Comparison of Lateral Flow Immunochromatography and Phenotypic Assays to PCR for the Detection of Carbapenemase-Producing Gram-Negative Bacteria, a Multicenter Experience in Mexico
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Manoukian, S.; Stewart, S.; Graves, N.; Mason, H.; Robertson, C.; Kennedy, S.; Pan, J.; Kavanagh, K.; Haahr, L.; Adil, M.; et al. Bed-days and costs associated with the inpatient burden of healthcare-associated infection in the UK. J. Hosp. Infect. 2021, 114, 43–50. [Google Scholar] [CrossRef]
- Bahrami, S.; Shafiee, F.; Hakamifard, A.; Fazeli, H.; Soltani, R. Antimicrobial susceptibility pattern of carbapenemase-producing Gram-negative nosocomial bacteria at Al Zahra hospital, Isfahan, Iran. Iran. J. Microbiol. 2021, 13, 50–57. [Google Scholar] [CrossRef]
- Nasiri, M.J.; Mirsaeidi, M.; Mousavi, S.M.J.; Arshadi, M.; Fardsanei, F.; Deihim, B.; Davoudabadi, S.; Zamani, S.; Hajikhani, B.; Goudarzi, H.; et al. Prevalence and Mechanisms of Carbapenem Resistance in Klebsiella pneumoniae and Escherichia coli: A Systematic Review and Meta-Analysis of Cross-Sectional Studies from Iran. Microb. Drug Resist. 2020, 26, 1491–1502. [Google Scholar] [CrossRef]
- Sivalingam, P.; Poté, J.; Prabakar, K. Environmental Prevalence of Carbapenem Resistance Enterobacteriaceae (CRE) in a Tropical Ecosystem in India: Human Health Perspectives and Future Directives. Pathogens 2019, 8, 174. [Google Scholar] [CrossRef]
- Li, Y.; Sun, Q.; Shen, Y.; Zhang, Y.; Yang, J.; Shu, L.; Zhou, H.; Wang, Y.; Wang, B.; Zhang, R.; et al. Rapid Increase in Prevalence of Carbapenem-Resistant Enterobacteriaceae (CRE) and Emergence of Colistin Resistance Gene mcr-1 in CRE in a Hospital in Henan, China. J. Clin. Microbiol. 2018, 56, e01932-17. [Google Scholar] [CrossRef]
- Kazmierczak, K.M.; Karlowsky, J.A.; Jonge, B.L.M.d.; Stone, G.G.; Sahm, D.F. Epidemiology of Carbapenem Resistance Determinants Identified in Meropenem-Nonsusceptible Enterobacterales Collected as Part of a Global Surveillance Program, 2012 to 2017. Antimicrob. Agents Chemother. 2021, 65, e02000-20. [Google Scholar] [CrossRef]
- Garza-González, E.; Franco-Cendejas, R.; Morfín-Otero, R.; Echaniz-Aviles, G.; Rojas-Larios, F.; Bocanegra-Ibarias, P.; Flores-Treviño, S.; Ponce-de-León, A.; Rodríguez-Noriega, E.; Alavez-Ramírez, N.; et al. The Evolution of Antimicrobial Resistance in Mexico During the Last Decade: Results from the INVIFAR Group. Microb. Drug Resist. 2020, 26, 1372–1382. [Google Scholar] [CrossRef]
- Garza-González, E.; Morfín-Otero, R.; Mendoza-Olazarán, S.; Bocanegra-Ibarias, P.; Flores-Treviño, S.; Rodríguez-Noriega, E.; Ponce-de-León, A.; Sanchez-Francia, D.; Franco-Cendejas, R.; Arroyo-Escalante, S.; et al. A snapshot of antimicrobial resistance in Mexico. Results from 47 centers from 20 states during a six-month period. PLoS ONE 2019, 14, e0209865. [Google Scholar] [CrossRef]
- Goldenberg, S.D.; Dodgson, A.R.; Barlow, G.; Parcell, B.J.; Jones, L.; Albur, M.; Wilson, A.P.R.; Enoch, D.A.; Marek, A.; Micallef, C.; et al. Epidemiology, Outcomes and Resource Utilisation in Patients with Carbapenem Non-susceptible Gram-Negative Bacteria in the UK: A Retrospective, Observational Study (CARBAR UK). Adv. Ther. 2022, 39, 3602–3615. [Google Scholar] [CrossRef]
- Humphries, R.M. CIM City: The Game Continues for a Better Carbapenemase Test. J. Clin. Microbiol. 2019, 57, e00353-19. [Google Scholar] [CrossRef]
- M100Ed32; Performance Standards for Antimicrobial Susceptibility Testing. 32nd ed. Clinical & Laboratory Standards Institute: Wayne, PA, USA, 2022.
