Evaluation of Antibiotic Resistance Mechanisms in Gram-Negative Bacteria
Abstract
:1. Introduction
Antimicrobial Resistance and Multidrug Resistance
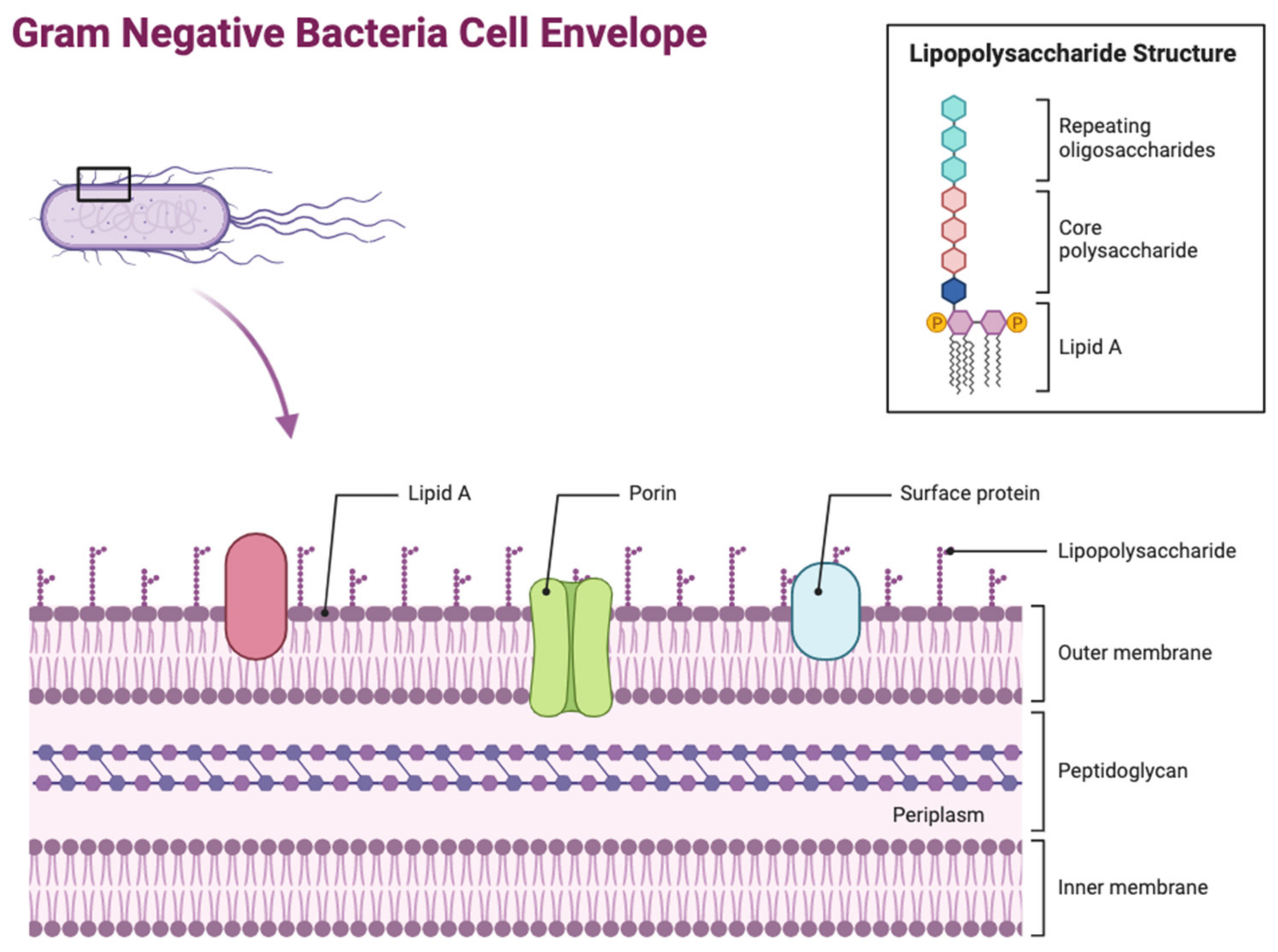
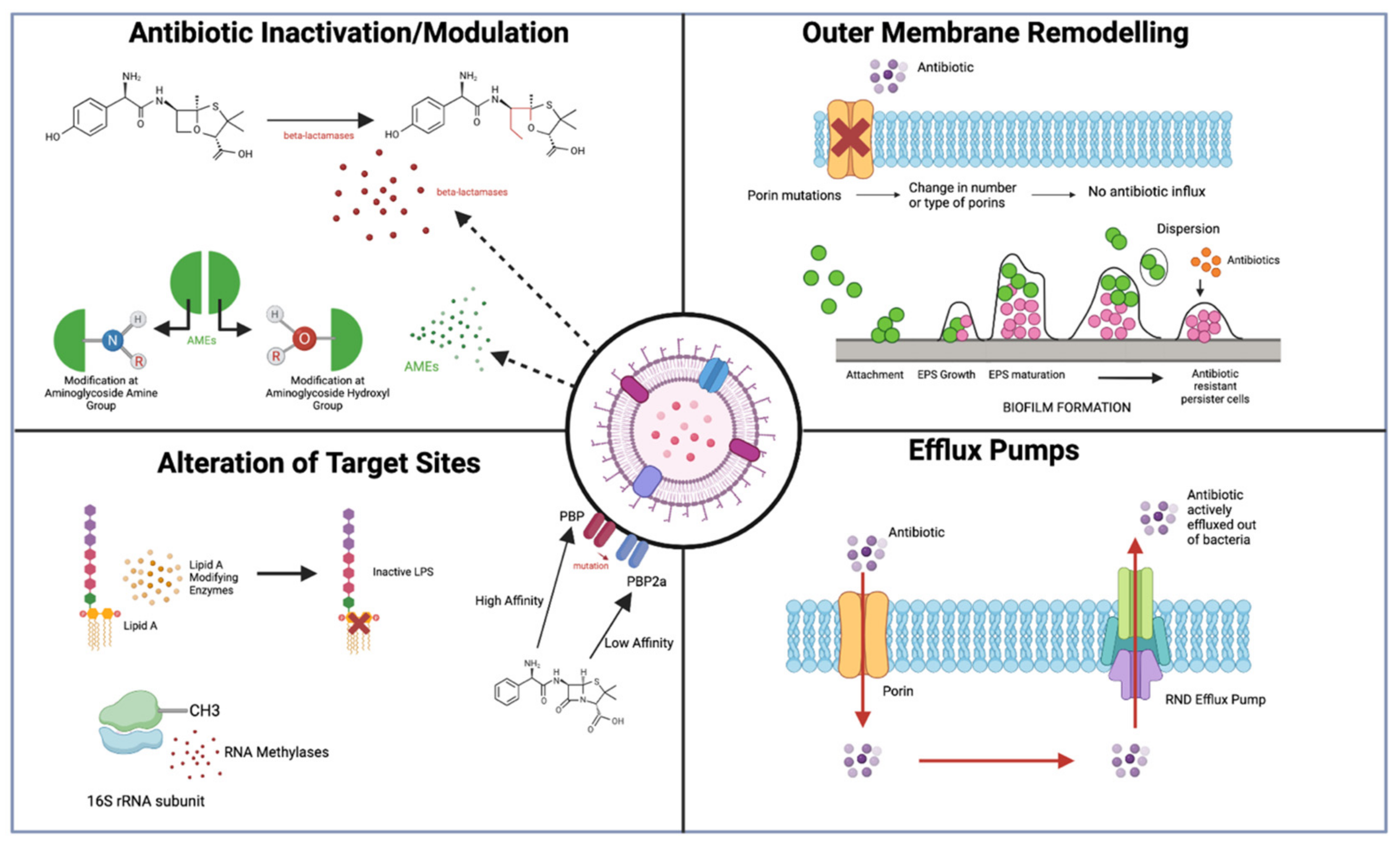
2. Antibiotic Resistance Mechanisms in Gram-Negative Bacteria
- (1)
- Drug inactivation;
- (2)
- Limiting drug uptake;
- (3)
- Altering drug target;
- (4)
- High levels of drug efflux.
2.1. Antibiotic Inactivation/Modulation
2.1.1. β-Lactamase-Antibiotic Inactivation
2.1.2. Aminoglycoside-Modifying Enzymes (AMEs)—Antibiotic Modification
2.2. Limiting Influx of Antibiotics
OM Remodelling
2.3. Modifications of Antibiotic Targets
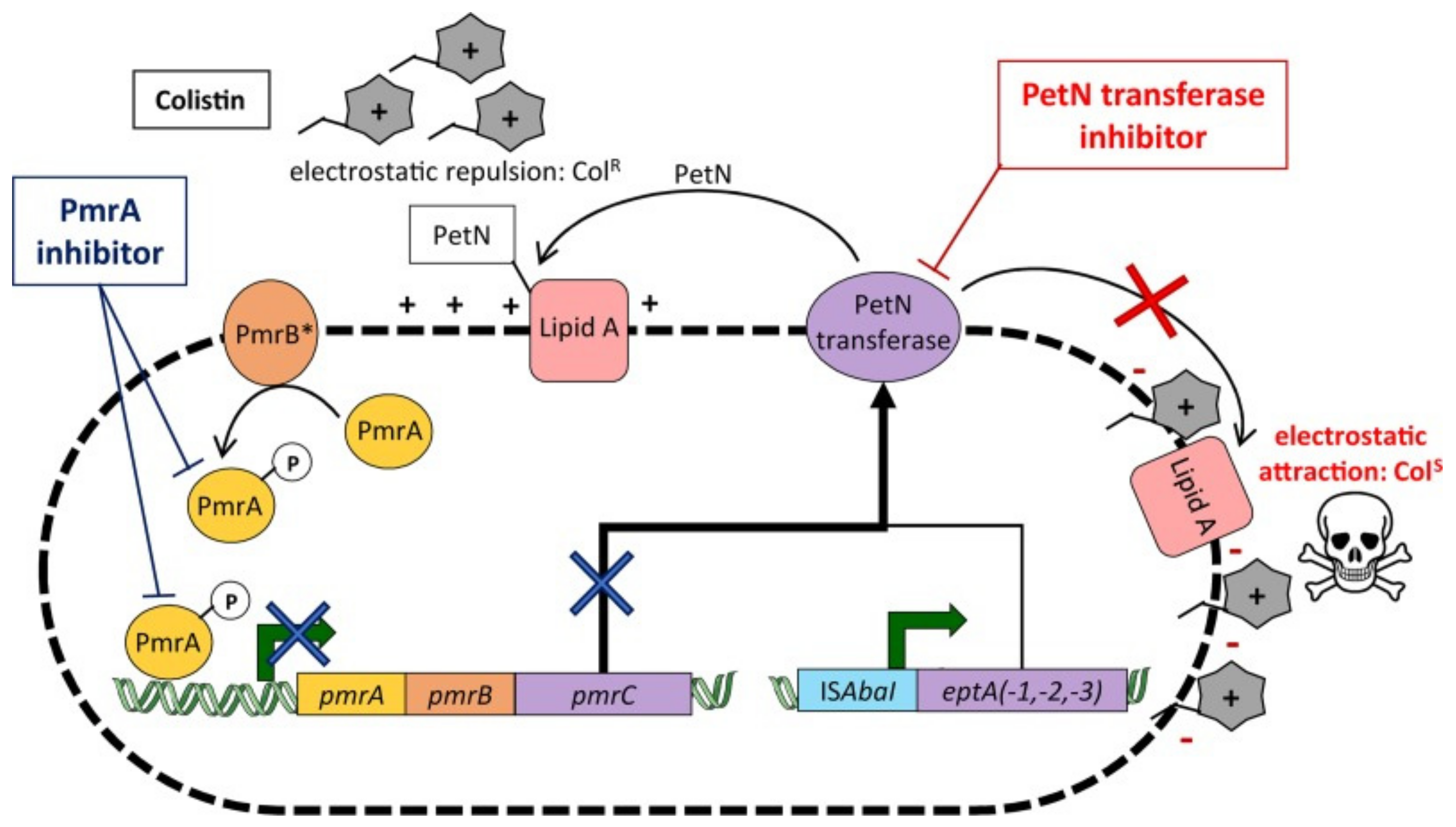
2.3.1. Lipid A Modifications
2.3.2. 16S Ribosomal RNA Methylation
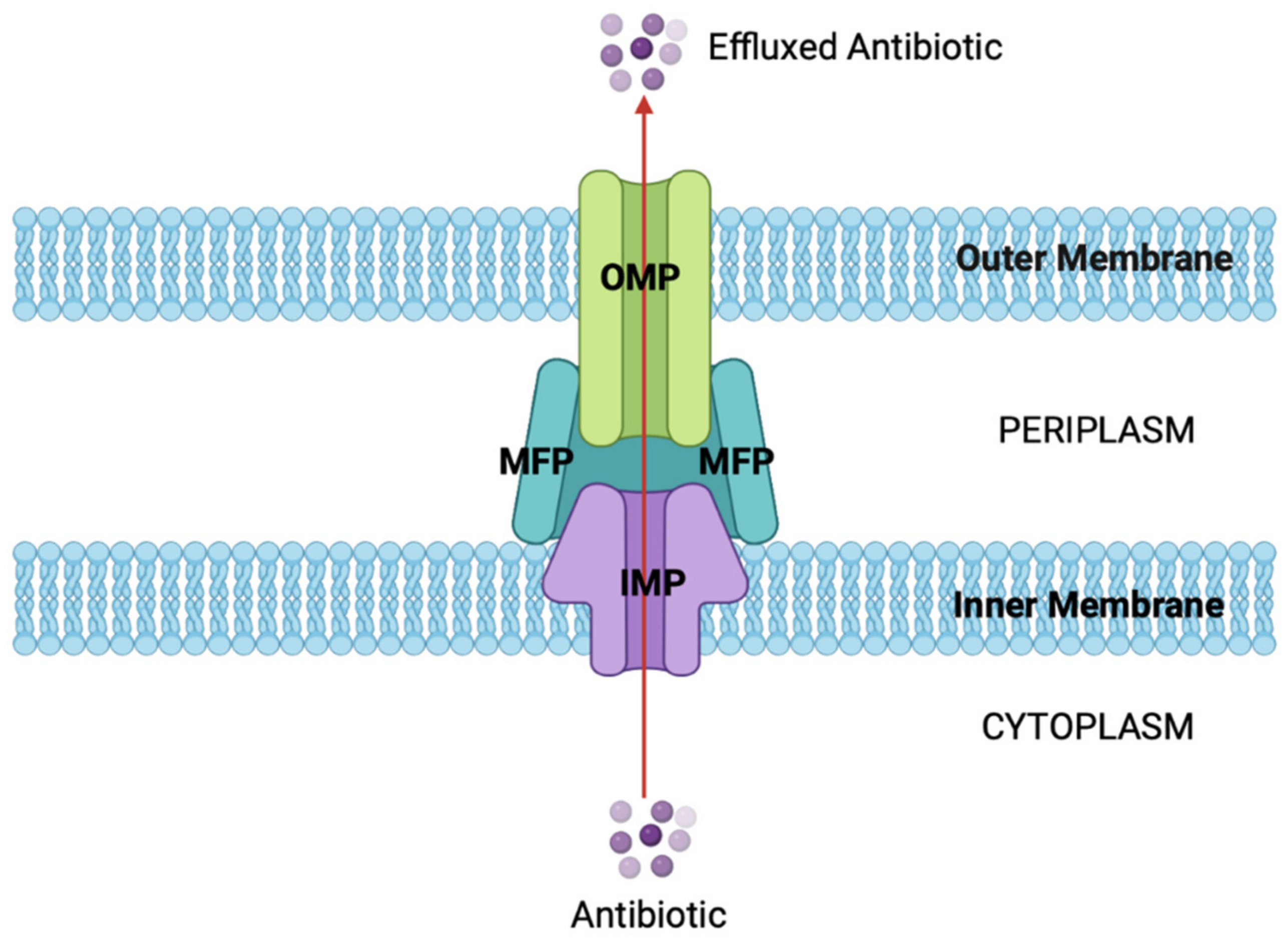
2.4. Increasing Efflux of Antibiotics
Efflux Pumps
3. Key Multidrug-Resistant Gram-Negative Bacteria
3.1. Escherichia coli
3.1.1. β-Lactamases
3.1.2. Aminoglycoside-Modifying Enzymes
3.1.3. OM Remodelling
3.1.4. Efflux Pumps
3.1.5. Alteration of Target Sites
3.1.6. Key Findings
3.2. Acinetobacter baumannii
3.2.1. β-Lactamases
3.2.2. Aminoglycoside-Modifying Enzymes
3.2.3. Efflux Pumps
3.2.4. OM Remodelling
3.2.5. Alteration of Target Sites
3.2.6. Key Findings
3.3. Klebsiella pneumoniae
3.3.1. β-Lactamases
3.3.2. Aminoglycoside-Modifying Enzymes
3.3.3. Efflux Pumps
3.3.4. OM Remodelling
3.3.5. Alterations of Target Sites
3.3.6. Key Findings
3.4. Pseudomonas aeruginosa
3.4.1. β-Lactamases
3.4.2. Aminoglycoside-Modifying Enzymes
3.4.3. Outer Membrane Remodelling
3.4.4. Efflux Pumps
3.4.5. Alteration of Target Sites
3.4.6. Key Findings
4. Future Directions and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Collet, J.F.; Cho, S.H.; Iorga, B.I.; Goemans, C.V. How the Assembly and Protection of the Bacterial Cell Envelope Depend on Cysteine Residues. J. Biol. Chem. 2020, 295, 11984–11994. [Google Scholar] [CrossRef] [PubMed]
- Silhavy, T.J.; Kahne, D.; Walker, S. The Bacterial Cell Envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef] [PubMed]
- El Rayes, J.; Rodríguez-Alonso, R.; Collet, J.F. Lipoproteins in Gram-Negative Bacteria: New Insights into Their Biogenesis, Subcellular Targeting and Functional Roles. Curr. Opin. Microbiol. 2021, 61, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Rollauer, S.E.; Sooreshjani, M.A.; Noinaj, N.; Buchanan, S.K. Outer Membrane Protein Biogenesis in Gram-Negative Bacteria. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20150023. [Google Scholar] [CrossRef] [PubMed]
- Bertani, B.; Ruiz, N. Function and Biogenesis of Lipopolysaccharides. EcoSal Plus 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Otten, C.; Brilli, M.; Vollmer, W.; Viollier, P.H.; Salje, J. Peptidoglycan in Obligate Intracellular Bacteria. Mol. Microbiol. 2018, 107, 142–163. [Google Scholar] [CrossRef]
- Garde, S.; Chodisetti, P.K.; Reddy, M. Peptidoglycan: Structure, Synthesis, and Regulation. EcoSal Plus 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Breijyeh, Z.; Jubeh, B.; Karaman, R. Resistance of Gram-Negative Bacteria to Current Antibacterial Agents and Approaches to Resolve It. Molecules 2020, 25, 1340. [Google Scholar] [CrossRef]
- Dalbey, R.E.; Wang, P.; Kuhn, A. Assembly of Bacterial Inner Membrane Proteins. Annu. Rev. Biochem. 2011, 80, 161–187. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial Resistance: A Global Multifaceted Phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef]
