Antimicrobial Susceptibility and Resistance Genes in Streptococcus uberis Isolated from Bovine Mastitis in the Czech Republic
Abstract
:1. Introduction
2. Results
2.1. Antimicrobial Susceptibility Testing
2.2. Detection of AMR Genes
3. Discussion
4. Materials and Methods
4.1. Bacterial Sampling
4.2. Bacterial Isolation and Identification
4.3. Antimicrobial Susceptibility Testing
4.4. Detection of AMR Genes
4.4.1. Nucleic Acid Extraction
4.4.2. Whole-Genome Sequencing
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Halasa, T.; Huijps, K.; Østerås, O.; Hogeveen, H. Economic effects of bovine mastitis and mastitis management: A review. Vet. Q. 2007, 29, 18–31. [Google Scholar] [CrossRef]
- Bradley, A.J.; Leach, K.A.; Breen, J.E.; Green, L.E.; Gree, M.J. Survey of the incidence and aetiology of mastitis on dairy farms in England and Wales. Vet. Rec. 2007, 60, 253–257. [Google Scholar] [CrossRef]
- Vezina, B.; Al-harbi, H.; Ramay, H.R.; Soust, M.; Moore, R.J.; Olchowy, T.W.; Alawneh, J.I. Sequence characterisation and novel insights into bovine mastitis-associated Streptococcus uberis in dairy herds. Sci. Rep. 2021, 11, 3046. [Google Scholar] [CrossRef]
- Zouharova, M.; Nedbalcova, K.; Slama, P.; Bzdil, J.; Masarikova, M.; Matiasovic, J. Occurrence of virulence associated genes in Streptococcus uberis and Streptococcus parauberis isolated from bovine mastitis. Vet. Med.-Czech. 2022, 67, 123–130. [Google Scholar] [CrossRef]
- Saini, V.; McClure, J.T.; Léger, D.; Dufour, S.; Sheldon, A.G.; Scholl, D.T.; Barkema, H.W. Antimicrobial use on Canadian dairy farms. J. Dairy Sci. 2012, 95, 1209–1221. [Google Scholar] [CrossRef]
- Boonyayatra, S. Treatment of bovine mastitis during lactating period. Chiang Mai Vet. J. 2012, 10, 87–107. [Google Scholar]
- Bolte, J.; Zhang, Y.; Wente, N.; Krömker, V. In Vitro Susceptibility of Mastitis Pathogens Isolated from Clinical Mastitis Cases on Northern German Dairy Farms. Vet. Sci. 2020, 7, 10. [Google Scholar] [CrossRef]
- Martins, L.; Gonçalves, J.L.; Leite, R.F.; Tomazi, T.; Rall, V.L.M.; Santos, M.V. Association between antimicrobial use and antimicrobial resistance of Streptococcus uberis causing clinical mastitis. J. Dairy Sci. 2021, 104, 12030–12041. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Niu, G.; Boonyayatra, S.; Pichpol, D. Antimicrobial Resistance Profiles and Genes in Streptococcus uberis Associated With Bovine Mastitis in Thailand. Front. Vet. Sci. 2021, 8, 705338. [Google Scholar] [CrossRef]
- Vezina, B.; Rosa, M.N.; Canu, A.; Tola, S. Genomic surveillance reveals antibiotic resistance gene transmission via phage recombinases within sheep mastitis-associated Streptococcus uberis. BMC Vet. Res. 2022, 18, 264. [Google Scholar] [CrossRef]
- de Jong, A.; Garch, F.E.; Simjee, S.; Moyaert, H.; Rose, M.; Youala, M.; Siegwart, E. VetPath Study Group. Monitoring of antimicrobial susceptibility of udder pathogens recovered from cases of clinical mastitis in dairy cows across Europe: VetPath results. Vet. Microbiol. 2018, 213, 73–81. [Google Scholar] [CrossRef]
- Constable, P.D.; Morin, D.E. Treatment of clinical mastitis: Using antimicrobial susceptibility profiles for treatment decisions. Vet. Clin. N. Am. Food Anim. Pract. 2003, 19, 139–155. [Google Scholar] [CrossRef]
