Epidemiological Analysis of Extended-Spectrum β-Lactamase-Producing Klebsiella pneumoniae Outbreak in a Neonatal Clinic in Poland
Abstract
1. Introduction
2. Results
2.1. Outbreak Description
2.2. Chronological Case Analysis
2.3. Data Analyzed
2.4. Environmental Contamination Control
2.5. Microbiology and Molecular Results
2.6. Pulsed-Filed Gel Electrophoresis (PFGE) Results
3. Discussion
4. Materials and Methods
4.1. Hospital Settings
4.2. Microbiological Diagnostic
4.3. Molecular Typing
4.4. PCR
4.5. Statistics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kontopoulou, K.; Iosifidis, E.; Antoniadou, E.; Tasioudis, P.; Petinaki, E.; Malli, E.; Metallidis, S.; Vatopoulos, A.; Malisiovas, N. The clinical significance of carbapenem-resistant Klebsiella pneumoniae rectal colonization in critically ill patients: From colonization to bloodstream infection. J. Med. Microbiol. 2019, 68, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Effah, C.Y.; Sun, T.; Liu, S.; Wu, Y. Klebsiella pneumoniae: An increasing threat to public health. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 1. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, C.A.; Turner, S.D.; Riley, M.F.; Petri, W.A.J.; Hewlett, E.L. Whole-genome sequencing in outbreak analysis. Clin. Microbiol. Rev. 2015, 28, 541–563. [Google Scholar] [CrossRef]
- Gajul, S.V.; Mohite, S.T.; Mangalgi, S.S.; Wavare, S.M.; Kakade, S.V. Klebsiella pneumoniae in septicemic neonates with special reference to extended spectrum β-lactamase, AmpC, metallo β-lactamase production and multiple drug resistance in Tertiary Care Hospital. J. Lab. Physicians 2015, 7, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Abdelwahab, R.; Alhammadi, M.M.; Hassan, E.A.; Ahmed, E.H.; Abu-Faddan, N.H.; Daef, E.A.; Busby, S.J.W.; Browning, D.F. Antimicrobial resistance and comparative genome analysis of Klebsiella pneumoniae strains isolated in Egypt. Microorganisms 2021, 9, 1880. [Google Scholar] [CrossRef]
- Corbella, M.; Caltagirone, M.; Gaiarsa, S.; Mariani, B.; Sassera, D.; Bitar, I.; Muzzi, A.; Migliavacca, R.; Scudeller, L.; Stronati, M.; et al. Characterization of an outbreak of extended-spectrum β-lactamase-producing Klebsiella pneumoniae in a Neonatal Intensive Care Unit in Italy. Microb. Drug Resist. 2018, 24, 1128–1136. [Google Scholar] [CrossRef] [PubMed]
- Haller, S.; Kramer, R.; Becker, K.; Bohnert, J.A.; Eckmanns, T.; Hans, J.B.; Hecht, J.; Heidecke, C.-D.; Hübner, N.-O.; Kramer, A.; et al. Extensively drug-resistant Klebsiella pneumoniae ST307 outbreak, north-eastern Germany, June to October 2019. Eurosurveillance 2019, 24, 1900734. [Google Scholar] [CrossRef]
- Gupta, A. Hospital-acquired infections in the neonatal intensive care unit—Klebsiella pneumoniae. Semin. Perinatol. 2002, 26, 340–345. [Google Scholar] [CrossRef]
- Garrett, W.S.; Gallini, C.A.; Yatsunenko, T.; Michaud, M.; DuBois, A.; Delaney, M.L.; Punit, S.; Karlsson, M.; Bry, L.; Glickman, J.N.; et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 2010, 8, 292–300. [Google Scholar] [CrossRef]
- Pope, J.L.; Yang, Y.; Newsome, R.C.; Sun, W.; Sun, X.; Ukhanova, M.; Neu, J.; Issa, J.-P.; Mai, V.; Jobin, C. Microbial colonization coordinates the pathogenesis of a Klebsiella pneumoniae infant isolate. Sci. Rep. 2019, 9, 3380. [Google Scholar] [CrossRef]
- Bor, M.; Ilhan, O. Carbapenem-resistant Klebsiella pneumoniae outbreak in a Neonatal Intensive Care Unit: Risk factors for mortality. J. Trop. Pediatr. 2020, 67, fmaa057. [Google Scholar] [CrossRef]
- Chakkarapani, A.; Ninan, B.; Sekar, U.; Amboiram, P.; Balakrishnan, U. Pattern and antimicrobial susceptibility of carbapenem resistant organisms in tertiary care Neonatal Intensive Care Unit, India. J. Clin. Neonatol. 2014, 3, 200–204. [Google Scholar] [CrossRef]