- Zhong, H.; Wu, M.-L.; Feng, W.-J.; Huang, S.-F.; Yang, P. Accuracy and applicability of different phenotypic methods for carbapenemase detection in Enterobacteriaceae: A systematic review and meta-analysis. J. Glob. Antimicrob. Resist. 2020, 21, 138–147. [Google Scholar] [CrossRef]
- Bogaerts, P.; Berger, A.-S.; Evrard, S.; Huang, T.-D. Comparison of two multiplex immunochromatographic assays for the rapid detection of major carbapenemases in Enterobacterales. J. Antimicrob. Chemother. 2020, 75, 1491–1494. [Google Scholar] [CrossRef]
- Jenkins, S.; Ledeboer, N.A.; Westblade, L.F.; Burnham, C.-A.D.; Faron, M.L.; Bergman, Y.; Yee, R.; Mesich, B.; Gerstbrein, D.; Wallace, M.A.; et al. Evaluation of NG-Test Carba 5 for Rapid Phenotypic Detection and Differentiation of Five Common Carbapenemase Families: Results of a Multicenter Clinical Evaluation. J. Clin. Microbiol. 2020, 58, e00344-20. [Google Scholar] [CrossRef]
- Boutal, H.; Vogel, A.; Bernabeu, S.; Devilliers, K.; Creton, E.; Cotellon, G.; Plaisance, M.; Oueslati, S.; Dortet, L.; Jousset, A.; et al. A multiplex lateral flow immunoassay for the rapid identification of NDM-, KPC-, IMP- and VIM-type and OXA-48-like carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 909–915. [Google Scholar] [CrossRef]
- Potron, A.; Fournier, D.; Emeraud, C.; Triponney, P.; Plésiat, P.; Naas, T.; Dortet, L. Evaluation of the Immunochromatographic NG-Test Carba 5 for Rapid Identification of Carbapenemase in Nonfermenters. Antimicrob. Agents Chemother. 2019, 63, e00968-19. [Google Scholar] [CrossRef]
- Garza-González, E.; Bocanegra-Ibarias, P.; Bobadilla-del-Valle, M.; Ponce-de-León-Garduño, L.A.; Esteban-Kenel, V.; Silva-Sánchez, J.; Garza-Ramos, U.; Barrios-Camacho, H.; López-Jácome, L.E.; Colin-Castro, C.A.; et al. Drug resistance phenotypes and genotypes in Mexico in representative gram-negative species: Results from the infivar network. PLoS ONE 2021, 16, e0248614. [Google Scholar] [CrossRef]
- M07; Dilution AST for Aerobically Grown Bacteria. Clinical & Laboratory Standards Institute: Wayne, PA, USA, 2018.
- Dimitriu, T. Evolution of horizontal transmission in antimicrobial resistance plasmids. Microbiology 2022, 168, 001214. [Google Scholar] [CrossRef]
- WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 27 February 2017).