- Pratova, H.; Pokludova, L.; Dubska, M.; Bures, J. Prevalence rezistence k antimikrobikům u vybraných původců mastitidy skotu. Veterinářství 2021, 71, 20–31. (In Czech) [Google Scholar]
- Nedbalcova, K.; Nechvatalova, K.; Pokludova, L.; Bures, J.; Kucerova, Z.; Koutecka, L.; Hera, A. Resistance to selected beta-lactam antibiotics. Vet. Microbiol. 2014, 171, 328–336. [Google Scholar] [CrossRef]
- Davies, P.L.; Leigh, J.A.; Bradley, A.J.; Archer, S.C.; Emes, R.D.; Green, M.J. Molecular Epidemiology of Streptococcus uberis Clinical Mastitis in Dairy Herds: Strain Heterogeneity and Transmission. J. Clin. Microbiol. 2016, 54, 68–74. [Google Scholar] [CrossRef]
- Pokludova, L.; Maxova, L.; Maskova, Z.; Novotna, P.; Chumchalova, J.; Bures, J. Léčiva používaná k prevenci a terapii mastitid–přehled, trendy spotřeb a důraz na zodpovědnější přístup k antimikrobikům. Medicines used to prevent and treat mastitis–overview, consumption trends and emphasis on a more responsible approach to antimicrobials. Veterinářství 2021, 71, 82–93. (In Czech) [Google Scholar]
- Käppeli, N.; Morach, M.; Zurfluh, K.; Corti, S.; Nüesch-Inderbinen, M.; Stephan, R. Sequence Types and Antimicrobial Resistance Profiles of Streptococcus uberis Isolated from Bovine Mastitis. Front. Vet. Sci. 2019, 16, 234. [Google Scholar] [CrossRef]
- Monistero, V.; Barberio, A.; Cremonesi, P.; Castiglioni, B.; Morandi, S.; Lassen, D.C.K.; Astrup, L.B.; Locatelli, C.; Piccinini, R.; Addis, M.F.; et al. Genotyping and Antimicrobial Susceptibility Profiling of Streptococcus uberis Isolated from a Clinical Bovine Mastitis Outbreak in a Dairy Farm. Antibiotics 2021, 10, 644. [Google Scholar] [CrossRef]
- Haenni, M.; Lupo, A.; Madec, J. Antimicrobial resistance in Streptococcus spp. Microbiol. Spectr. 2018, 6, 1–25. [Google Scholar] [CrossRef]
- Haenni, M.; Galofaro, L.; Ythier, M.; Giddey, M.; Majcherczyk, P.; Moreillon, P.; Madec, J.Y. Penicillin-binding protein gene alterations in Streptococcus uberis isolates presenting decreased susceptibility to penicillin. Antimicrob. Agents Chemother. 2010, 54, 1140–1145. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, F.; Li, X.P.; Luo, J.Y.; Wang, L.; Zhou, Y.L.; Yan, Y.; Wang, X.R.; Li, H.S. Detection of antimicrobial resistence and virulence-related genes in Streptococcus uberis and Streptococcus parauberis isolated from clinical bovine mastitis cases in northwestern China. J. Integr. Agric. 2020, 19, 2784–2791. [Google Scholar] [CrossRef]
- Tomazi, T.; Freu, G.; Alves, B.G.; de Souza Filho, A.F.; Heinemann, M.B.; Veiga Dos Santos, M. Genotyping and antimicrobial resistance of Streptococcus uberis isolated from bovine clinical mastitis. PLoS ONE 2019, 14, e0223719. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Aziz, N.K.; Ammar, A.M.; El Damaty, H.M.; Abd Elkader, R.A.; Saad, H.A.; El-Kazzaz, W.; Khalifa, E. Environmental Streptococcus uberis Associated with Clinical Mastitis in Dairy Cows: Virulence Traits, Antimicrobial and Biocide Resistance, and Epidemiological Typing. Animals 2021, 11, 1849. [Google Scholar] [CrossRef] [PubMed]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12.0. 2022. Available online: http://www.eucast.org (accessed on 5 May 2023).
