Occurrence, Typing, and Resistance Genes of ESBL/AmpC-Producing Enterobacterales in Fresh Vegetables Purchased in Central Israel
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Study Design and Sample
4.2. Laboratory Methods
4.2.1. Specimen Processing
4.2.2. Disk Diffusion
4.2.3. FT-IR Typing
4.2.4. WGS and Bioinformatics Analysis
4.2.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Livermore, D.M.; Woodford, N. The β-lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. Trends Microbiol. 2006, 14, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Biggest Threats and Data: 2019 AR Threats Report; CDC: Atlanta, GA, USA, 2019.
- Namikawa, H.; Imoto, W.; Yamada, K.; Tochino, Y.; Kaneko, Y.; Kakeya, H.; Shuto, T. Predictors of mortality from extended-spectrum beta-lactamase-producing Enterobacteriaceae bacteremia. Emerg. Microbes Infect. 2023, 12, 2217951. [Google Scholar] [CrossRef] [PubMed]
- Tansarli, G.S.; Karageorgopoulos, D.E.; Kapaskelis, A.; Falagas, M.E. Impact of antimicrobial multidrug resistance on inpatient care cost: An evaluation of the evidence. Expert Rev. Anti-Infect. Ther. 2013, 11, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Tamma, P.D.; Girdwood, S.C.T.; Gopaul, R.; Tekle, T.; Roberts, A.A.; Harris, A.D.; Cosgrove, S.E.; Carroll, K.C. The use of cefepime for treating AmpC β-lactamase-oroducing Enterobacteriaceae. Clin. Infect. Dis. 2013, 57, 781–788. [Google Scholar] [CrossRef]
- Castanheira, M.; Simner, P.; Bradford, P. Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC-Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef]
- Wang, S.; Xie, H.; Chen, Y.; Liu, L.; Fang, M.; Sun, D.; Xu, L.; Bi, Z.; Sun, G.; Li, Y.; et al. Intestinal colonization with ESBL-producing Klebsiella pneumoniae in healthy rural villager: A genomic surveillance study in China, 2015-2017. Front. Public Health 2022, 10, 1017050. [Google Scholar] [CrossRef]
- Villa, L.; García-Fernández, A.; Fortini, D.; Carattoli, A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 2010, 65, 2518–2529. [Google Scholar] [CrossRef]
- Jacoby, G.A. AmpC b-lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef]
- Paumier, A.; Asquier-Khati, A.; Thibaut, S.; Coeffic, T.; Lemenand, O.; Larramendy, S.; Leclère, B.; Caillon, J.; Boutoille, D.; Birgand, G. Assessment of factors associated with community-acquired extended-spectrum β-lactamase–producing Escherichia coli urinary tract infections in France. JAMA Netw. Open 2022, 5, e2232679. [Google Scholar] [CrossRef]
- Hölzel, C.S.; Tetens, J.L.; Schwaiger, K. Unraveling the role of vegetables in spreading antimicrobial-resistant bacteria: A need for quantitative risk assessment. Foodborne Pathog. Dis. 2018, 15, 671–688. [Google Scholar] [CrossRef]
- Rahman, M.; Alam, M.U.; Luies, S.K.; Kamal, A.; Ferdous, S.; Lin, A.; Sharior, F.; Khan, R.; Rahman, Z.; Parvez, S.M.; et al. Contamination of fresh produce with antibiotic-resistant bacteria and associated risks to human health: A scoping review. Int. J. Environ. Res. Public Health 2022, 19, 360. [Google Scholar] [CrossRef] [PubMed]
- Feldman, S.F.; Temkin, E.; Wullfhart, L.; Nutman, A.; Schechner, V.; Shitrit, P.; Shvartz, R.; Schwaber, M.J.; Andremont, A.; Carmeli, Y. A nationwide population-based study of Escherichia coli bloodstream infections: Incidence, antimicrobial resistance and mortality. Clin. Microbiol. Infect. 2022, 28, 879.E1–879.E7. [Google Scholar] [CrossRef] [PubMed]