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- García-Betancur, J.C.; Appel, T.M.; Esparza, G.; Gales, A.C.; Levy-Hara, G.; Cornistein, W.; Vega, S.; Nuñez, D.; Cuellar, L.; Bavestrello, L.; et al. Update on the epidemiology of carbapenemases in Latin America and the Caribbean. Expert Rev. Anti-Infect. Ther. 2021, 19, 197–213. [Google Scholar] [CrossRef]
- Shanmugakani, R.K.; Srinivasan, B.; Glesby, M.J.; Westblade, L.F.; Cárdenas, W.B.; Raj, T.; Erickson, D.; Mehta, S. Current state of the art in rapid diagnostics for antimicrobial resistance. Lab A Chip 2020, 20, 2607–2625. [Google Scholar] [CrossRef]
- Ben-Haim, O.; Azrad, M.; Saleh, N.; Tkhawkho, L.; Peretz, A. Evaluation of the NG-Test CARBA 5 Kit for Rapid Detection of Carbapenemase Resistant Enterobacteriaceae. Lab. Med. 2021, 52, 375–380. [Google Scholar] [CrossRef]
- Ratnayake, L.; Ang, H.Z.; Ong, C.H.; Chan, D.S.G. An optimized algorithm with improved turnaround time for detection of carbapenemase-producing Enterobacterales using the NG Test CARBA 5 in a routine laboratory. J. Med. Microbiol. 2020, 69, 228–232. [Google Scholar] [CrossRef]
- Bianco, G.; Boattini, M.; van Asten, S.a.V.; Iannaccone, M.; Zanotto, E.; Zaccaria, T.; Bernards, A.T.; Cavallo, R.; Costa, C. RESIST-5 O.O.K.N.V. and NG-Test Carba 5 assays for the rapid detection of carbapenemase-producing Enterobacterales from positive blood cultures: A comparative study. J. Hosp. Infect. 2020, 105, 162–166. [Google Scholar] [CrossRef]
- Takissian, J.; Bonnin, R.A.; Naas, T.; Dortet, L. NG-Test Carba 5 for Rapid Detection of Carbapenemase-Producing Enterobacterales from Positive Blood Cultures. Antimicrob. Agents Chemother. 2019, 63, e00011-19. [Google Scholar] [CrossRef]
- Baer, D.; Azrad, M.; Saleh, N.; Peretz, A. Detection of Carbapenem-Resistant Enterobacterales in Simulated Blood Culture in 15 Minutes. Life 2021, 11, 145. [Google Scholar] [CrossRef]
- Kuchibiro, T.; Komatsu, M.; Yamasaki, K.; Nakamura, T.; Nishio, H.; Nishi, I.; Kimura, K.; Niki, M.; Ono, T.; Sueyoshi, N.; et al. Evaluation of the modified carbapenem inactivation method for the detection of carbapenemase-producing Enterobacteriaceae. J. Infect. Chemother. 2018, 24, 262–266. [Google Scholar] [CrossRef]
