Poland. Int. J. Mol. Sci. 2021, 22, 9381. [Google Scholar] [CrossRef]
- Eun, Y.; Jeong, H.; Kim, S.; Park, W.; Ahn, B.; Kim, D.; Kim, E.; Park, E.; Park, S.; Hwang, I.; et al. A Large Outbreak of Salmonella enterica Serovar Thompson Infections Associated with Chocolate Cake in Busan, Korea. Epidemiol. Health. 2019, 41, e2019002. [Google Scholar] [CrossRef]
- Post, A.S.; Diallo, S.N.; Guiraud, I.; Lompo, P.; Tahita, M.C.; Maltha, J.; Van Puyvelde, S.; Mattheus, W.; Ley, B.; Thriemer, K.; et al. Supporting Evidence for A Human Reservoir of Invasive Non-Typhoidal Salmonella from Household Samples in Burkina Faso. PLoS Negl. Trop. Dis. 2019, 13, e0007782. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Li, T.; Liu, F.; Cheng, Y.; Guo, X.; Wen, G.; Luo, Q.; Shao, H.; Pan, Z.; et al. Characterization of Salmonella spp. Isolated from Chickens in Central China. BMC Vet. Res. 2020, 16, 299. [Google Scholar] [CrossRef]
- Lee, S.K.; Chon, J.W.; Song, K.Y.; Hyeon, J.Y.; Moon, J.S.; Seo, K.H. Prevalence, Characterization, and Antimicrobial Susceptibility of Salmonella Gallinarum Isolated from Eggs Produced in Conventional or Organic Farms in South Korea. Poult. Sci. 2013, 92, 2789–2797. [Google Scholar] [CrossRef]
- Mechesso, A.F.; Moon, D.C.; Kim, S.J.; Song, H.J.; Kang, H.Y.; Na, S.H.; Choi, J.H.; Kim, H.Y.; Yoon, S.S.; Lim, S.K. Nationwide Surveillance on Serotype Distribution and Antimicrobial Resistance Profiles of Non-Typhoidal Salmonella Serovars Isolated from Food-Producing Animals in South Korea. Int. J. Food Microbiol. 2020, 335, 108893. [Google Scholar] [CrossRef]
- Chen, H.; Qiu, H.; Zhong, H. Non-Typhoidal Salmonella Infections Among Children in Fuzhou, Fujian, China: A 10-Year Retrospective Review from 2012 to 2021. Infect. Drug Resist. 2023, 16, 2737–2749. [Google Scholar] [CrossRef]
- Thung, T.Y.; Radu, S.; Mahyudin, N.A.; Rukayadi, Y.; Zakaria, Z.; Mazlan, N.; Tan, B.H.; Lee, E.; Yeoh, S.L.; Chin, Y.Z.; et al. Prevalence, Virulence Genes and Antimicrobial Resistance Profiles of Salmonella Serovars from Retail Beef in Selangor, Malaysia. Front. Microbiol. 2017, 8, 2697. [Google Scholar] [CrossRef]
- Wang, Z.; Duan, L.; Liu, F.; Hu, Y.; Leng, C.; Kan, Y.; Yao, L.; Shi, H. First Report of Enterobacter Hormaechei with Respiratory Disease in Calves. BMC Vet. Res. 2020, 16, 1. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, X.; Ding, X.; Bin, P.; Zhu, G. The Vertical Transmission of Salmonella Enteritidis in A One-Health context. One Health 2023, 16, 100469. [Google Scholar] [CrossRef]
- Eng, S.-K.; Pusparajah, P.; Ab Mutalib, N.-S.; Ser, H.-L.; Chan, K.-G.; Lee, L.-H. Salmonella: A Review on Pathogenesis, Epidemiology and Antibiotic Resistance. Front. Life Sci. 2015, 8, 284–293. [Google Scholar] [CrossRef]
- Tang, B.; Elbediwi, M.; Nambiar, R.B.; Yang, H.; Lin, J.; Yue, M. Genomic Characterization of Antimicrobial-Resistant Salmonella enterica in Duck, Chicken, and Pig Farms and Retail Markets in Eastern China. Microbiol. Spectr. 2022, 10, e0125722. [Google Scholar] [CrossRef] [PubMed]
- Vo, A.T.; van Duijkeren, E.; Gaastra, W.; Fluit, A.C. Antimicrobial Resistance, Class 1 Integrons, and Genomic Island 1 in Salmonella Isolates from Vietnam. PLoS ONE 2010, 5, e9440. [Google Scholar] [CrossRef] [PubMed]