- Yang, X.; Ye, W.; Qi, Y.; Ying, Y.; Xia, Z. Overcoming Multidrug Resistance in Bacteria through Antibiotics Delivery in Surface-Engineered Nano-Cargos: Recent Developments for Future Nano-Antibiotics. Front. Bioeng. Biotechnol. 2021, 9, 696514. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.K.; Buroni, S.; Swain, S.S.; Bonacorsi, A.; da Fonseca Amorim, E.A.; Kulshrestha, M.; da Silva, L.C.N.; Tiwari, V. Recent Advances to Combat ESKAPE Pathogens with Special Reference to Essential Oils. Front. Microbiol. 2022, 13, 1029098. [Google Scholar] [CrossRef] [PubMed]
- Rémy, V.; Largeron, N.; Quilici, S.; Carroll, S. The Economic Value of Vaccination: Why Prevention Is Wealth. J. Mark. Access Health Policy 2015, 3, 29284. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. In Review on Antimicrobial Resistance; Wellcome Trust: London, UK, 2014; pp. 1–16. [Google Scholar]
- O’Neill, J. Securing New Drugs for Future Generations: The Pipeline of Antibiotics. In Review on Antimicrobial Resistance; Wellcome Trust: London, UK, 2015; pp. 1–40. [Google Scholar]
- Miller, S.I. Antibiotic Resistance and Regulation of the Gram-Negative Bacterial Outer Membrane Barrier by Host Innate Immune Molecules. mBio 2016, 7, e01541-16. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.; Chen, H.; Li, N.; Wang, T.; Liang, W. The Spread of Antibiotic Resistance Genes In Vivo Model. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 3348695. [Google Scholar] [CrossRef] [PubMed]
- Dell’Annunziata, F.; Folliero, V.; Giugliano, R.; De Filippis, A.; Santarcangelo, C.; Izzo, V.; Daglia, M.; Galdiero, M.; Arciola, C.R.; Franci, G. Gene Transfer Potential of Outer Membrane Vesicles of Gram-Negative Bacteria. Int. J. Mol. Sci. 2021, 22, 5985. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Jeannot, K.; Xiao, Y.; Knapp, C.W. Editorial: Horizontal Gene Transfer Mediated Bacterial Antibiotic Resistance. Front. Microbiol. 2019, 10, 1933. [Google Scholar] [CrossRef]
- Reygaert, W.C. An Overview of the Antimicrobial Resistance Mechanisms of Bacteria. AIMS Microbiol. 2018, 4, 482–501. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular Mechanisms of Antibiotic Resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Bush, K.; Jacoby, G.A. Updated Functional Classification of β-Lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar] [CrossRef]
- Sawa, T.; Kooguchi, K.; Moriyama, K. Molecular Diversity of Extended-Spectrum β-Lactamases and Carbapenemases, and Antimicrobial Resistance. J. Intensive Care 2020, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside Modifying Enzymes. Drug Resist. Updates 2010, 13, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 464–473. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Gupta, V.; Pathania, R. Efflux Pump Inhibitors for Bacterial Pathogens: From Bench to Bedside. Indian J. Med. Res. 2019, 149, 129. [Google Scholar] [CrossRef] [PubMed]
- Rosas, N.C.; Lithgow, T. Targeting Bacterial Outer-Membrane Remodelling to Impact Antimicrobial Drug Resistance. Trends Microbiol. 2022, 30, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Leyton, D.L.; Belousoff, M.J.; Lithgow, T. The β-Barrel Assembly Machinery Complex. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2015; pp. 1–16. [Google Scholar]