- Haenni, M.; Saras, E.; Chaussière, S.; Treilles, M.; Madec, J.Y. ermB mediated erythromycin resistance in Streptococcus uberis from bovine mastitis. Vet. J. 2011, 189, 356–358. [Google Scholar] [CrossRef]
- Cheng, J.; Qu, W.; Barkema, H.W.; Nobrega, D.B.; Gao, J.; Liu, G.; De Buck, J.; Kastelic, J.P.; Sun, H.; Han, B. Antimicrobial resistance profiles of 5 common bovine mastitis pathogens in large Chinese dairy herds. J. Dairy Sci. 2019, 102, 2416–2426. [Google Scholar] [CrossRef]
- Minst, K.; Märtlbauer, E.; Miller, T.; Meyer, C. Short communication: Streptococcus species isolated from mastitis milk samples in Germany and their resistance to antimicrobial agents. J. Dairy Sci. 2012, 95, 6957–6962. [Google Scholar] [CrossRef]
- Cameron, M.; Saab, M.; Heider, L.; McClure, J.T.; Rodriguez-Lecompte, J.C.; Sanchez, J. Antimicrobial susceptibility patterns of environmental streptococci recovered from bovine milk samples in the Maritime provinces of Canada. Front. Vet. Sci. 2016, 3, 79. [Google Scholar] [CrossRef]
- Gruet, P.; Maincent, P.; Berthelot, X.; Kaltsatos, V. Bovine mastitis and intramammary drug delivery: Review and perspectives. Adv. Drug Deliv. Rev. 2001, 50, 245–259. [Google Scholar] [CrossRef]
- Persson, Y.; Nyman, A.K.J.; Grönlund-Andersson, U. Etiology and antimicrobial susceptibility of udder pathogens from cases of subclinical mastitis in dairy cows in Sweden. Acta Vet. Scand. 2011, 53, 36. [Google Scholar] [CrossRef]
- Tian, X.Y.; Zheng, N.; Han, R.W.; Ho, H.; Wang, J.; Wang, Y.T.; Wang, S.Q.; Li, H.G.; Liu, H.W.; Yu, Z.N. Antimicrobial resistence and virulence genes of Streptococcus isolated from dairy cows with mastitis in China. Microb. Pathog. 2019, 131, 33–39. [Google Scholar] [CrossRef]
- Stasiak, M.; Maćkiw, E.; Kowalska, J.; Kucharek, K.; Postupolski, J. Silent Genes: Antimicrobial Resistance and Antibiotic Production. Pol. J. Microbiol. 2021, 70, 421–429. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 5th ed.; CLSI supplement VET01S; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- CA-SFM-VET. Comité de l’Antibiogramme de la Société Francaise de Microbiologie—Recommandations Vétérinaries; Société Francaise de Microbiologie: Paris, France, 2021; p. 15. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals: Second Informational Supplement, 4th ed.; CLSI Document VET01-S2; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2013; p. 70. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Quijada, N.M.; Rodríguez-Lázaro, D.; Eiros, J.M.; Hernández, M. TORMES: An automated pipeline for whole bacterial genome analysis. Bioinformatics 2019, 35, 4207–4212. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Tange, O. GNU Parallel-The Command-Line Power Tool. USENIX Mag. 2011, 36, 42–47. [Google Scholar]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Seemann, T. ABRicate Github. Available online: https://github.com/tseemann/abricate (accessed on 8 June 2021).