- Stanton, R.A.; Campbell, D.; McAllister, G.A.; Breaker, E.; Adamczyk, M.; Daniels, J.B.; Lutgring, J.D.; Karlsson, M.; Schutz, K.; Jacob, J.T.; et al. Whole-Genome Sequencing Reveals Diversity of Carbapenem-Resistant Pseudomonas aeruginosa Collected through CDC’s Emerging Infections Program, United States, 2016–2018. Antimicrob. Agents Chemother. 2022, 66, e0049622. [Google Scholar] [CrossRef]
- Nishida, S.; Ihashi, Y.; Yoshino, Y.; Ono, Y. Evaluation of an immunological assay for the identification of multiple carbapenemase-producing Gram-negative bacteria. Pathology 2022, 54, 917–921. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, D.; Li, Y.; Liu, Y.; Qin, X. Comparison of the Performance of Phenotypic Methods for the Detection of Carbapenem-Resistant Enterobacteriaceae (CRE) in Clinical Practice. Front. Cell. Infect. Microbiol. 2022, 12, 165. [Google Scholar] [CrossRef]
- Ding, L.; Shi, Q.; Han, R.; Yin, D.; Wu, S.; Yang, Y.; Guo, Y.; Zhu, D.; Hu, F. Comparison of Four Carbapenemase Detection Methods for blaKPC-2 Variants. Microbiol. Spectr. 2021, 9, e0095421. [Google Scholar] [CrossRef]
- Zhu, Y.; Jia, P.; Li, X.; Wang, T.; Zhang, J.; Zhang, G.; Duan, S.; Kang, W.; Xu, Y.; Yang, Q. Carbapenemase detection by NG-Test CARBA 5—A rapid immunochromatographic assay in carbapenem-resistant Enterobacterales diagnosis. Ann. Transl. Med. 2021, 9, 769. [Google Scholar] [CrossRef]
- Hopkins, K.L.; Meunier, D.; Naas, T.; Volland, H.; Woodford, N. Evaluation of the NG-Test CARBA 5 multiplex immunochromatographic assay for the detection of KPC, OXA-48-like, NDM, VIM and IMP carbapenemases. J. Antimicrob. Chemother. 2018, 73, 3523–3526. [Google Scholar] [CrossRef]
- Volland, H.; Girlich, D.; Laguide, M.; Gonzalez, C.; Paris, V.; Laroche, M.; Oueslati, S.; Dortet, L.; Simon, S.; Naas, T. Improvement of the Immunochromatographic NG-Test Carba 5 Assay for the Detection of IMP Variants Previously Undetected. Antimicrob. Agents Chemother. 2019, 64, e01940-19. [Google Scholar] [CrossRef]
- Josa, M.D.; Leal, R.; Rojas, J.; Torres, M.I.; Cortés-Muñoz, F.; Esparza, G.; Reyes, L.F. Comparative Evaluation of Phenotypic Synergy Tests versus RESIST-4 O.K.N.V. and NG Test Carba 5 Lateral Flow Immunoassays for the Detection and Differentiation of Carbapenemases in Enterobacterales and Pseudomonas aeruginosa. Microbiol. Spectr. 2022, 10, e01080-21. [Google Scholar] [CrossRef]