- Anandan, A.; Vrielink, A. Structure and Function of Lipid A–Modifying Enzymes. Ann. N. Y. Acad. Sci. 2020, 1459, 19–37. [Google Scholar] [CrossRef]
- Doi, Y.; Arakawa, Y. 16S Ribosomal RNA Methylation: Emerging Resistance Mechanism against Aminoglycosides. Clin. Infect. Dis. 2007, 45, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.; Richmond, G.E.; Piddock, L.J. Multidrug Efflux Pumps in Gram-Negative Bacteria and Their Role in Antibiotic Resistance. Future Microbiol. 2014, 9, 1165–1177. [Google Scholar] [CrossRef]
- Lorusso, A.B.; Carrara, J.A.; Barroso, C.D.N.; Tuon, F.F.; Faoro, H. Role of Efflux Pumps on Antimicrobial Resistance in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2022, 23, 15779. [Google Scholar] [CrossRef]
- Amaral, L.; Martins, A.; Spengler, G.; Molnar, J. Efflux Pumps of Gram-Negative Bacteria: What They Do, How They Do It, with What and How to Deal with Them. Front. Pharmacol. 2014, 4, 168. [Google Scholar] [CrossRef] [PubMed]
- Auda, I.G.; Ali Salman, I.M.; Odah, J.G. Efflux Pumps of Gram-Negative Bacteria in Brief. Gene Rep. 2020, 20, 100666. [Google Scholar] [CrossRef]
- Mueller, M.T.C.R. Escherichia coli. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Reygaert, W.C. Antimicrobial Mechanisms of Escherichia coli. In Recent Advances on Physiology, Pathogenesis and Biotechnological Applications; InTech: Vienna, Austria, 2017. [Google Scholar]
- Galindo-Méndez, M. Antimicrobial Resistance in Escherichia coli. In E. coli Infections—Importance of Early Diagnosis and Efficient Treatment; IntechOpen: Rijeka, Croatia, 2020. [Google Scholar]
- Zhang, L.; Levy, K.; Trueba, G.; Cevallos, W.; Trostle, J.; Foxman, B.; Marrs, C.F.; Eisenberg, J.N.S. Effects of Selection Pressure and Genetic Association on the Relationship between Antibiotic Resistance and Virulence in Escherichia coli. Antimicrob. Agents Chemother. 2015, 59, 6733–6740. [Google Scholar] [CrossRef] [PubMed]
- Bradford, P.A. Extended-Spectrum β-Lactamases in the 21st Century: Characterization, Epidemiology, and Detection of This Important Resistance Threat. Clin. Microbiol. Rev. 2001, 14, 933–951. [Google Scholar] [CrossRef] [PubMed]
- Palzkill, T. Structural and Mechanistic Basis for Extended-Spectrum Drug-Resistance Mutations in Altering the Specificity of TEM, CTX-M, and KPC β-Lactamases. Front. Mol. Biosci. 2018, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, P.; Singh, N.S.; Virdi, J.S. Escherichia coli β-Lactamases: What Really Matters. Front. Microbiol. 2016, 7, 417. [Google Scholar] [CrossRef] [PubMed]
- Koga, V.L.; Maluta, R.P.; da Silveira, W.D.; Ribeiro, R.A.; Hungria, M.; Vespero, E.C.; Nakazato, G.; Kobayashi, R.K.T. Characterization of CMY-2-Type Beta-Lactamase-Producing Escherichia coli Isolated from Chicken Carcasses and Human Infection in a City of South Brazil. BMC Microbiol. 2019, 19, 174. [Google Scholar] [CrossRef]
- Shen, Z.; Hu, Y.; Sun, Q.; Hu, F.; Zhou, H.; Shu, L.; Ma, T.; Shen, Y.; Wang, Y.; Li, J.; et al. Emerging Carriage of NDM-5 and MCR-1 in Escherichia coli from Healthy People in Multiple Regions in China: A Cross Sectional Observational Study. eClinicalMedicine 2018, 6, 11–20. [Google Scholar] [CrossRef]
- Poirel, L.; Madec, J.-Y.; Lupo, A.; Schink, A.-K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia coli. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Zou, W.; Li, C.; Yang, X.; Wang, Y.; Cheng, G.; Zeng, J.; Zhang, X.; Chen, Y.; Cai, R.; Huang, Q.; et al. Frequency of Antimicrobial Resistance and Integron Gene Cassettes in Escherichia coli Isolated from Giant Pandas (Ailuropoda melanoleuca) in China. Microb. Pathog. 2018, 116, 173–179. [Google Scholar] [CrossRef]