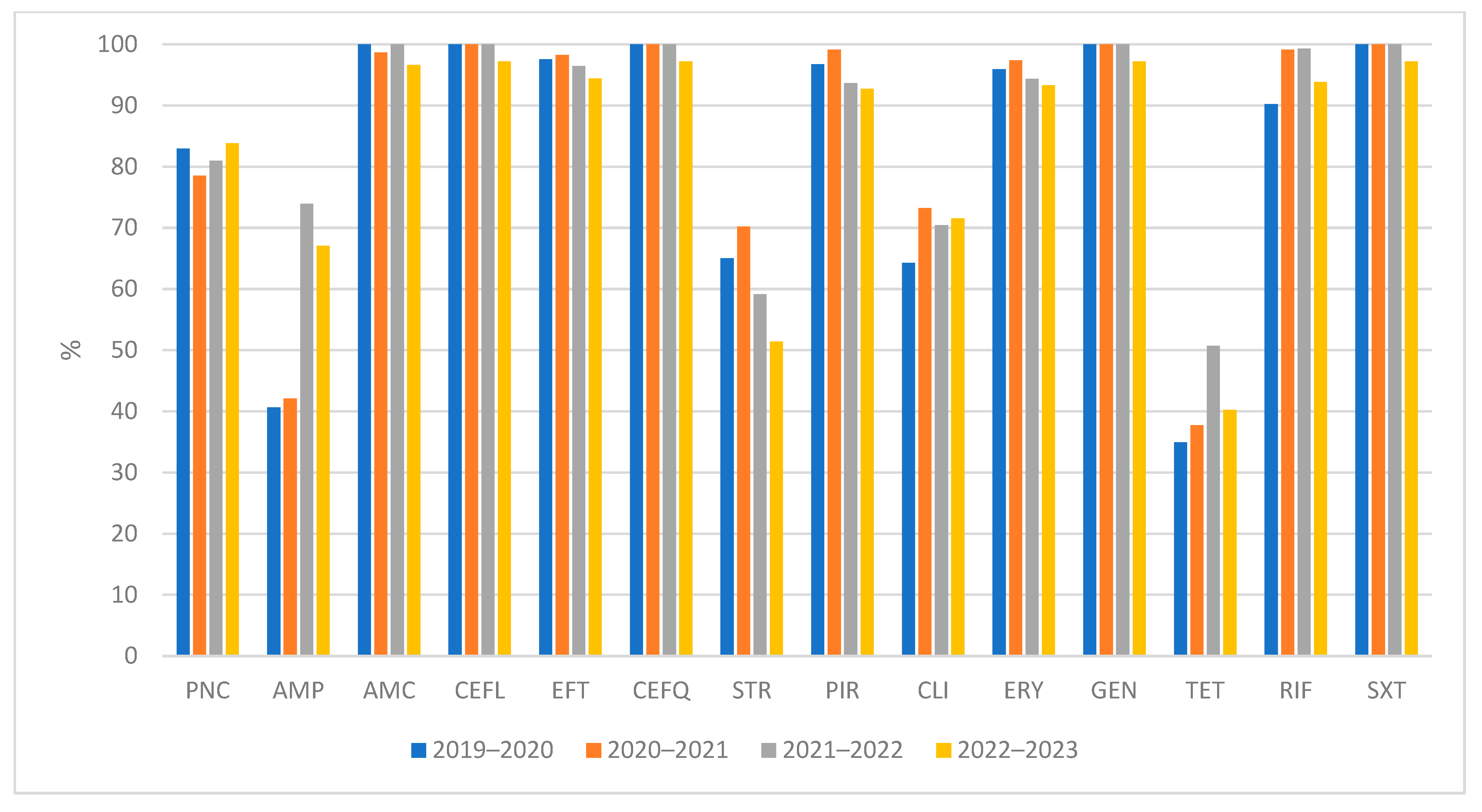
| MIC (mg/L) | R | MIC50 | MIC90 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.03 | 0.06 | 0.13 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | 1024 | (%) | (mg/L) | (mg/L) | |
| May 2019–April 2020: n = 123 (from 72 farms) | |||||||||||||||||||
| PNC | 27 | 12 | 63 | 21 | 0 | 0.25 | 0.5 | ||||||||||||
| AMP | 11 | 17 | 22 | 73 | 0 | 1 | 1 | ||||||||||||
| AMC | 24 | 4 | 6 | 83 | 6 | 0 | 0.25 | 0.25 | |||||||||||
| CEFL | 123 | 0 | ≤2 | ≤2 | |||||||||||||||
| CEF | 103 | 17 | 3 | 0 | ≤1 | 2 | |||||||||||||
| CEFQ | 123 | 0 | ≤0.5 | ≤0.5 | |||||||||||||||
| STR | 15 | 29 | 36 | 2 | 41 | 35 | 256 | >512 | |||||||||||
| PIR | 80 | 25 | 14 | 3 | 1 | 3 | ≤0.5 | 2 | |||||||||||
| CLI | 76 | 1 | 2 | 1 | 15 | 27 | 1 | 36 | ≤0.06 | 4 | |||||||||
| ERY | 117 | 1 | 1 | 1 | 2 | 1 | 3 | ≤0.06 | ≤0.06 | ||||||||||
| GEN | 6 | 19 | 23 | 75 | 0 | 32 | 32 | ||||||||||||
| TET | 42 | 1 | 1 | 15 | 28 | 36 | 65 | 32 | >32 | ||||||||||
| RIF | 24 | 87 | 10 | 2 | 2 | 0.06 | 0.06 | ||||||||||||
| SXT | 123 | 0 | ≤0.125 | ≤0.125 | |||||||||||||||
| May 2020–April 2021: n= 228 (from 119 farms) | |||||||||||||||||||
| PNC | 45 | 134 | 48 | 1 | 0 | 0.25 | 0.5 | ||||||||||||
| AMP | 15 | 31 | 50 | 124 | 8 | 0 | 1 | 1 | |||||||||||
| AMC | 23 | 23 | 7 | 106 | 66 | 3 | 0 | 0.25 | 0.5 | ||||||||||
| CEFL | 225 | 3 | 0 | ≤2 | ≤2 | ||||||||||||||
| CEF | 146 | 78 | 4 | 0 | ≤1 | 2 | |||||||||||||
| CEFQ | 225 | 3 | 0 | ≤0.5 | ≤0.5 | ||||||||||||||
| STR | 48 | 71 | 41 | 15 | 53 | 29 | 128 | >512 | |||||||||||
| PIR | 173 | 30 | 23 | 1 | 1 | 1 | ≤0.5 | 2 | |||||||||||
| CLI | 164 | 3 | 1 | 15 | 43 | 2 | 27 | ≤0.06 | 4 | ||||||||||
| ERY | 221 | 1 | 1 | 1 | 1 | 1 | 2 | 3 | ≤0.06 | ≤0.06 | |||||||||
| GEN | 11 | 18 | 57 | 138 | 4 | 0 | 32 | 32 | |||||||||||