- Lopes, R.; Fuentes-Castillo, D.; Fontana, H.; Rodrigues, L.; Dantas, K.; Cerdeira, L.; Henriques, I.; Lincopan, N. Endophytic lifestyle of global clones of extended-spectrum β-lactamase-producing priority pathogens in fresh vegetables: A trojan horse strategy favoring human colonization? mSystems 2021, 6, e01125-20. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.H.; Udaondo, Z.; Abram, K.Z.; Li, X.; Yang, X.; DiCaprio, E.L.; Jun, S.R.; Huang, E. Isolation of AmpC- and extended spectrum β-lactamase-producing Enterobacterales from fresh vegetables in the United States. Food Control 2022, 132, 108559. [Google Scholar] [CrossRef] [PubMed]
- Huizinga, P.; Schrauwen, E.; García-Cobos, S.; Willemsen, I.; Verhulst, C.; Friedrich, A.W.; Savelkoul, P.H.M.; Rossen, J.W.; Kluytmans, J. Extended-spectrum beta-lactamase producing Enterobacteriaceae (ESBL-E) isolated from bean sprouts in the Netherlands. PLoS ONE 2018, 13, e0203338. [Google Scholar] [CrossRef]
- van Hoek, A.H.A.M.; Veenman, C.; van Overbeek, W.M.; Lynch, G.; de Roda Husman, A.M.; Blaak, H. Prevalence and characterization of ESBL- and AmpC-producing Enterobacteriaceae on retail vegetables. Int. J. Food Microbiol. 2015, 204, 1–8. [Google Scholar] [CrossRef]
- Zurfluh, K.; Nüesch-Inderbinen, M.; Morach, M.; Berner, A.Z.; Hächler, H.; Stephan, R. Extended-spectrum-β-lactamase-producing Enterobacteriaceae isolated from vegetables imported from the Dominican Republic, India, Thailand, and Vietnam. Appl. Environ. Microbiol. 2015, 81, 3115–3120. [Google Scholar] [CrossRef]
- Pintor-Cora, A.; Álvaro-Llorente, L.; Otero, A.; Rodríguez-Calleja, J.M.; Santos, J.A. Extended-spectrum beta-lactamase-producing Enterobacteriaceae in fresh produce. Foods 2021, 10, 2609. [Google Scholar] [CrossRef]
- Colosi, I.A.; Baciu, A.M.; Opriș, R.V.; Peca, L.; Gudat, T.; Simon, L.M.; Colosi, H.A.; Costache, C. Prevalence of ESBL, AmpC and carbapenemase-producing Enterobacterales isolated from raw vegetables retailed in Romania. Foods 2020, 9, 1726. [Google Scholar] [CrossRef]
- Al-Kharousi, Z.S.; Guizani, N.; Al-Sadi, A.M.; Al-Bulushi, I.M. Antibiotic resistance of Enterobacteriaceae isolated from fresh fruits and vegetables and characterization of their AmpC b-lactamases. J. Food Prot. 2019, 82, 1857–1863. [Google Scholar] [CrossRef]
- Israel Fruits and Vegetables Maket Size & Share Analysis-Growth Trends & Forecasts (2023–2028). 2023. Available online: https://www.mordorintelligence.com/industry-reports/israel-fruits-and-vegetables-market (accessed on 7 August 2023).
- Chelaghma, W.; Loucif, L.; Bendahou, M.; Rolain, J.M. Vegetables and fruit as a reservoir of β-lactam and colistin-resistant Gram-negative bacteria: A review. Microorganisms 2021, 9, 2534. [Google Scholar] [CrossRef] [PubMed]
- National Water Reuse Action Plan. Delegation to Isreal-Summary Report; WRAP: Alexandria, VA, USA, 2022. [Google Scholar]
- Cravo Oliveira, T.; Padget, M. Chapter 3. OECD Trends in Antimicrobial Resistance in Countries. 2023. Available online: https://www.oecd-ilibrary.org/sites/9789264307599-6-en/index.html?itemId=/content/component/9789264307599-6-en#wrapper (accessed on 15 August 2023).