- Zhang, W.-J.; Xu, X.-R.; Schwarz, S.; Wang, X.-M.; Dai, L.; Zheng, H.-J.; Liu, S. Characterization of the IncA/C Plasmid PSCEC2 from Escherichia coli of Swine Origin That Harbours the Multiresistance Gene cfr. J. Antimicrob. Chemother. 2014, 69, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Chetri, S.; Singha, M.; Bhowmik, D.; Nath, K.; Chanda, D.D.; Chakravarty, A.; Bhattacharjee, A. Transcriptional Response of OmpC and OmpF in Escherichia coli against Differential Gradient of Carbapenem Stress. BMC Res. Notes 2019, 12, 138. [Google Scholar] [CrossRef] [PubMed]
- Morè, N.; Martorana, A.M.; Biboy, J.; Otten, C.; Winkle, M.; Serrano, C.K.G.; Montón Silva, A.; Atkinson, L.; Yau, H.; Breukink, E.; et al. Peptidoglycan Remodeling Enables Escherichia coli to Survive Severe Outer Membrane Assembly Defect. mBio 2019, 10, e02729-18. [Google Scholar] [CrossRef] [PubMed]
- Tsang, M.-J.; Yakhnina, A.A.; Bernhardt, T.G. NlpD Links Cell Wall Remodeling and Outer Membrane Invagination during Cytokinesis in Escherichia coli. PLoS Genet. 2017, 13, e1006888. [Google Scholar] [CrossRef] [PubMed]
- Redgrave, L.S.; Sutton, S.B.; Webber, M.A.; Piddock, L.J.V. Fluoroquinolone Resistance: Mechanisms, Impact on Bacteria, and Role in Evolutionary Success. Trends Microbiol. 2014, 22, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.-T.; Liao, X.-P.; Yang, S.-S.; Wang, X.-M.; Li, L.-L.; Sun, J.; Yang, Y.-R.; Fang, L.-X.; Li, L.; Zhao, D.-H.; et al. Detection of Mutations in the gyrA and parC Genes in Escherichia coli Isolates Carrying Plasmid-Mediated Quinolone Resistance Genes from Diseased Food-Producing Animals. J. Med. Microbiol. 2012, 61, 1591–1599. [Google Scholar] [CrossRef] [PubMed]
- Johnning, A.; Kristiansson, E.; Fick, J.; Weijdegård, B.; Larsson, D.G.J. Resistance Mutations in gyrA and parC Are Common in Escherichia Communities of Both Fluoroquinolone-Polluted and Uncontaminated Aquatic Environments. Front. Microbiol. 2015, 6, 1355. [Google Scholar] [CrossRef] [PubMed]
- Al-Tamimi, M.; Abu-Raideh, J.; Albalawi, H.; Shalabi, M.; Saleh, S. Effective Oral Combination Treatment for Extended-Spectrum Beta-Lactamase-Producing Escherichia coli. Microb. Drug Resist. 2019, 25, 1132–1141. [Google Scholar] [CrossRef]
- Harris, P.N.A.; Yin, M.; Jureen, R.; Chew, J.; Ali, J.; Paynter, S.; Paterson, D.L.; Tambyah, P.A. Comparable Outcomes for β-Lactam/β-Lactamase Inhibitor Combinations and Carbapenems in Definitive Treatment of Bloodstream Infections Caused by Cefotaxime-Resistant Escherichia coli or Klebsiella pneumoniae. Antimicrob. Resist. Infect. Control 2015, 4, 14. [Google Scholar] [CrossRef]
- Arbab, S.; Ullah, H.; Wang, W.; Zhang, J. Antimicrobial Drug Resistance against Escherichia coli and Its Harmful Effect on Animal Health. Vet. Med. Sci. 2022, 8, 1780–1786. [Google Scholar] [CrossRef]
- Lee, C.-R.; Lee, J.H.; Park, M.; Park, K.S.; Bae, I.K.; Kim, Y.B.; Cha, C.-J.; Jeong, B.C.; Lee, S.H. Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front. Cell. Infect. Microbiol. 2017, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a Successful Pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef] [PubMed]