- Viswanathan, R.; Singh, A.K.; Mukherjee, S.; Mukherjee, R.; DAS, P.; Basu, S. An outbreak of neonatal sepsis presenting with exanthematous rash caused by Klebsiella pneumoniae. Epidemiol. Infect. 2011, 139, 226–228. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, G.; Ma, X.; Yang, Q.; Yi, J. Outbreak of colonization by carbapenemase-producing Klebsiella pneumoniae in a Neonatal Intensive Care Unit: Investigation, control measures and assessment. Am. J. Infect. Control 2015, 43, 1122–1124. [Google Scholar] [CrossRef]
- Nordberg, V.; Jonsson, K.; Giske, C.G.; Iversen, A.; Aspevall, O.; Jonsson, B.; Camporeale, A.; Norman, M.; Navér, L. Neonatal intestinal colonization with extended-spectrum β-lactamase-producing Enterobacteriaceae—A 5-year follow-up study. Clin. Microbiol. Infect. 2018, 24, 1004–1009. [Google Scholar] [CrossRef] [PubMed]
- Stapleton, P.J.M.; Murphy, M.; McCallion, N.; Brennan, M.; Cunney, R.; Drew, R.J. Outbreaks of extended spectrum beta-lactamase-producing Enterobacteriaceae in Neonatal Intensive Care Units: A systematic review. Arch. Dis. Child. 2016, 101, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Flannery, D.D.; Chiotos, K.; Gerber, J.S.; Puopolo, K.M. Neonatal multidrug-resistant gram-negative infection: Epidemiology, mechanisms of resistance, and management. Pediatr. Res. 2022, 91, 380–391. [Google Scholar] [CrossRef]
- Hubbard, A.T.M.; Mason, J.; Roberts, P.; Parry, C.M.; Corless, C.; van Aartsen, J.; Howard, A.; Bulgasim, I.; Fraser, A.J.; Adams, E.R.; et al. Piperacillin/tazobactam resistance in a clinical isolate of Escherichia coli due to IS26-mediated amplification of blaTEM-1B. Nat. Commun. 2020, 11, 4915. [Google Scholar] [CrossRef]
- Fasciana, T.; Gentile, B.; Aquilina, M.; Ciammaruconi, A.; Mascarella, C.; Anselmo, A.; Fortunato, A.; Fillo, S.; Petralito, G.; Lista, F.; et al. Co-existence of virulence factors and antibiotic resistance in new Klebsiella pneumoniae clones emerging in south of Italy. BMC Infect. Dis. 2019, 19, 928. [Google Scholar] [CrossRef]
- Åttman, E.; Korhonen, P.; Tammela, O.; Vuento, R.; Aittoniemi, J.; Syrjänen, J.; Mattila, E.; Österblad, M.; Huttunen, R. A Serratia marcescens outbreak in a Neonatal Intensive Care Unit was successfully managed by rapid hospital hygiene interventions and screening. Acta Paediatr. 2018, 107, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Hashemizadeh, Z.; Hosseinzadeh, Z.; Azimzadeh, N.; Motamedifar, M. Dissemination pattern of multidrug resistant carbapenemase producing Klebsiella pneumoniae isolates using Pulsed-Field Gel Electrophoresis in Southwestern Iran. Infect. Drug Resist. 2020, 13, 921–929. [Google Scholar] [CrossRef]
- Frenk, S.; Rakovitsky, N.; Temkin, E.; Schechner, V.; Cohen, R.; Kloyzner, B.S.; Schwaber, M.J.; Solter, E.; Cohen, S.; Stepansky, S.; et al. Investigation of outbreaks of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in three Neonatal Intensive Care Units using whole genome sequencing. Antibiotics 2020, 9, 705. [Google Scholar] [CrossRef] [PubMed]
- Dorota, P.; Chmielarczyk, A.; Katarzyna, L.; Piotr, M.; Jan, L.; Renata, R.; Mach Jadwiga, W. Klebsiella pneumoniae in breast milk—A cause of sepsis in neonate. Arch. Med. 2017, 9, 1–4. [Google Scholar] [CrossRef]
- Adegboye, M.B.; Zakari, S.; Ahmed, B.A.; Olufemi, G.H. Knowledge, awareness and practice of infection control by health care workers in the intensive care units of a tertiary hospital in Nigeria. Afr. Health Sci. 2018, 18, 72–78. [Google Scholar] [CrossRef]
- Basso, M.; Zago, D.; Pozzetto, I.; De Canale, E.; Scaggiante, R.; Biasolo, M.A.; Peracchi, M.; Onelia, F.; Baldasso, E.; Palù, G.; et al. Intra-hospital acquisition of colonization and infection by Klebsiella pneumoniae strains producing carbapenemases and carriage evolution: A longitudinal analysis in an Italian teaching hospital from January 2017 to August 2019. Int. J. Infect. Dis. 2020, 92, 81–88. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 9.0. Available online: http://www.eucast.org (accessed on 1 July 2022).