| TET | 84 | 1 | 1 | 1 | 21 | 50 | 70 | 62 | 32 | >32 | |||||||||
| RIF | 9 | 178 | 39 | 2 | 1 | 0.06 | 0.125 | ||||||||||||
| SXT | 227 | 1 | 0 | ≤0.125 | ≤0.125 | ||||||||||||||
| May 2021–April 2022: n = 142 (from 75 farms) | |||||||||||||||||||
| PNC | 27 | 11 | 77 | 27 | 0 | 0.25 | 0.5 | ||||||||||||
| AMP | 18 | 10 | 60 | 17 | 36 | 1 | 0 | 0.25 | 1 | ||||||||||
| AMC | 20 | 7 | 10 | 88 | 17 | 0 | 0.25 | 0.5 | |||||||||||
| CEFL | 141 | 1 | 0 | ≤2 | ≤2 | ||||||||||||||
| EFT | 111 | 26 | 5 | 0 | ≤1 | 2 | |||||||||||||
| CEFQ | 140 | 2 | 0 | ≤0.5 | ≤0.5 | ||||||||||||||
| STR | 21 | 27 | 36 | 22 | 36 | 41 | 256 | >512 | |||||||||||
| PIR | 104 | 18 | 11 | 7 | 2 | 6 | ≤0.5 | 2 | |||||||||||
| CLI | 95 | 4 | 1 | 1 | 13 | 19 | 7 | 2 | 30 | ≤0.06 | 4 | ||||||||
| ERY | 126 | 8 | 4 | 1 | 1 | 2 | 6 | ≤0.06 | 0.125 | ||||||||||
| GEN | 7 | 11 | 48 | 62 | 7 | 7 | 0 | 32 | 32 | ||||||||||
| TET | 72 | 1 | 1 | 16 | 26 | 26 | 49 | ≤0.25 | >32 | ||||||||||
| RIF | 26 | 92 | 23 | 1 | 1 | 0.06 | 0.125 | ||||||||||||
| SXT | 142 | 0 | ≤0.125 | ≤0.125 | |||||||||||||||
| May 2022–April 2023: n= 174 (from 94 farms) | |||||||||||||||||||
| PNC | 39 | 9 | 102 | 23 | 1 | 0 | 0.25 | 0.5 | |||||||||||
| AMP | 14 | 13 | 60 | 33 | 52 | 2 | 0 | 0.25 | 1 | ||||||||||
| AMC | 36 | 4 | 34 | 87 | 12 | 1 | 0 | 0.25 | 0.25 | ||||||||||
| CEFL | 173 | 1 | 0 | ≤2 | ≤2 | ||||||||||||||
| EFT | 129 | 40 | 5 | 0 | ≤1 | 2 | |||||||||||||
| CEFQ | 171 | 3 | 0 | ≤0.5 | ≤0.5 | ||||||||||||||
| STR | 53 | 11 | 28 | 6 | 76 | 46 | 256 | >512 | |||||||||||
| PIR | 130 | 21 | 15 | 4 | 4 | 4 | ≤0.5 | 2 | |||||||||||
| CLI | 124 | 3 | 1 | 14 | 27 | 2 | 3 | 26 | ≤0.06 | 4 | |||||||||
| ERY | 165 | 2 | 1 | 6 | 4 | ≤0.06 | ≤0.06 | ||||||||||||
| GEN | 6 | 28 | 52 | 74 | 14 | 0 | 32 | 32 | |||||||||||