- Howard, A.; O’Donoghue, M.; Feeney, A.; Sleator, R.D. Acinetobacter baumannii . Virulence 2012, 3, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Maragakis, L.L.; Perl, T.M. Antimicrobial Resistance: Acinetobacter baumannii: Epidemiology, Antimicrobial Resistance, and Treatment Options. Clin. Infect. Dis. 2008, 46, 1254–1263. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, M.S.; Bonomo, R.A.; Tolmasky, M.E. Carbapenemases: Transforming Acinetobacter baumannii into a Yet More Dangerous Menace. Biomolecules 2020, 10, 720. [Google Scholar] [CrossRef] [PubMed]
- Traglia, G.M.; Quinn, B.; Schramm, S.T.J.; Soler-Bistue, A.; Ramirez, M.S. Serum Albumin and Ca2+ Are Natural Competence Inducers in the Human Pathogen Acinetobacter baumannii. Antimicrob. Agents Chemother. 2016, 60, 4920–4929. [Google Scholar] [CrossRef] [PubMed]
- Nikibakhsh, M.; Firoozeh, F.; Badmasti, F.; Kabir, K.; Zibaei, M. Molecular Study of Metallo-β-Lactamases and Integrons in Acinetobacter baumannii Isolates from Burn Patients. BMC Infect. Dis. 2021, 21, 782. [Google Scholar] [CrossRef] [PubMed]
- Alkasaby, N.M.; El Sayed Zaki, M. Molecular Study of Acinetobacter baumannii Isolates for Metallo-β-Lactamases and Extended-Spectrum-β-Lactamases Genes in Intensive Care Unit, Mansoura University Hospital, Egypt. Int. J. Microbiol. 2017, 2017, 3925868. [Google Scholar] [CrossRef]
- Colquhoun, J.M.; Farokhyfar, M.; Hutcheson, A.R.; Anderson, A.; Bethel, C.R.; Bonomo, R.A.; Clarke, A.J.; Rather, P.N. OXA-23 β-Lactamase Overexpression in Acinetobacter baumannii Drives Physiological Changes Resulting in New Genetic Vulnerabilities. mBio 2021, 12, e0313721. [Google Scholar] [CrossRef]
- Ribeiro, P.C.S.; Monteiro, A.S.; Marques, S.G.; Monteiro, S.G.; Monteiro-Neto, V.; Coqueiro, M.M.M.; Marques, A.C.G.; de Jesus Gomes Turri, R.; Santos, S.G.; Bomfim, M.R.Q. Phenotypic and Molecular Detection of the BlaKPC Gene in Clinical Isolates from Inpatients at Hospitals in São Luis, MA, Brazil. BMC Infect. Dis. 2016, 16, 737. [Google Scholar] [CrossRef]
- Tahbaz, S.V.; Azimi, L.; Lari, A.R. Characterization of Aminoglycoside Resistance Mechanisms in Acinetobacter baumannii Isolates from Burn Wound Colonization. Ann. Burn. Fire Disasters 2019, 32, 115–121. [Google Scholar]
- Garneau-Tsodikova, S.; Labby, K.J. Mechanisms of Resistance to Aminoglycoside Antibiotics: Overview and Perspectives. Medchemcomm 2016, 7, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Jouybari, M.A.; Ahanjan, M.; Mirzaei, B.; Goli, H.R. Role of Aminoglycoside-Modifying Enzymes and 16S RRNA Methylase (ArmA) in Resistance of Acinetobacter baumannii Clinical Isolates against Aminoglycosides. Rev. Soc. Bras. Med. Trop. 2021, 54, e05992020. [Google Scholar] [CrossRef]
- Abdi, S.N.; Ghotaslou, R.; Ganbarov, K.; Mobed, A.; Tanomand, A.; Yousefi, M.; Asgharzadeh, M.; Kafil, H.S. Acinetobacter baumannii- Efflux Pumps and Antibiotic Resistance. Infect. Drug Resist. 2020, 13, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.F.; Bilya, S.R.; Xu, W. AdeABC Efflux Gene in Acinetobacter baumannii. New Microbes New Infect. 2019, 30, 100549. [Google Scholar] [CrossRef] [PubMed]
- Coyne, S.; Courvalin, P.; Périchon, B. Efflux-Mediated Antibiotic Resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 2011, 55, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Dou, Q.; Zou, M.; Li, J.; Wang, H.; Hu, Y.; Liu, W. AdeABC Efflux Pump and Resistance of Acinetobacter baumannii against Carbapenem. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2017, 42, 426–433. [Google Scholar] [CrossRef]