- Chantziaras, I.; Boyen, F.; Callens, B.; Dewulf, J. Correlation between Veterinary Antimicrobial Use and Antimicrobial Resistance in Food-Producing Animals: A Report on Seven Countries. J. Antimicrob. Chemother. 2014, 69, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Lekagul, A.; Tangcharoensathien, V.; Yeung, S. Patterns of Antibiotic Use in Global Pig Production: A Systematic Review. Vet. Anim. Sci. 2019, 7, 100058. [Google Scholar] [CrossRef] [PubMed]
- Diaz, D.; Hernandez-Carreño, P.E. Prevalence, Main Serovars and Anti-Microbial Resistance Profiles of Non-Typhoidal Salmonella in Poultry Samples from The Americas: A Systematic Review and Meta-Analysis. Transbound. Emerg. Dis. 2022, 69, 2544–2558. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.F.; Li, H.Q.; Yang, Q.; Zhang, F.F.; Tan, J.; Zeng, Y.B.; Wei, Q.P.; Huang, J.N.; Wu, C.C.; Li, N.; et al. Prevalence and Antimicrobial Resistance Profile of Bacterial Pathogens Isolated from Poultry in Jiangxi Province, China from 2020 to 2022. Poult. Sci. 2023, 102, 102830. [Google Scholar] [CrossRef]
- Van, T.T.; Nguyen, H.N.; Smooker, P.M.; Coloe, P.J. The Antibiotic Resistance Characteristics of Non-Typhoidal Salmonella enterica Isolated from Food-Producing Animals, Retail Meat and Humans in South East Asia. Int. J. Food Microbiol. 2012, 154, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Boonmar, S.; Markvichitr, K.; Chaunchom, S.; Chanda, C.; Bangtrakulnonth, A.; Pornrunangwong, S.; Yamamoto, S.; Suzuki, D.; Kozawa, K.; Kimura, H.; et al. Salmonella Prevalence in Slaughtered Buffaloes and Pigs and Antimicrobial Susceptibility of Isolates in Vientiane, Lao People’s Democratic Republic. J. Vet. Med. Sci. 2008, 70, 1345–1348. [Google Scholar] [CrossRef] [PubMed]
- Heider, L.C.; Hoet, A.E.; Wittum, T.E.; Khaitsa, M.L.; Love, B.C.; Huston, C.L.; Morley, P.S.; Funk, J.A.; Gebreyes, W.A. Genetic and Phenotypic Characterization of The Bla(Cmy) Gene from Escherichia coli and Salmonella enterica Isolated from Food-Producing Animals, Humans, The Environment, and Retail Meat. Foodborne Pathog. Dis. 2009, 6, 1235–1240. [Google Scholar] [CrossRef]
- Dahshan, H.; Chuma, T.; Shahada, F.; Akiba, M.; Fujimoto, H.; Akasaka, K.; Kamimura, Y.; Okamoto, K. Characterization of Antibiotic Resistance and the Emergence of AmpC-Producing Salmonella Infantis from Pigs. J. Vet. Med. Sci. 2010, 72, 1437–1442. [Google Scholar] [CrossRef] [PubMed]
- Glenn, L.M.; Lindsey, R.L.; Frank, J.F.; Meinersmann, R.J.; Englen, M.D.; Fedorka-Cray, P.J.; Frye, J.G. Analysis of Antimicrobial Resistance Genes Detected in Multidrug-Resistant Salmonella enterica Serovar Typhimurium Isolated from Food Animals. Microb. Drug Resist. 2011, 17, 407–418. [Google Scholar] [CrossRef]
- Sugawara, M.; Komori, J.; Kawakami, M.; Izumiya, H.; Watanabe, H.; Akiba, M. Molecular and Phenotypic Characteristics of CMY-2 &beta-Lactamase-Producing Salmonella enterica Serovar Typhimurium Isolated from Cattle in Japan. J. Vet. Med. Sci. 2011, 73, 345–349. [Google Scholar] [PubMed]
- Campioni, F.; Zoldan, M.M.; Falcão, J.P. Characterization of Salmonella Enteritidis Strains Isolated from Poultry and Farm Environments in Brazil. Epidemiol. Infect. 2014, 142, 1403–1410. [Google Scholar] [CrossRef] [PubMed]