- Giufrè, M.; Errico, G.; Monaco, M.; Del Grosso, M.; Sabbatucci, M.; Pantosti, A.; Cerquetti, M.; Pagnotta, M.; Marra, M.; Carollo, M.; et al. Whole Genome Sequencing and Molecular Analysis of Carbapenemase-Producing Escherichia coli from Intestinal Carriage in Elderly Inpatients. Microorganisms 2022, 10, 1561. [Google Scholar] [CrossRef]
- Karlsson, M.; Lutgring, J.D.; Ansari, U.; Lawsin, A.; Albrecht, V.; McAllister, G.; Daniels, J.; Lonsway, D.; McKay, S.; Beldavs, Z.; et al. Molecular Characterization of Carbapenem-Resistant Enterobacterales Collected in the United States. Microb. Drug Resist. 2022, 28, 389–397. [Google Scholar] [CrossRef]
- Martins, A.F.; Zavascki, A.P.; Gaspareto, P.B.; Barth, A.L. Dissemination of Pseudomonas aeruginosa Producing SPM-1-like and IMP-1-like Metallo-β-lactamases in Hospitals from Southern Brazil. Infection 2007, 35, 457–460. [Google Scholar] [CrossRef]
- Torres-González, P.; Bobadilla-del Valle, M.; Tovar-Calderón, E.; Leal-Vega, F.; Hernández-Cruz, A.; Martínez-Gamboa, A.; Niembro-Ortega, M.D.; Sifuentes-Osornio, J.; Ponce-de-León, A. Outbreak Caused by Enterobacteriaceae Harboring NDM-1 Metallo-β-Lactamase Carried in an IncFII Plasmid in a Tertiary Care Hospital in Mexico City. Antimicrob. Agents Chemother. 2015, 59, 7080–7083. [Google Scholar] [CrossRef]
- Gomez, S.A.; Rapoport, M.; Piergrossi, N.; Faccone, D.; Pasteran, F.; De Belder, D.; Re, L.G.; Petroni, A.; Corso, A. Performance of a PCR assay for the rapid identification of the Klebsiella pneumoniae ST258 epidemic clone in Latin American clinical isolates. Infect. Genet. Evol. 2016, 44, 145–146. [Google Scholar] [CrossRef] [PubMed]
- Atrissi, J.; Milan, A.; Bressan, R.; Lucafò, M.; Petix, V.; Busetti, M.; Dolzani, L.; Lagatolla, C. Interplay of OpdP Porin and Chromosomal Carbapenemases in the Determination of Carbapenem Resistance/Susceptibility in Pseudomonas aeruginosa. Microbiol. Spectr. 2021, 9, e0118621. [Google Scholar] [CrossRef]
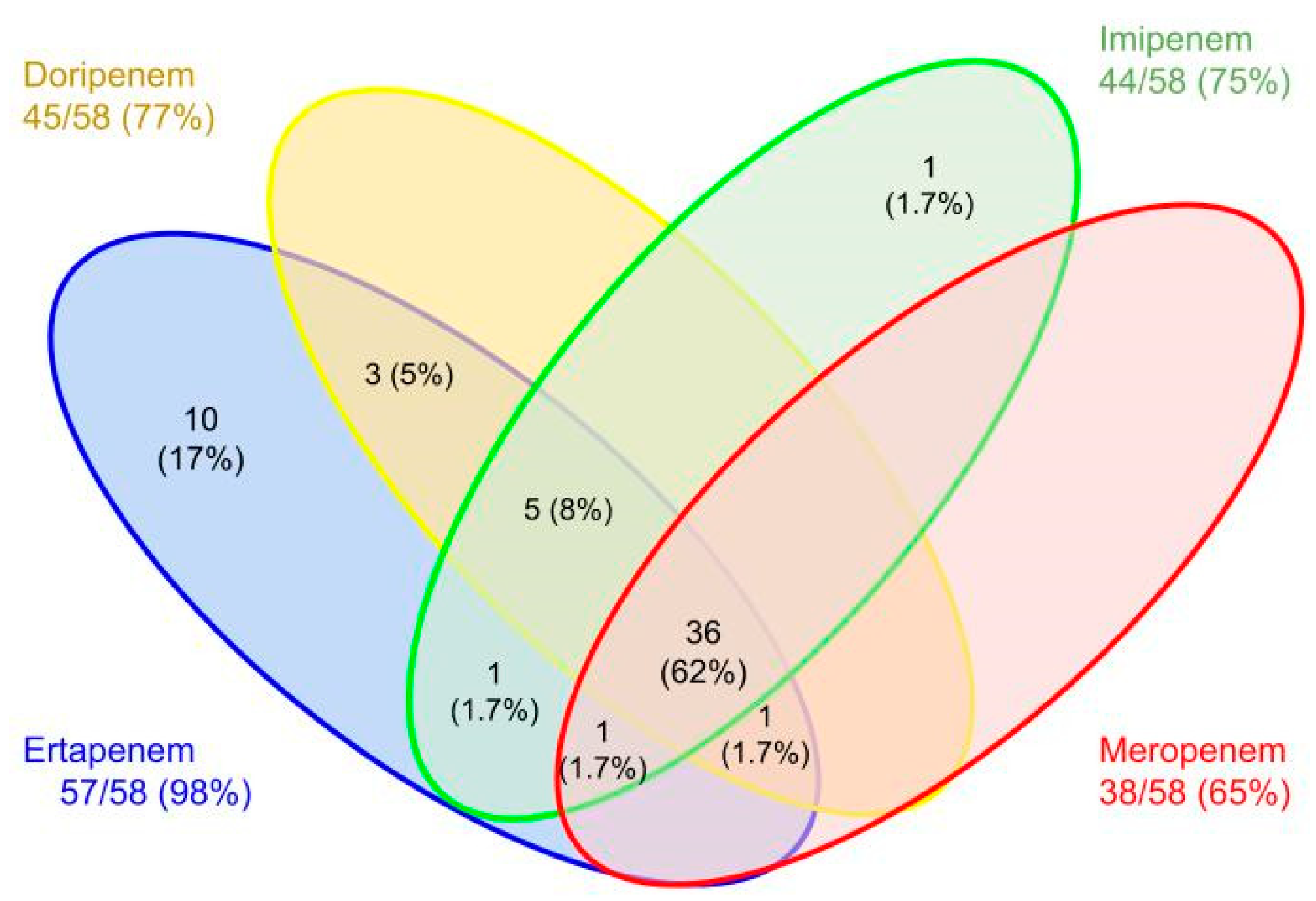
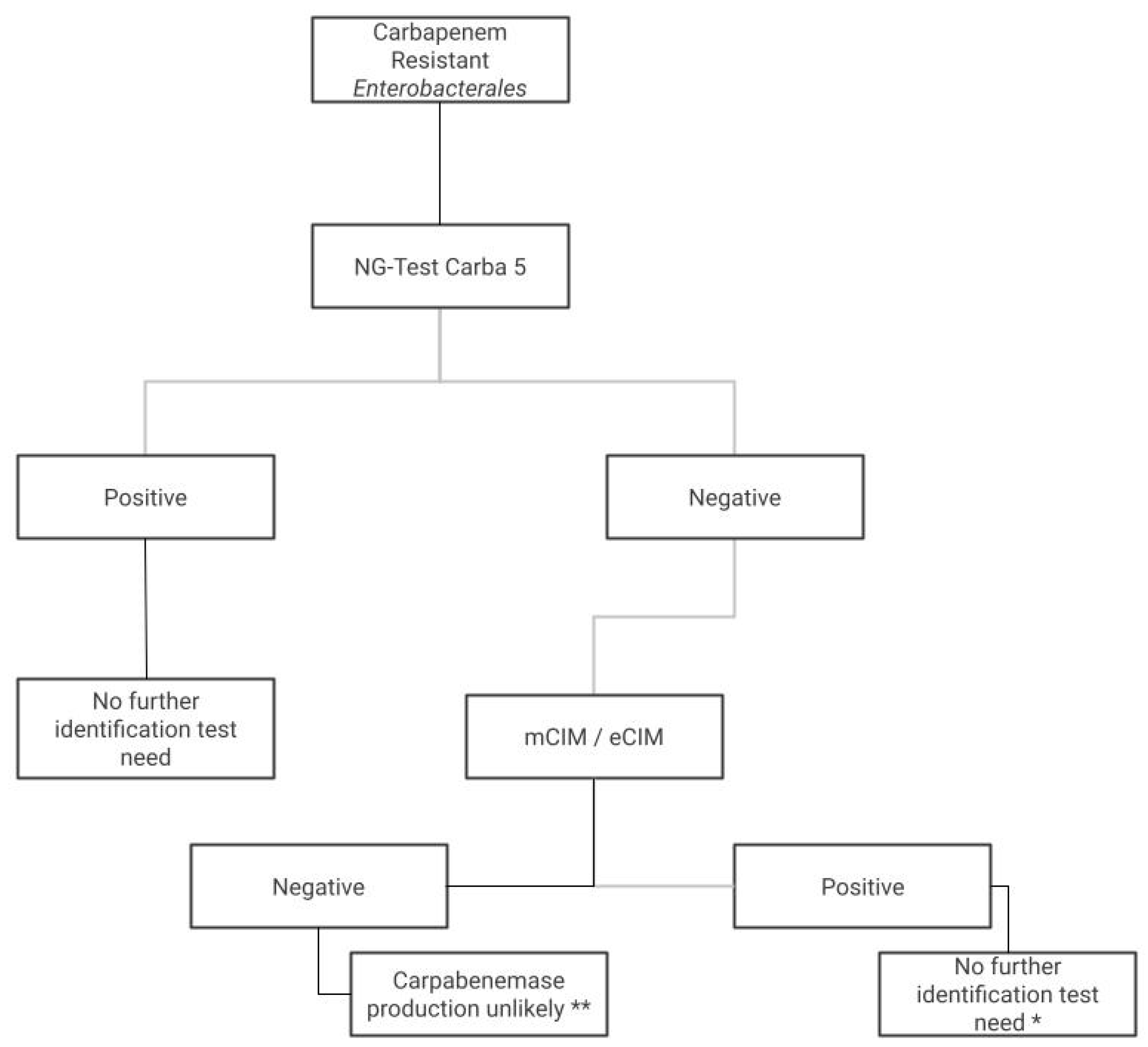