- Uppalapati, S.R.; Sett, A.; Pathania, R. The Outer Membrane Proteins OmpA, CarO, and OprD of Acinetobacter baumannii Confer a Two-Pronged Defense in Facilitating Its Success as a Potent Human Pathogen. Front. Microbiol. 2020, 11, 589234. [Google Scholar] [CrossRef]
- Nie, D.; Hu, Y.; Chen, Z.; Li, M.; Hou, Z.; Luo, X.; Mao, X.; Xue, X. Outer Membrane Protein A (OmpA) as a Potential Therapeutic Target for Acinetobacter baumannii Infection. J. Biomed. Sci. 2020, 27, 26. [Google Scholar] [CrossRef]
- Sariyer, E. The Role of Acinetobacter baumannii CarO Outer Membrane Protein in Carbapenems Influx. Res. Microbiol. 2022, 173, 103966. [Google Scholar] [CrossRef]
- Royer, S.; de Campos, P.A.; Araújo, B.F.; Ferreira, M.L.; Gonçalves, I.R.; da Fonseca Batistão, D.W.; e Silva Brígido, R.T.; Cerdeira, L.T.; Machado, L.G.; de Brito, C.S.; et al. Molecular Characterization and Clonal Dynamics of Nosocomial BlaOXA-23 Producing XDR Acinetobacter baumannii. PLoS ONE 2018, 13, e0198643. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Trejo, A.; Ruiz-Ruiz, J.M.; Gonzalez-Avila, L.U.; Saldaña-Padilla, A.; Hernández-Cortez, C.; Loyola-Cruz, M.A.; Bello-López, J.M.; Castro-Escarpulli, G. Evasion of Antimicrobial Activity in Acinetobacter baumannii by Target Site Modifications: An Effective Resistance Mechanism. Int. J. Mol. Sci. 2022, 23, 6582. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ling, B.; Zhou, L. Prevalence of 16S RRNA Methylase, Modifying Enzyme, and Extended-Spectrum Beta-Lactamase Genes among Acinetobacter baumannii Isolates. J. Chemother. 2015, 27, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Trebosc, V.; Gartenmann, S.; Tötzl, M.; Lucchini, V.; Schellhorn, B.; Pieren, M.; Lociuro, S.; Gitzinger, M.; Tigges, M.; Bumann, D.; et al. Dissecting Colistin Resistance Mechanisms in Extensively Drug-Resistant Acinetobacter baumannii Clinical Isolates. mBio 2019, 10, e01083-19. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, M.; D’Andrea, M.M.; Pelegrin, A.C.; Perrot, N.; Mirande, C.; Blanc, B.; Legakis, N.; Goossens, H.; Rossolini, G.M.; van Belkum, A. Abundance of Colistin-Resistant, OXA-23- and ArmA-Producing Acinetobacter baumannii Belonging to International Clone 2 in Greece. Front. Microbiol. 2020, 11, 668. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhao, G.; Chao, X.; Xie, L.; Wang, H. The Characteristic of Virulence, Biofilm and Antibiotic Resistance of Klebsiella pneumoniae. Int. J. Environ. Res. Public Health 2020, 17, 6278. [Google Scholar] [CrossRef]
- Wyres, K.L.; Lam, M.M.C.; Holt, K.E. Population Genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020, 18, 344–359. [Google Scholar] [CrossRef] [PubMed]
- Choby, J.E.; Howard-Anderson, J.; Weiss, D.S. Hypervirulent Klebsiella pneumoniae—Clinical and Molecular Perspectives. J. Intern. Med. 2020, 287, 283–300. [Google Scholar] [CrossRef]
- Ashurst, J.V.; Dawson, A. Klebsiella pneumonia. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Xiang, T.; Chen, C.; Wen, J.; Liu, Y.; Zhang, Q.; Cheng, N.; Wu, X.; Zhang, W. Resistance of Klebsiella pneumoniae Strains Carrying BlaNDM–1 Gene and the Genetic Environment of BlaNDM–1. Front. Microbiol. 2020, 11, 700. [Google Scholar] [CrossRef]
- Abdul-Mutakabbir, J.C.; Kebriaei, R.; Jorgensen, S.C.J.; Rybak, M.J. Teaching an Old Class New Tricks: A Novel Semi-Synthetic Aminoglycoside, Plazomicin. Infect. Dis. Ther. 2019, 8, 155–170. [Google Scholar] [CrossRef]