- Voss-Rech, D.; Kramer, B.; Silva, V.S.; Rebelatto, R.; Abreu, P.G.; Coldebella, A.; Vaz, C.S.L. Longitudinal Study Reveals Persistent Environmental Salmonella Heidelberg in Brazilian Broiler Farms. Vet. Microbiol. 2019, 233, 118–123. [Google Scholar] [CrossRef]
- Cui, L.; Liu, Q.; Jiang, Z.; Song, Y.; Yi, S.; Qiu, J.; Hao, G.; Sun, S. Characteristics of Salmonella from Chinese Native Chicken Breeds Fed on Conventional or Antibiotic-Free Diets. Front. Vet. Sci. 2021, 8, 607491. [Google Scholar] [CrossRef]
- Le Hello, S.; Harrois, D.; Bouchrif, B.; Sontag, L.; Elhani, D.; Guibert, V.; Zerouali, K.; Weill, F.X. Highly Drug-Resistant Salmonella enterica Serotype Kentucky St198-X1: A Microbiological Study. Lancet Infect. Dis. 2013, 13, 672–679. [Google Scholar] [CrossRef]
- Ladely, S.R.; Meinersmann, R.J.; Ball, T.A.; Fedorka-Cray, P.J. Antimicrobial Susceptibility and Plasmid Replicon Typing of Salmonella enterica Serovar Kentucky Isolates Recovered from Broilers. Foodborne Pathog. Dis. 2016, 13, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Bai, J.; Zhang, X.; Wang, S.; Chen, K.; Lin, Q.; Xu, C.; Qu, X.; Zhang, H.; Liao, M.; et al. Highly Prevalent Multidrug Resistance and Qrdr Mutations in Salmonella Isolated from Chicken, Pork and Duck Meat in Southern China, 2018–2019. Int. J. Food Microbiol. 2021, 340, 109055. [Google Scholar] [CrossRef]
- Schatz, A.; Bugie, E.; Waksman, S.A. Streptomycin, A Substance Exhibiting Antibiotic Activity against Gram-Positive and Gram-Negative Bacteria. 1944. Clin. Orthop. Relat. Res. 2005, 437, 3–6. [Google Scholar] [CrossRef]
- Mengistu, G.; Dejenu, G.; Tesema, C.; Arega, B.; Awoke, T.; Alemu, K.; Moges, F. Epidemiology of Streptomycin Resistant Salmonella from Humans and Animals in Ethiopia: A Systematic Review and Meta-Analysis. PLoS ONE 2020, 15, e0244057. [Google Scholar]
- Tadesse, D.A.; Singh, A.; Zhao, S.; Bartholomew, M.; Womack, N.; Ayers, S.; Fields, P.I.; McDermott, P.F. Antimicrobial Resistance in Salmonella in the United States from 1948 to 1995. Antimicrob. Agents Chemother. 2016, 60, 2567–2571. [Google Scholar] [CrossRef] [PubMed]
- Agustín, A.I.; Carramiñana, J.J.; Rota, C.; Herrera, A. Antimicrobial Resistance of Salmonella spp. from Pigs at Slaughter in Spain in 1993 and 2001. Lett. Appl. Microbiol. 2005, 41, 39–44. [Google Scholar] [CrossRef]
- Ryder, R.W.; Blake, P.A.; Murlin, A.C.; Carter, G.P.; Pollard, R.A.; Merson, M.H.; Allen, S.D.; Brenner, D.J. Increase in Antibiotic Resistance among Isolates of Salmonella in The United States, 1967–1975. J. Infect. Dis. 1980, 142, 485–491. [Google Scholar] [CrossRef]
- Kim, M.S.; Lim, T.H.; Jang, J.H.; Lee, D.H.; Kim, B.Y.; Kwon, J.H.; Choi, S.W.; Noh, J.Y.; Hong, Y.H.; Lee, S.B.; et al. Prevalence and Antimicrobial Resistance of Salmonella Species Isolated from Chicken Meats Produced by Different Integrated Broiler Operations in Korea. Poult. Sci. 2012, 91, 2370–2375. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, I.; Ishihara, T.; Teranishi, H.; Saito, S.; Yatsuyanagi, J.; Wada, E.; Kumagai, Y.; Takahashi, S.; Konno, T.; Kashio, H.; et al. Prevalence and Characteristics of Salmonella and Campylobacter in Retail Poultry Meat in Japan. Jpn. J. Infect. Dis. 2017, 70, 239–247. [Google Scholar] [CrossRef]