- Song, J.; Oh, S.S.; Kim, J.; Shin, J. Extended-spectrum β-lactamase-producing Escherichia coli isolated from raw vegetables in South Korea. Sci. Rep. 2020, 10, 19721. [Google Scholar] [CrossRef] [PubMed]
- Sapkota, S.; Adhikari, S.; Khadka, S.; Adhikari, M.; Kandel, H.; Pathak, S.; Pandey, A.; Pandey, A. multi-drug resistant extended-spectrum beta-lactamase producing E. coli and Salmonella on raw vegetable salads served at hotels and restaurants in Bharatpur, Nepal. BMC Res. Notes 2019, 12, 516. [Google Scholar] [CrossRef]
- Randall, L.P.; Lodge, M.P.; Elviss, N.C.; Lemma, F.L.; Hopkins, K.L.; Teale, C.J.; Woodford, N. Evaluation of meat, fruit and vegetables from retail stores in five United Kingdom regions as sources of extended-spectrum beta-lactamase (ESBL)-producing and carbapenem-resistant Escherichia coli. Int. J. Food Microbiol. 2017, 241, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Reuland, E.A.; al Naiemi, N.; Raadsen, S.A.; Savelkoul, P.H.M.; Kluytmans, J.A.J.W.; Vandenbroucke-Grauls, C.M.J.E. Prevalence of ESBL-producing Enterobacteriaceae in raw vegetables. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1843–1846. [Google Scholar] [CrossRef]
- Jones, K.; Bradshaw, S.B. Biofilm formation by the Enterobacteriaceae: A comparison between Salmonella enteritidis, Escherich Ia Coli and nitrogenfixing strain of Klebsiella pneumoniae. J. Appl. Bacteriol. 1996, 80, 458–464. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, K.; Zheng, B.; Zhao, L.; Shen, P.; Ji, J.; Wei, Z.; Li, L.; Zhou, J.; Xiao, Y. High prevalence of ESBL-producing Klebsiella pneumoniae causing community-onset infections in China. Front. Microbiol. 2016, 7, 1830. [Google Scholar] [CrossRef]