- Moya, C.; Maicas, S. Antimicrobial Resistance in Klebsiella pneumoniae Strains: Mechanisms and Outbreaks. In Proceedings of the 1st International Electronic Conference on Microbiology, Online, 2–30 November 2020; MDPI: Basel, Switzerland, 2020; p. 11. [Google Scholar]
- Almaghrabi, R.; Clancy, C.J.; Doi, Y.; Hao, B.; Chen, L.; Shields, R.K.; Press, E.G.; Iovine, N.M.; Townsend, B.M.; Wagener, M.M.; et al. Carbapenem-Resistant Klebsiella pneumoniae Strains Exhibit Diversity in Aminoglycoside-Modifying Enzymes, Which Exert Differing Effects on Plazomicin and Other Agents. Antimicrob. Agents Chemother. 2014, 58, 4443–4451. [Google Scholar] [CrossRef] [PubMed]
- Latifi, B.; Tajbakhsh, S.; Ahadi, L.; Yousefi, F. Coexistence of Aminoglycoside Resistance Genes in CTX-M-Producing Isolates of Klebsiella pneumoniae in Bushehr Province, Iran. Iran. J. Microbiol. 2021, 13, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Morgan, C.E.; Bonomo, R.A.; Yu, E.W. Cryo-EM Structures of the Klebsiella pneumoniae AcrB Multidrug Efflux Pump. mBio 2023, 14, e0065923. [Google Scholar] [CrossRef]
- Li, J.; Zhang, H.; Ning, J.; Sajid, A.; Cheng, G.; Yuan, Z.; Hao, H. The Nature and Epidemiology of OqxAB, a Multidrug Efflux Pump. Antimicrob. Resist. Infect. Control 2019, 8, 44. [Google Scholar] [CrossRef] [PubMed]
- Rocker, A.; Lacey, J.A.; Belousoff, M.J.; Wilksch, J.J.; Strugnell, R.A.; Davies, M.R.; Lithgow, T. Global Trends in Proteome Remodeling of the Outer Membrane Modulate Antimicrobial Permeability in Klebsiella pneumoniae. mBio 2020, 11, e00603-20. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, H.; Ko, K.S. Reduced Virulence in Tigecycline-Resistant Klebsiella pneumoniae Caused by Overexpression of OmpR and down-Regulation of OmpK35. J. Biomed. Sci. 2023, 30, 22. [Google Scholar] [CrossRef] [PubMed]
- David, S.; Wong, J.L.C.; Sanchez-Garrido, J.; Kwong, H.-S.; Low, W.W.; Morecchiato, F.; Giani, T.; Rossolini, G.M.; Brett, S.J.; Clements, A.; et al. Widespread Emergence of OmpK36 Loop 3 Insertions among Multidrug-Resistant Clones of Klebsiella pneumoniae. PLoS Pathog. 2022, 18, e1010334. [Google Scholar] [CrossRef]
- Kidd, T.J.; Mills, G.; Sá-Pessoa, J.; Dumigan, A.; Frank, C.G.; Insua, J.L.; Ingram, R.; Hobley, L.; Bengoechea, J.A. A Klebsiella pneumoniae Antibiotic Resistance Mechanism That Subdues Host Defences and Promotes Virulence. EMBO Mol. Med. 2017, 9, 430–447. [Google Scholar] [CrossRef]
- Kot, B.; Piechota, M.; Szweda, P.; Mitrus, J.; Wicha, J.; Grużewska, A.; Witeska, M. Virulence Analysis and Antibiotic Resistance of Klebsiella pneumoniae Isolates from Hospitalised Patients in Poland. Sci. Rep. 2023, 13, 4448. [Google Scholar] [CrossRef]
- Wilson, M.G.; Pandey, S. Pseudomonas aeruginosa. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Qin, S.; Xiao, W.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, Virulence Factors, Antibiotic Resistance, Interaction with Host, Technology Advances and Emerging Therapeutics. Signal Transduct. Target. Ther. 2022, 7, 199. [Google Scholar] [CrossRef]
- Bassetti, M.; Vena, A.; Croxatto, A.; Righi, E.; Guery, B. How to Manage Pseudomonas aeruginosa Infections. Drugs Context 2018, 7, 212527. [Google Scholar] [CrossRef] [PubMed]