| TET | 70 | 2 | 2 | 27 | 31 | 42 | 57 | 16 | >32 | ||||||||||
| RIF | 43 | 93 | 32 | 1 | 5 | 3 | 0.06 | 0.125 | |||||||||||
| SXT | 173 | 1 | 0 | ≤0.125 | ≤0.125 | ||||||||||||||
| May 2019–April 2023: n = 667 (from 216 farms) | |||||||||||||||||||
| PNC | 138 | 32 | 376 | 119 | 1 | 1 | 0 | 0 | 0.25 | 0.5 | |||||||||
| AMP | 32 | 49 | 168 | 122 | 285 | 11 | 0 | 0 | 0.5 | 1 | |||||||||
| AMC | 103 | 38 | 57 | 364 | 101 | 4 | 0 | 0 | 0.25 | 0.5 | |||||||||
| CEFL | 662 | 4 | 1 | 0 | 0 | ≤2 | ≤2 | ||||||||||||
| EFT | 489 | 161 | 17 | 0 | 0 | ≤1 | 2 | ||||||||||||
| CEFQ | 659 | 8 | 0 | ≤0.5 | ≤0.5 | ||||||||||||||
| STR | 137 | 138 | 141 | 45 | 206 | 38 | 256 | >512 | |||||||||||
| PIR | 487 | 94 | 63 | 15 | 8 | 3 | ≤0.5 | 2 | |||||||||||
| CLI | 459 | 11 | 4 | 0 | 3 | 57 | 116 | 9 | 8 | 29 | ≤0.06 | 4 | |||||||
| ERY | 629 | 11 | 1 | 2 | 6 | 3 | 4 | 1 | 10 | 4 | ≤0.06 | ≤0.06 | |||||||
| GEN | 30 | 76 | 180 | 349 | 25 | 7 | 0 | 32 | 32 | ||||||||||
| TET | 268 | 1 | 4 | 0 | 1 | 5 | 79 | 135 | 174 | 59 | 16 | >32 | |||||||
| RIF | 102 | 450 | 104 | 1 | 0 | 10 | 1 | 0.06 | 0.125 | ||||||||||
| SXT | 665 | 2 | 0 | ≤0.125 | ≤0.125 | ||||||||||||||
| No. of Antimicrobial Groups | Phenotype Profile of Resistance | Percentage of Resistant Isolates |
|---|---|---|
| 0 | susceptible | 28.8 |
| 1 | TET | 31.3 |
| 1 | STR | 4.3 |
| 1 | PIR | 0.3 |
| 1 | CLI | 0.6 |
| 1 | ERY | 0.7 |
| 2 | STR, CLI | 4.8 |
| 2 | CLI, TET | 1.0 |
| 2 | STR, TET | 5.1 |
| 2 | ERY, TET | 1.2 |
| 2 | TET, RIF | 0.9 |
| 2 | STR, PIR, CLI | 1.3 |
| 3 | STR, CLI, TET | 16.6 |
| 3 | STR, TET, RIF | 0.1 |
| 3 | STR, CLI, RIF | 0.1 |
| 3 | STR, ERY, TET | 0.3 |
| 3 | CLI, ERY, TET | 0.1 |
| 3 | STR, PIR, CLI, TET | 0.6 |
| 3 | PIR, CLI, ERY, TET | 0.3 |
| 4 | STR, CLI, ERY, TET | 0.1 |
| 4 | STR, CLI, TET, RIF | 0.1 |