- Toh, B.E.W.; Bokhari, O.; Kutbi, A.; Haroon, M.F.; Mantilla-Calderon, D.; Zowawi, H.; Hong, P.Y. Varying occurrence of extended-spectrum beta-lactamase bacteria among three produce types. J. Food Saf. 2018, 38, e12373. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Microbiological Hazards in Fresh Leafy Vegetables and Herbs: Meeting Report; Microbiological Risk Assessment Series; WHO: Geneva, Switzerland, 2008.
- Coque, T.M.; Baquero, F.; Canton, R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Eurosurveillance 2008, 13, 19044. [Google Scholar] [CrossRef]
- Wiener, J.; Quinn, J.P.; Bradford, P.A.; Goering, R.V.; Nathan, C.; Bush, K.; Weinstein, R.A. Multiple antibiotic-resistant Klebsiella and Escherichia coli in nursing homes. JAMA 1999, 281, 517–523. [Google Scholar] [CrossRef]
- Pitout, J.D.; Laupland, K.B. Extended-spectrum β-lactamase-producing Enterobacteriaceae: An emerging public-health concern. Lancet Infect. Dis. 2008, 8, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Morosini, M.I.; García-Castillo, M.; Coque, T.M.; Valverde, A.; Novais, Â.; Loza, E.; Baquero, F.; Cantón, R. Antibiotic coresistance in extended-spectrum-β-lactamase-producing Enterobacteriaceae and in vitro activity of tigecycline. Antimicrob. Agents Chemother. 2006, 50, 2695–2699. [Google Scholar] [CrossRef] [PubMed]
- Rocha, F.R.; Pinto, V.P.T.; Barbosa, F.C.B. The spread of CTX-M-type extended-spectrum β-lactamases in Brazil: A systematic review. Microb. Drug Resist. 2016, 22, 301–311. [Google Scholar] [CrossRef]