- Long, L.; You, L.; Wang, D.; Wang, M.; Wang, J.; Bai, G.; Li, J.; Wei, X. Highly Prevalent MDR, Frequently Carrying Virulence Genes and Antimicrobial Resistance Genes in Salmonella enterica Serovar 4,[5],12:I:-Isolates from Guizhou Province, China. PLoS ONE 2022, 17, e0266443. [Google Scholar]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M. The Global Burden of Nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef]
- Rao, S.; Linke, L.; Doster, E.; Hyatt, D.; Burgess, B.A.; Magnuson, R.; Pabilonia, K.L.; Morley, P.S. Genomic Diversity of Class I Integrons from Antimicrobial Resistant Strains of Salmonella Typhimurium Isolated from Livestock, Poultry and Humans. PLoS ONE 2020, 15, e0243477. [Google Scholar]
- Mina, S.A.; Hasan, M.Z.; Hossain, A.; Barua, A.; Mirjada, M.R.; Chowdhury, A. The Prevalence of Multi-Drug Resistant Salmonella Typhi Isolated from Blood Sample. Microbiol. Insights. 2023, 16, 11786361221150760. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, J.; Chen, C.; Wang, Y.; Chen, X.; Li, X.; Xu, F. Resistance and Pathogenicity of Salmonella Thompson Isolated from Incubation End of a Poultry Farm. Vet. Sci. 2022, 9, 349. [Google Scholar]
- Le Hello, S.; Hendriksen, R.S.; Doublet, B.; Fisher, I.; Nielsen, E.M.; Whichard, J.M.; Bouchrif, B.; Fashae, K.; Granier, S.A.; Jourdan-Da Silva, N.; et al. International Spread of An Epidemic Population of Salmonella enterica Serotype Kentucky ST198 Resistant to Ciprofloxacin. J. Infect. Dis. 2011, 204, 675–684. [Google Scholar]
- ESFA. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA J. 2018, 16, e05182. [Google Scholar]
- Coipan, C.E.; Westrell, T.; van Hoek, A.; Alm, E. Genomic Epidemiology of Emerging ESBL-Producing Salmonella Kentucky bla (CTX-M-14b) in Europe. Emerg. Microbes. Infect. 2020, 9, 2124–2135. [Google Scholar] [PubMed]
- Samper-Cativiela, C.; Diéguez-Roda, B.; Trigo da Roza, F.; Ugarte-Ruiz, M.; Elnekave, E.; Lim, S.; Hernández, M.; Abad, D.; Collado, S.; Sáez, J.L.; et al. Genomic Characterization of Multidrug-Resistant Salmonella Serovar Kentucky ST198 Isolated in Poultry Flocks in Spain (2011–2017). Microb. Genom. 2022, 8, 773. [Google Scholar]
- Liu, Y.; Jiang, J.; Ed-Dra, A.; Li, X.; Peng, X.; Xia, L.; Guo, Q.; Yao, G.; Yue, M. Prevalence and Genomic Investigation of Salmonella Isolates Recovered from Animal Food-Chain in Xinjiang, China. Food Res. Int. 2021, 142, 110198. [Google Scholar] [PubMed]
- Liu, Q.; Chen, W.; Elbediwi, M.; Pan, H.; Wang, L.; Zhou, C.; Zhao, B.; Xu, X.; Li, D.; Yan, X.; et al. Characterization of Salmonella Resistome and Plasmidome in Pork Production System in Jiangsu, China. Front. Vet. Sci. 2020, 7, 617. [Google Scholar] [PubMed]
- Zhao, W.H.; Hu, Z.Q. Epidemiology and Genetics of CTX-M Extended-Spectrum Β-Lactamases in Gram-Negative Bacteria. Crit. Rev. Microbiol. 2013, 39, 79–101. [Google Scholar]
- de Jong, A.; Smet, A.; Ludwig, C.; Stephan, B.; De Graef, E.; Vanrobaeys, M.; Haesebrouck, F. Antimicrobial Susceptibility of Salmonella Isolates from Healthy Pigs and Chickens (2008–2011). Vet. Microbiol. 2014, 171, 298–306. [Google Scholar]