| 4 | STR, PIR, CLI, ERY, TET | 0.7 |
| 4 | STR, PIR, CLI, TET, RIF | 0.1 |
| Multi-resistant isolates | 19.5 | |
| Substance | Genes Detected | No. of Isolates with AMR Gene | No. of Isolates Phenotypic Resistant | Gene+/Phen− | Gene−/Phen+ |
|---|---|---|---|---|---|
| streptomycin | ant(6)-Ia | 76 | 48 | 28 | 0 |
| clindamycin | lnu(B) + lsa(E); erm(B) | 47 | 43 | 4 | 0 |
| pirlimycin | lnu(B) + lsa(E); erm(B) | 9 | 9 | 0 | 0 |
| tetracyklin | tet(M); tet(L); tet(O); tet(S) | 81 | 80 | 1 | 0 |
| erythromycin | erm(B) | 6 | 9 | 0 | 3 |
| rifaximin | - | 0 | 2 | 0 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zouharova, M.; Nedbalcova, K.; Matiaskova, K.; Slama, P.; Matiasovic, J. Antimicrobial Susceptibility and Resistance Genes in Streptococcus uberis Isolated from Bovine Mastitis in the Czech Republic. Antibiotics 2023, 12, 1527. https://doi.org/10.3390/antibiotics12101527
Zouharova M, Nedbalcova K, Matiaskova K, Slama P, Matiasovic J. Antimicrobial Susceptibility and Resistance Genes in Streptococcus uberis Isolated from Bovine Mastitis in the Czech Republic. Antibiotics. 2023; 12(10):1527. https://doi.org/10.3390/antibiotics12101527
Chicago/Turabian StyleZouharova, Monika, Katerina Nedbalcova, Katarina Matiaskova, Petr Slama, and Jan Matiasovic. 2023. "Antimicrobial Susceptibility and Resistance Genes in Streptococcus uberis Isolated from Bovine Mastitis in the Czech Republic" Antibiotics 12, no. 10: 1527. https://doi.org/10.3390/antibiotics12101527
APA StyleZouharova, M., Nedbalcova, K., Matiaskova, K., Slama, P., & Matiasovic, J. (2023). Antimicrobial Susceptibility and Resistance Genes in Streptococcus uberis Isolated from Bovine Mastitis in the Czech Republic. Antibiotics, 12(10), 1527. https://doi.org/10.3390/antibiotics12101527