- Kurittu, P.; Khakipoor, B.; Aarnio, M.; Nykäsenoja, S.; Brouwer, M.; Myllyniemi, A.L.; Vatunen, E.; Heikinheimo, A. Plasmid-borne and chromosomal ESBL/AmpC genes in Escherichia coli and Klebsiella pneumoniae in global food products. Front. Microbiol. 2021, 12, 592291. [Google Scholar] [CrossRef] [PubMed]
- Hassen, B.; Abbassi, M.S.; Benlabidi, S.; Ruiz-Ripa, L.; Mama, O.M.; Ibrahim, C.; Hassen, A.; Hammami, S.; Torres, C. Genetic characterization of ESBL-producing Escherichia coli and Klebsiella pneumoniae isolated from wastewater and river water in Tunisia: Predominance of CTX-M-15 and high genetic diversity. Environ. Sci. Pollut. Res. 2020, 27, 44368–44377. [Google Scholar] [CrossRef]
- del Carmen Rocha-Gracia, R.; Lozano-Zarain, P.; Cázarez, Z.G.; Alonso, C.A.; Brambila, E.; Torres, C.; Cortés-Cortés, G. IncFIB plasmids carrying the resistance gene blaCTX-M-15 in ESBL-producing Escherichia coli clones from pediatric patients. J. Infect. Dev. Ctries 2022, 16, 500–506. [Google Scholar] [CrossRef]
- Zhao, H.; He, Z.; Li, Y.; Sun, B. Epidemiology of carbapenem-resistant Klebsiella pneumoniae ST15 of producing KPC-2, SHV-106 and CTX-M-15 in Anhui, China. BMC Microbiol. 2022, 22, 262. [Google Scholar] [CrossRef]
- De Angelis, G.; Del Giacomo, P.; Posteraro, B.; Sanguinetti, M.; Tumbarello, M. Molecular mechanisms, epidemiology, and clinical importance of β-lactam resistance in Enterobacteriaceae. Int. J. Mol. Sci. 2020, 21, 5090. [Google Scholar] [CrossRef]