- Torrens, G.; Hernández, S.B.; Ayala, J.A.; Moya, B.; Juan, C.; Cava, F.; Oliver, A. Regulation of AmpC-Driven β-Lactam Resistance in Pseudomonas aeruginosa: Different Pathways, Different Signaling. mSystems 2019, 4, e00524-19. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, S.; Bouffartigues, E.; Bodilis, J.; Maillot, O.; Lesouhaitier, O.; Feuilloley, M.G.J.; Orange, N.; Dufour, A.; Cornelis, P. Structure, Function and Regulation of Pseudomonas aeruginosa Porins. FEMS Microbiol. Rev. 2017, 41, 698–722. [Google Scholar] [CrossRef] [PubMed]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef] [PubMed]
- Pachori, P.; Gothalwal, R.; Gandhi, P. Emergence of Antibiotic Resistance Pseudomonas aeruginosa in Intensive Care Unit; a Critical Review. Genes Dis. 2019, 6, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Ciofu, O.; Tolker-Nielsen, T. Tolerance and Resistance of Pseudomonas aeruginosa Biofilms to Antimicrobial Agents—How P. Aeruginosa Can Escape Antibiotics. Front. Microbiol. 2019, 10, 913. [Google Scholar] [CrossRef] [PubMed]
- Vetrivel, A.; Ramasamy, M.; Vetrivel, P.; Natchimuthu, S.; Arunachalam, S.; Kim, G.-S.; Murugesan, R. Pseudomonas aeruginosa Biofilm Formation and Its Control. Biologics 2021, 1, 312–336. [Google Scholar] [CrossRef]
- Wang, Z.; Koirala, B.; Hernandez, Y.; Zimmerman, M.; Park, S.; Perlin, D.S.; Brady, S.F. A Naturally Inspired Antibiotic to Target Multidrug-Resistant Pathogens. Nature 2022, 601, 606–611. [Google Scholar] [CrossRef]
- Liu, G.; Catacutan, D.B.; Rathod, K.; Swanson, K.; Jin, W.; Mohammed, J.C.; Chiappino-Pepe, A.; Syed, S.A.; Fragis, M.; Rachwalski, K.; et al. Deep Learning-Guided Discovery of an Antibiotic Targeting Acinetobacter baumannii. Nat. Chem. Biol. 2023, 19, 1342–1350. [Google Scholar] [CrossRef]
- Laws, M.; Shaaban, A.; Rahman, K.M. Antibiotic Resistance Breakers: Current Approaches and Future Directions. FEMS Microbiol. Rev. 2019, 43, 490–516. [Google Scholar] [CrossRef]
- Corbett, D.; Wise, A.; Langley, T.; Skinner, K.; Trimby, E.; Birchall, S.; Dorali, A.; Sandiford, S.; Williams, J.; Warn, P.; et al. Potentiation of Antibiotic Activity by a Novel Cationic Peptide: Potency and Spectrum of Activity of SPR741. Antimicrob. Agents Chemother. 2017, 61, e00200-17. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Garau, J. Current and Future Perspectives in the Treatment of Multidrug-Resistant Gram-Negative Infections. J. Antimicrob. Chemother. 2021, 76, iv23–iv37. [Google Scholar] [CrossRef] [PubMed]
| Beta-Lactamases | AMEs | Efflux Pumps | Altered Target Sites | OM Remodelling | |
|---|---|---|---|---|---|
| E. coli |
|
|
|
|
|
| A. baumannii |
|
|
|
|
|
| K. pneumoniae |
|
|
|
|
|
| P. aeruginosa |
|
|
|
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gauba, A.; Rahman, K.M. Evaluation of Antibiotic Resistance Mechanisms in Gram-Negative Bacteria. Antibiotics 2023, 12, 1590. https://doi.org/10.3390/antibiotics12111590
Gauba A, Rahman KM. Evaluation of Antibiotic Resistance Mechanisms in Gram-Negative Bacteria. Antibiotics. 2023; 12(11):1590. https://doi.org/10.3390/antibiotics12111590
Chicago/Turabian StyleGauba, Anusha, and Khondaker Miraz Rahman. 2023. "Evaluation of Antibiotic Resistance Mechanisms in Gram-Negative Bacteria" Antibiotics 12, no. 11: 1590. https://doi.org/10.3390/antibiotics12111590
APA StyleGauba, A., & Rahman, K. M. (2023). Evaluation of Antibiotic Resistance Mechanisms in Gram-Negative Bacteria. Antibiotics, 12(11), 1590. https://doi.org/10.3390/antibiotics12111590