| Microorganisms | n | Test | Sensitivity | Specificity | PPV | NPV | Kappa (CI 95%) | |
|---|---|---|---|---|---|---|---|---|
| Enterobacterales | 58 | mCIM | 100 | 100 | 100 | 100 | 1 (1–1) | p < 0.001 |
| mCIM/eCIM | 86 | 100 | 100 | 50 | 0.603 (0.27–0.85) | p < 0.001 | ||
| CARBA NP | 67 | 100 | 100 | 30 | 0.347 (0.15–0.55) | p < 0.001 | ||
| CARBA NP +EDTA | 63 | 100 | 100 | 27 | 0.29 (0.1–0.48) | p < 0.001 | ||
| CARBA 5 | 98 | 100 | 100 | 87 | 0.923 (0.661–1) | p < 0.001 | ||
| P. aeruginosa | 26 | mCIM | 100 | 100 | 100 | 100 | 1 (1–1) | p < 0.001 |
| mCIM/eCIM | 83 | 100 | 100 | 43 | 0.523 (0.001–0.9) | p = 0.002 | ||
| CARBA NP | 61 | 100 | 100 | 25 | 0.264 (0.001–0.58) | p = 0.047 | ||
| CARBA 5 | 82.6 | 100 | 100 | 42.9 | 0.523 (0.14–0.9) | p = 0.002 |
| Microorganisms | Genomic Sequencing | mCIM/eCIM | Carba NP/EDTA * | PCR | Carba 5® | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Enterobacterales | N | Detected | Non Detected | Detected | Non Detected | Detected | Non Detected | Detected | Non Detected | |
| KPC (10) | 8 | KPC-2 | 8 | 0 | 8 | 0 | 8 | 0 | 8 | 0 |
| 1 | KPC-82 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | |
| 1 | KPC-3 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | |
| NDM (17) | 16 | NDM-1 | 12 | 4 | 16 | 0 | 16 | 0 | 16 | 0 |
| 1 | NDM-5 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | |
| VIM (3) | 2 | VIM-2 | 1 | 1 | 1 | 1 | 2 | 0 | 1 | 1 |
| 1 | VIM-67 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | |
| OXA-48 like (19) | 14 | OXA-232 | 14 | 0 | 3 | 11 | 14 | 0 | 14 | 0 |
| 3 | OXA-48 | 3 | 0 | 1 | 2 | 3 | 0 | 3 | 0 | |
| 2 | OXA-181 | 2 | 0 | 0 | 2 | 2 | 0 | 2 | 0 | |
| Co-producers | 2 | NDM-1 KPC-2 | 0 | 2 | 0 | 2 | 2 | 0 | 2 | 0 |
| No-carbapenemases producers | 7 | - | 0 | 7 | 0 | 7 | 0 | 7 | 0 | 7 |
| Total | 58 | 44 | 14 | 32 | 26 | 51 | 7 | 50 | 8 | |
| P. aeruginosa | N | Detected | Non detected | Detected | Non detected | Detected | Non detected | Detected | Non detected | |
| IMP (8) | 4 | IMP-75 | 2 | 2 | 4 | 0 | 4 | 0 | 1 | 3 |
| 3 | IMP-62 | 3 | 0 | 2 | 1 | 3 | 0 | 3 | 0 | |
| 1 | IMP-15 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | |
| VIM | 13 | VIM-2 | 11 | 2 | 5 | 8 | 13 | 0 | 13 | 0 |
| NDM | 1 | NDM-1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 |