- Djordjevic, S.P.; Stokes, H.W.; Chowdhury, P.R. Mobile elements, zoonotic pathogens and commensal bacteria: Conduits for the delivery of resistance genes into humans, production animals and soil microbiota. Front. Microbiol. 2013, 4, 86. [Google Scholar] [CrossRef]
- Humeniuk, C.; Arlet, G.; Gautier, V.; Grimont, P.; Labia, R.; Philippon, A. β-lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 2002, 46, 3045–3049. [Google Scholar] [CrossRef]
- Rodríguez, M.M.; Power, P.; Radice, M.; Vay, C.; Famiglietti, A.; Galleni, M.; Ayala, J.A.; Gutkind, G. Chromosome-encoded CTX-M-3 from Kluyvera ascorbata: A possible origin of plasmid-borne CTX-M-1-derived cefotaximases. Antimicrob. Agents Chemother. 2004, 48, 4895–4897. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Food and Drug Administration (FDA). Guidance for Industry: Guide to Minimize Microbial Food Safety Hazards for Fresh Fruits and Vegetables; U.S. Food and Drug Administratio: College Park, MD, USA, 1998.
- Clinical and Laboratory Standards Institute. M100 Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed.; CLSI: Wayne, PA, USA, 2023; Volume 8, ISBN 0956-4624. [Google Scholar]
- Kon, H.; Temkin, E.; Elmalih, P.; Keren-Paz, A.; Ben-David, D.; Najjar-Debbiny, R.; Gottesman, T.; Carmeli, Y. Analysis of four carbapenem-resistant Acinetobacter baumannii outbreaks using Fourier-transform infrared spectroscopy. Infect. Control Hosp. Epidemiol. 2022, 44, 991–993. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Li, P.; Liu, H.; Lü, D.; Liang, H.; Dou, Y. Phenotypic and molecular characterization of multidrug resistant Klebsiella pneumoniae isolated from a university teaching hospital, China. PLoS ONE 2014, 9, e95181. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, J.; Navon-Venezia, S.; Chmelnitsky, I.; Hammer-Münz, O.; Leavitt, A.; Gold, H.S.; Schwaber, M.J.; Carmeli, Y. Extended-spectrum beta-lactamases among Enterobacter isolates obtained in Tel Aviv, Israel. Antimicrob. Agents Chemother. 2005, 49, 1150–1156. [Google Scholar] [CrossRef]
- Ejaz, H.; Younas, S.; Abosalif, K.; Junaid, K.; Alzahrani, B.; Alsrhani, A.; Abdalla, A.; Ullah, M.; Qamar, M.; Hamam, S. Molecular analysis of blaSHV, blaTEM, and blaCTX-M in Extended-spectrum β-lactamase producing Enterobacteriaceae recovered from fecal specimens of animals. PLoS ONE 2021, 16, e0245126. [Google Scholar] [CrossRef]

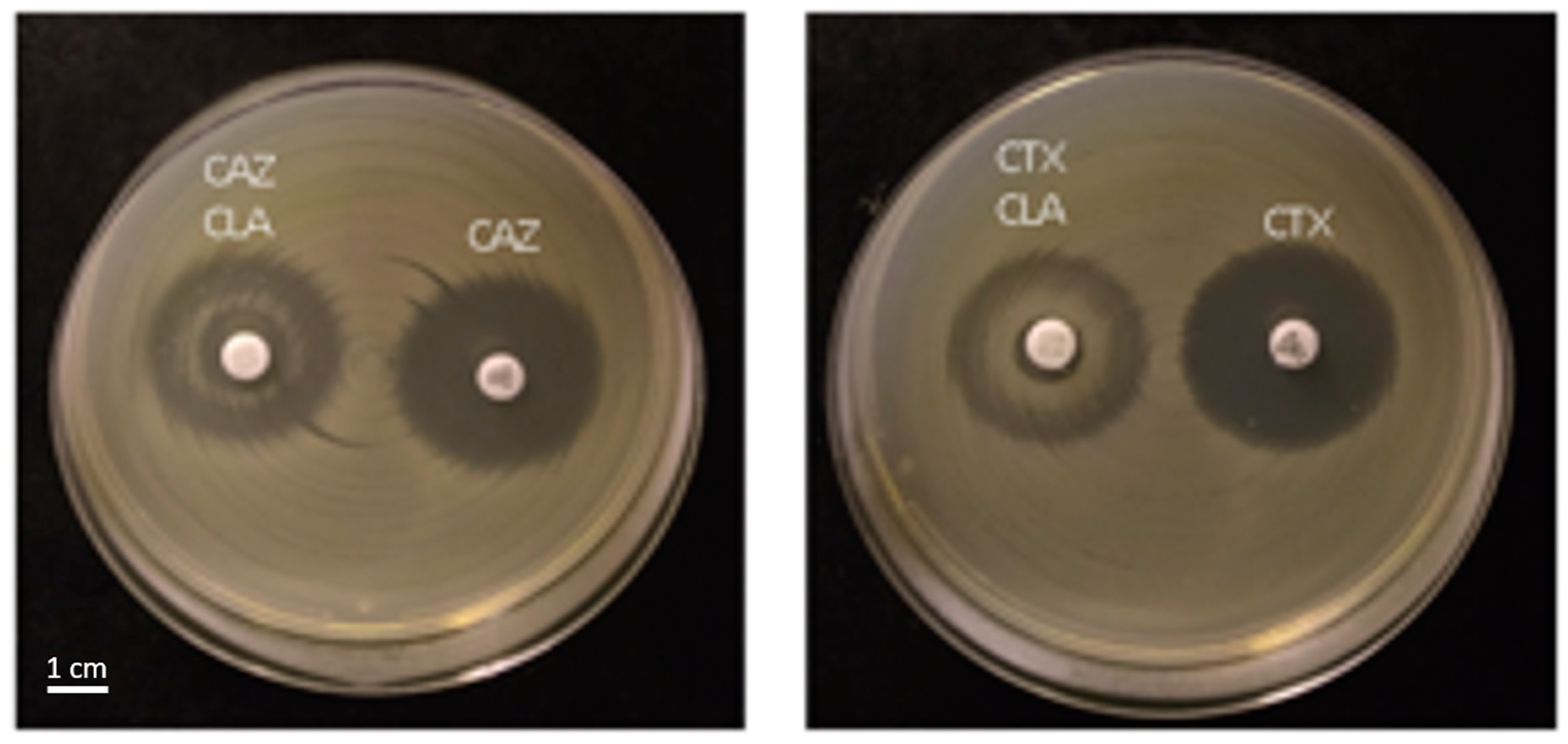

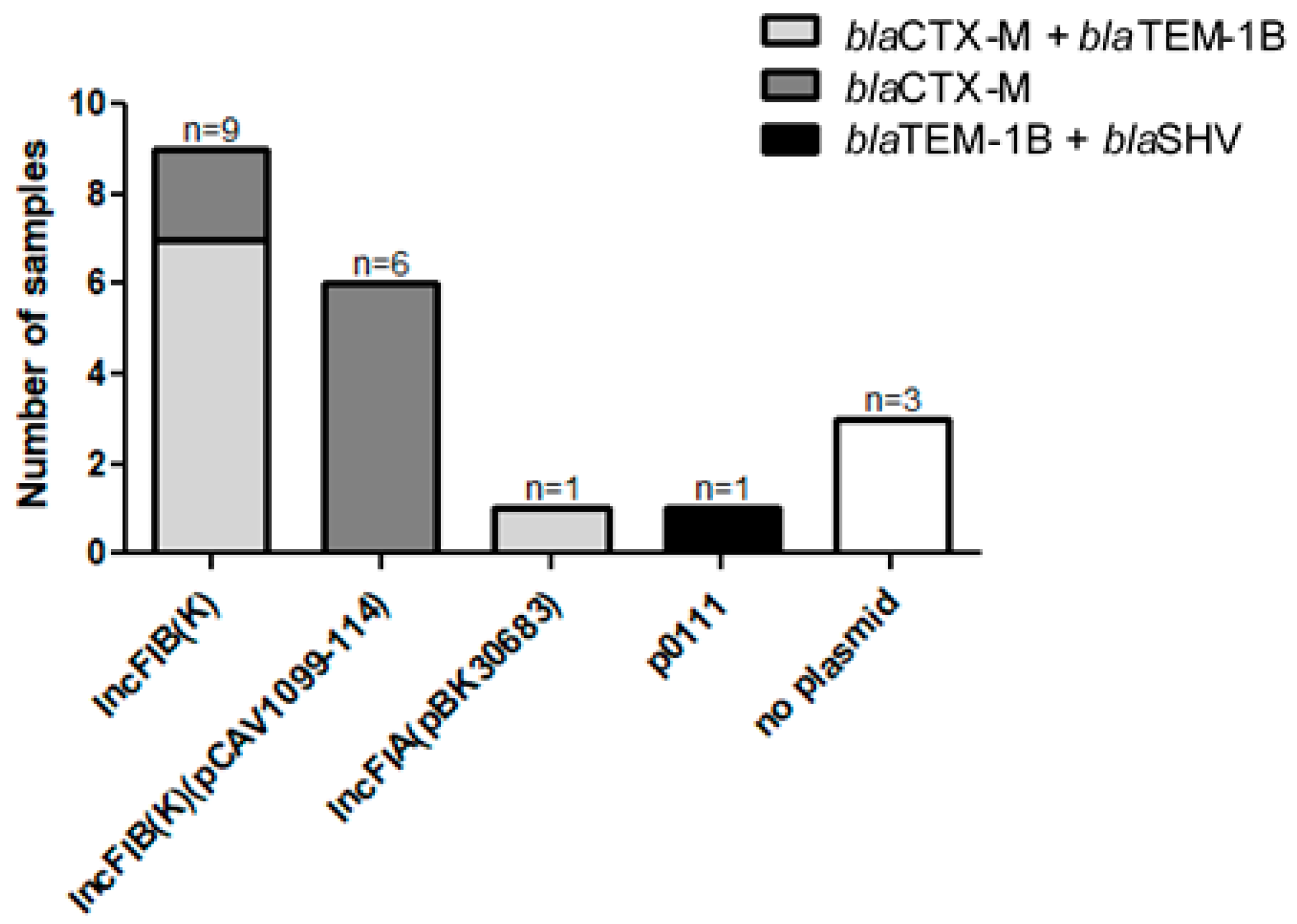
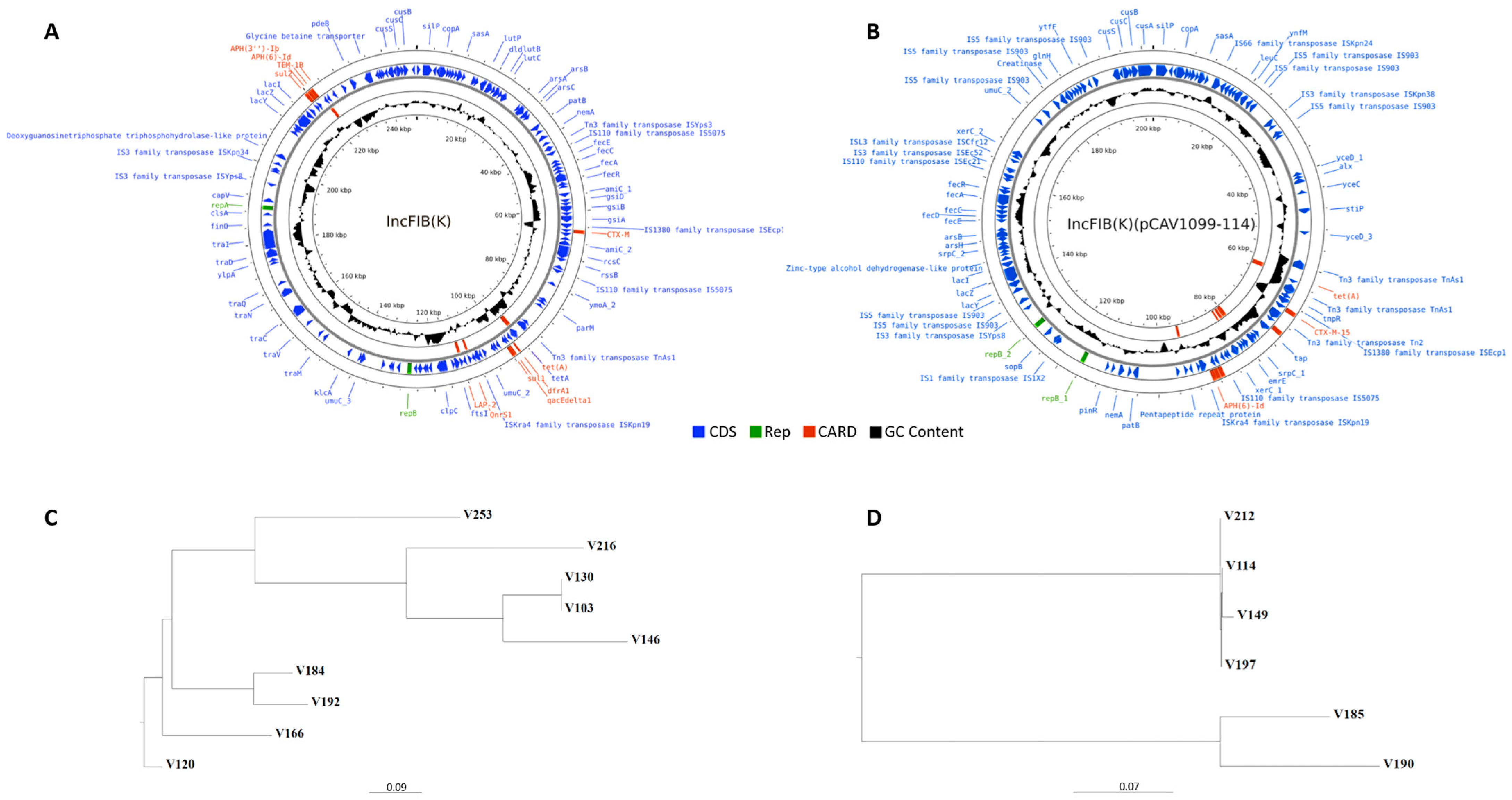
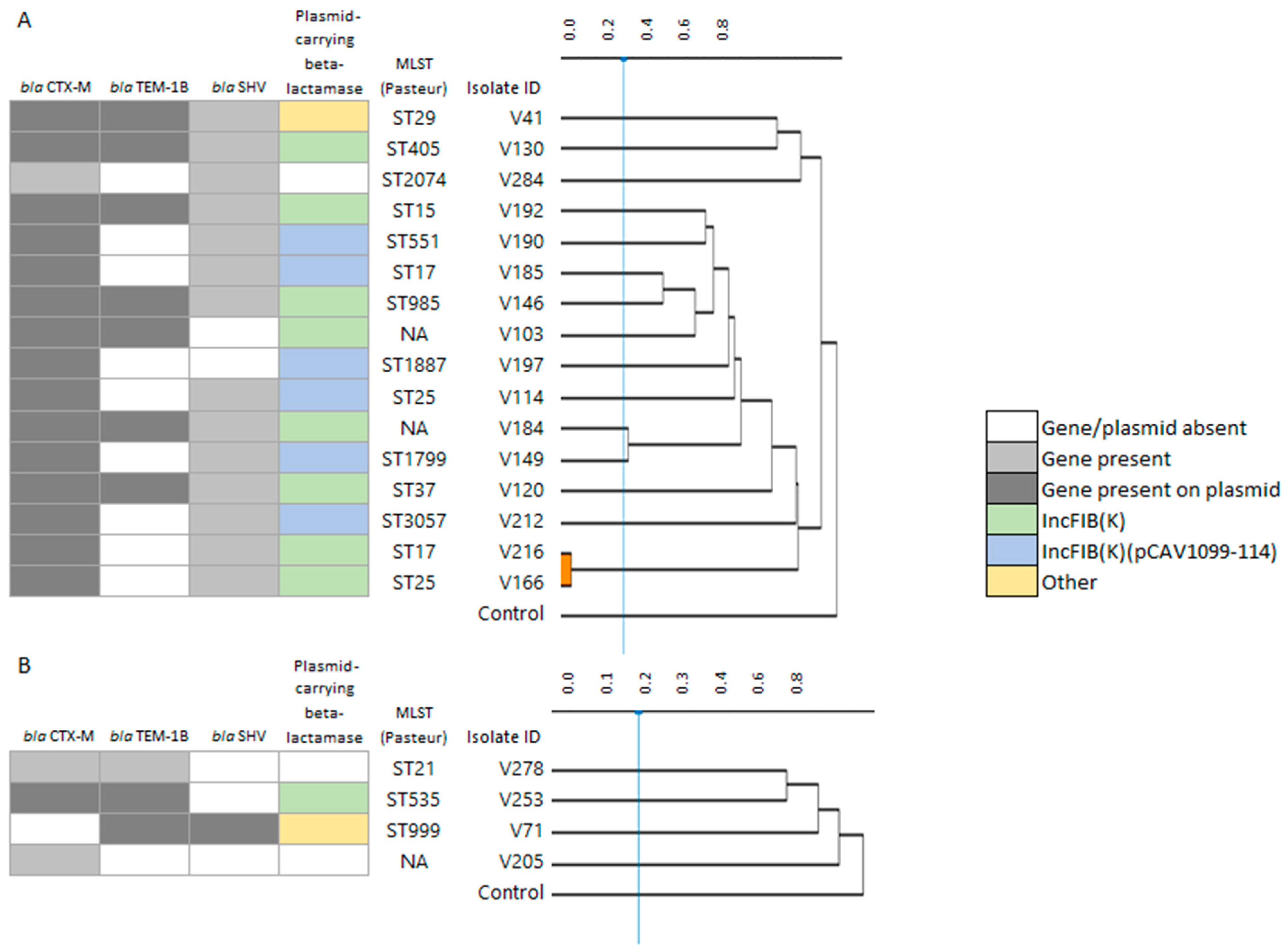
| AmpC-Positive | ESBL-Positive | |||
|---|---|---|---|---|
| N (%) | Odds Ratio (95% CI) | N (%) | Odds Ratio (95% CI) | |
| Vegetable type | ||||
| Climbing vegetables | 18/123 (14.6%) | Reference | 3/123 (2.4%) | Reference |
| Leafy greens | 40/121 (33.1%) | 2.88 (1.54–5.39) | 12/121 (9.9%) | 4.40 (1.21–16.02) |
| Environment-controlled beds | 5/57 (8.8%) | 0.56 (0.20–1.59) | 5/57 (8.8%) | 3.85 (0.89–16.69) |
| Packaging | ||||
| Unpackaged | 34/169 (20.1%) | Reference | 7/169 (4.1%) | Reference |
| Packaged | 29/132 (22.0%) | 1.12 (0.64–1.95) | 13/132 (9.8%) | 2.53 (0.98–6.53) |
| Market type | ||||