- Liu, B.T.; Yang, Q.E.; Li, L.; Sun, J.; Liao, X.P.; Fang, L.X.; Yang, S.S.; Deng, H.; Liu, Y.H. Dissemination and Characterization of Plasmids Carrying Oqxab-BlaCtx-M Genes in Escherichia Coli Isolates from Food-Producing Animals. PLoS ONE 2013, 8, e73947. [Google Scholar]
- Chen, W.; Fang, T.; Zhou, X.; Zhang, D.; Shi, X.; Shi, C. IncHI2 Plasmids Are Predominant in Antibiotic-Resistant Salmonella Isolates. Front. Microbiol. 2016, 7, 1566. [Google Scholar]
- Teng, L.; Liao, S.; Zhou, X.; Jia, C.; Feng, M.; Pan, H.; Ma, Z.; Yue, M. Prevalence and Genomic Investigation of Multidrug-Resistant Salmonella Isolates from Companion Animals in Hangzhou, China. Antibiotics 2022, 11, 625. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Liu, X.; Li, C.; Jiang, Z.; Li, D.; Zhu, L. Genomic Virulence Genes Profile Analysis of Salmonella enterica Isolates from Animal and Human in China from 2004 to 2019. Microb. Pathog. 2022, 173, 105808. [Google Scholar] [CrossRef] [PubMed]
- Guiney, D.G.; Fierer, J. The Role of The spv Genes in Salmonella Pathogenesis. Front. Microbiol. 2011, 2, 129. [Google Scholar] [CrossRef]
- Shi, W.; Tang, W.; Li, Y.; Han, Y.; Cui, L.; Sun, S. Comparative Analysis between Salmonella enterica Isolated from Imported and Chinese Native Chicken Breeds. Microorganisms 2023, 11, 390. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; CLSI Supplement M100; Clinical & Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, C.E.; Kruczkiewicz, P.; Laing, C.R.; Lingohr, E.J.; Gannon, V.P.; Nash, J.H.; Taboada, E.N. The Salmonella In Silico Typing Resource (SISTR): An Open Web-Accessible Tool for Rapidly Typing and Subtyping Draft Salmonella Genome Assemblies. PLoS ONE 2016, 11, e0147101. [Google Scholar]
- Bharat, A.; Petkau, A.; Avery, B.P.; Chen, J.C.; Folster, J.P.; Carson, C.A.; Kearney, A.; Nadon, C.; Mabon, P.; Thiessen, J.; et al. Correlation between Phenotypic and In Silico Detection of Antimicrobial Resistance in Salmonella enterica in Canada Using Staramr. Microorganisms 2022, 10, 292. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Papid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ju, Z.; Cui, L.; Lei, C.; Song, M.; Chen, X.; Liao, Z.; Zhang, T.; Wang, H. Whole-Genome Sequencing Analysis of Non-Typhoidal Salmonella Isolated from Breeder Poultry Farm Sources in China, 2020–2021. Antibiotics 2023, 12, 1642. https://doi.org/10.3390/antibiotics12111642
Ju Z, Cui L, Lei C, Song M, Chen X, Liao Z, Zhang T, Wang H. Whole-Genome Sequencing Analysis of Non-Typhoidal Salmonella Isolated from Breeder Poultry Farm Sources in China, 2020–2021. Antibiotics. 2023; 12(11):1642. https://doi.org/10.3390/antibiotics12111642
Chicago/Turabian StyleJu, Zijing, Lulu Cui, Changwei Lei, Mengze Song, Xuan Chen, Ziwei Liao, Tiejun Zhang, and Hongning Wang. 2023. "Whole-Genome Sequencing Analysis of Non-Typhoidal Salmonella Isolated from Breeder Poultry Farm Sources in China, 2020–2021" Antibiotics 12, no. 11: 1642. https://doi.org/10.3390/antibiotics12111642
APA StyleJu, Z., Cui, L., Lei, C., Song, M., Chen, X., Liao, Z., Zhang, T., & Wang, H. (2023). Whole-Genome Sequencing Analysis of Non-Typhoidal Salmonella Isolated from Breeder Poultry Farm Sources in China, 2020–2021. Antibiotics, 12(11), 1642. https://doi.org/10.3390/antibiotics12111642