| Co-producers | 1 | IMP-75 + NDM-1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 |
| No-carbapenemases producers | 3 | 0 | 3 | 0 | 3 | 0 | 3 | 0 | 3 | |
| Total | 26 | 19 | 7 | 14 | 12 | 23 | 3 | 19 | 7 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendez-Sotelo, B.J.; López-Jácome, L.E.; Colín-Castro, C.A.; Hernández-Durán, M.; Martínez-Zavaleta, M.G.; Rivera-Buendía, F.; Velázquez-Acosta, C.; Rodríguez-Zulueta, A.P.; Morfín-Otero, M.d.R.; Franco-Cendejas, R. Comparison of Lateral Flow Immunochromatography and Phenotypic Assays to PCR for the Detection of Carbapenemase-Producing Gram-Negative Bacteria, a Multicenter Experience in Mexico. Antibiotics 2023, 12, 96. https://doi.org/10.3390/antibiotics12010096
Mendez-Sotelo BJ, López-Jácome LE, Colín-Castro CA, Hernández-Durán M, Martínez-Zavaleta MG, Rivera-Buendía F, Velázquez-Acosta C, Rodríguez-Zulueta AP, Morfín-Otero MdR, Franco-Cendejas R. Comparison of Lateral Flow Immunochromatography and Phenotypic Assays to PCR for the Detection of Carbapenemase-Producing Gram-Negative Bacteria, a Multicenter Experience in Mexico. Antibiotics. 2023; 12(1):96. https://doi.org/10.3390/antibiotics12010096
Chicago/Turabian StyleMendez-Sotelo, Braulio Josue, Luis Esaú López-Jácome, Claudia A. Colín-Castro, Melissa Hernández-Durán, Maria Guadalupe Martínez-Zavaleta, Frida Rivera-Buendía, Consuelo Velázquez-Acosta, Ana Patricia Rodríguez-Zulueta, Maria del Rayo Morfín-Otero, and Rafael Franco-Cendejas. 2023. "Comparison of Lateral Flow Immunochromatography and Phenotypic Assays to PCR for the Detection of Carbapenemase-Producing Gram-Negative Bacteria, a Multicenter Experience in Mexico" Antibiotics 12, no. 1: 96. https://doi.org/10.3390/antibiotics12010096
APA StyleMendez-Sotelo, B. J., López-Jácome, L. E., Colín-Castro, C. A., Hernández-Durán, M., Martínez-Zavaleta, M. G., Rivera-Buendía, F., Velázquez-Acosta, C., Rodríguez-Zulueta, A. P., Morfín-Otero, M. d. R., & Franco-Cendejas, R. (2023). Comparison of Lateral Flow Immunochromatography and Phenotypic Assays to PCR for the Detection of Carbapenemase-Producing Gram-Negative Bacteria, a Multicenter Experience in Mexico. Antibiotics, 12(1), 96. https://doi.org/10.3390/antibiotics12010096