| Outdoor market | 21/89 (23.6%) | Reference | 10/89 (11.2%) | Reference |
| Supermarket | 17/114 (14.9%) | 0.57 (0.28–1.16) | 8/114 (7.0%) | 0.60 (0.23–1.58) |
| Minimarket | 25/98 (25.5%) | 1.11 (0.57–2.16) | 2/98 (2.0%) | 0.16 (0.04–0.77) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kon, H.; Lurie-Weinberger, M.; Cohen, A.; Metsamber, L.; Keren-Paz, A.; Schwartz, D.; Carmeli, Y.; Schechner, V. Occurrence, Typing, and Resistance Genes of ESBL/AmpC-Producing Enterobacterales in Fresh Vegetables Purchased in Central Israel. Antibiotics 2023, 12, 1528. https://doi.org/10.3390/antibiotics12101528
Kon H, Lurie-Weinberger M, Cohen A, Metsamber L, Keren-Paz A, Schwartz D, Carmeli Y, Schechner V. Occurrence, Typing, and Resistance Genes of ESBL/AmpC-Producing Enterobacterales in Fresh Vegetables Purchased in Central Israel. Antibiotics. 2023; 12(10):1528. https://doi.org/10.3390/antibiotics12101528
Chicago/Turabian StyleKon, Hadas, Mor Lurie-Weinberger, Adi Cohen, Liat Metsamber, Alona Keren-Paz, David Schwartz, Yehuda Carmeli, and Vered Schechner. 2023. "Occurrence, Typing, and Resistance Genes of ESBL/AmpC-Producing Enterobacterales in Fresh Vegetables Purchased in Central Israel" Antibiotics 12, no. 10: 1528. https://doi.org/10.3390/antibiotics12101528
APA StyleKon, H., Lurie-Weinberger, M., Cohen, A., Metsamber, L., Keren-Paz, A., Schwartz, D., Carmeli, Y., & Schechner, V. (2023). Occurrence, Typing, and Resistance Genes of ESBL/AmpC-Producing Enterobacterales in Fresh Vegetables Purchased in Central Israel. Antibiotics, 12(10), 1528. https://doi.org/10.3390/antibiotics12101528