- Liu, Y.; Liu, C.; Zheng, W.; Zhang, X.; Yu, J.; Gao, Q.; Hou, Y.; Huang, X. PCR detection of Klebsiella pneumoniae in infant formula based on 16S-23S internal transcribed spacer. Int. J. Food Microbiol. 2008, 125, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Nahar, N.; Bin Rashid, R. Phylogenetic analysis of antibiotic resistance genes and virulence genes of Klebsiella species in silico. Dhaka Univ. J. Pharm. Sci. 2017, 16, 119–127. [Google Scholar] [CrossRef][Green Version]
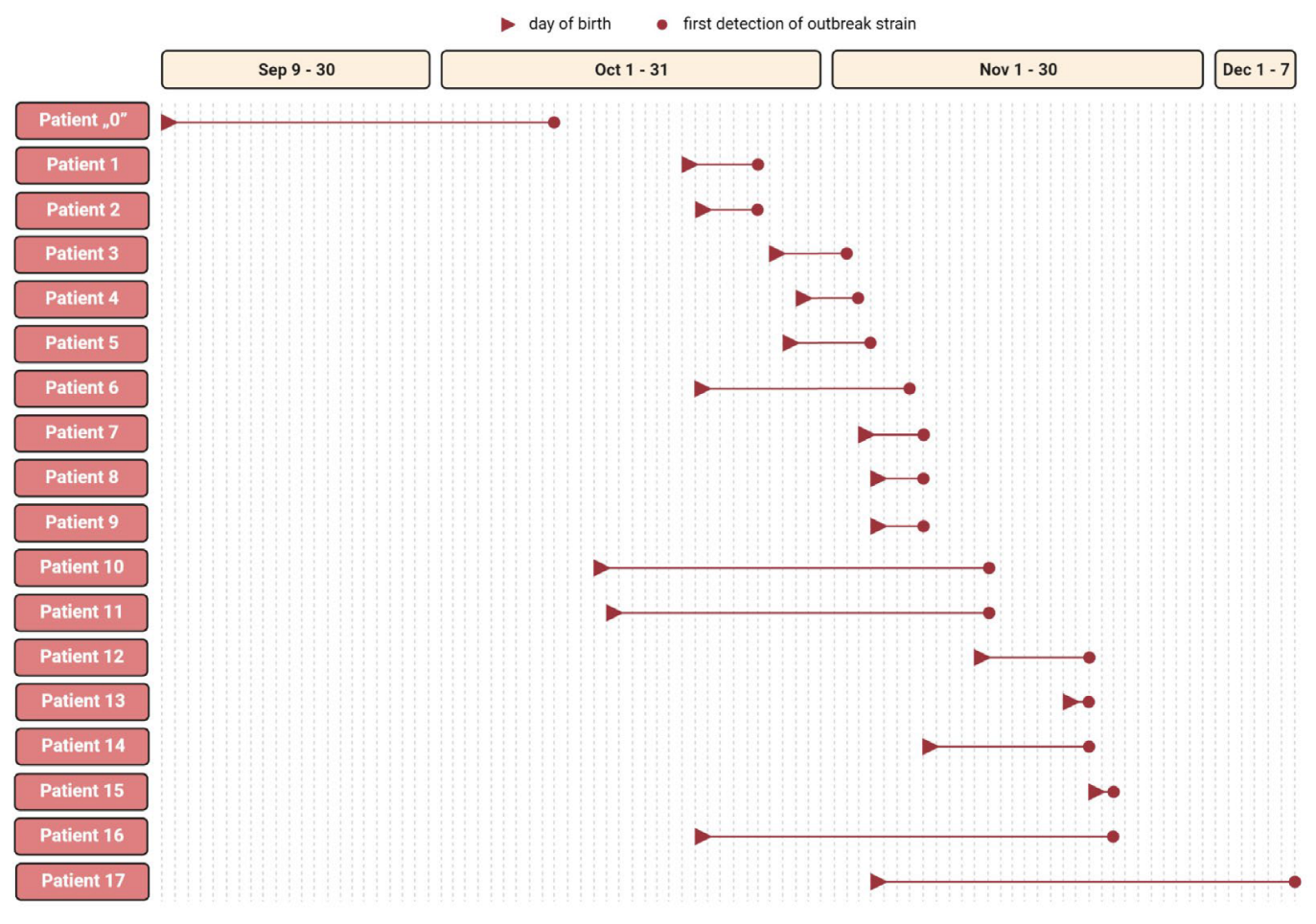
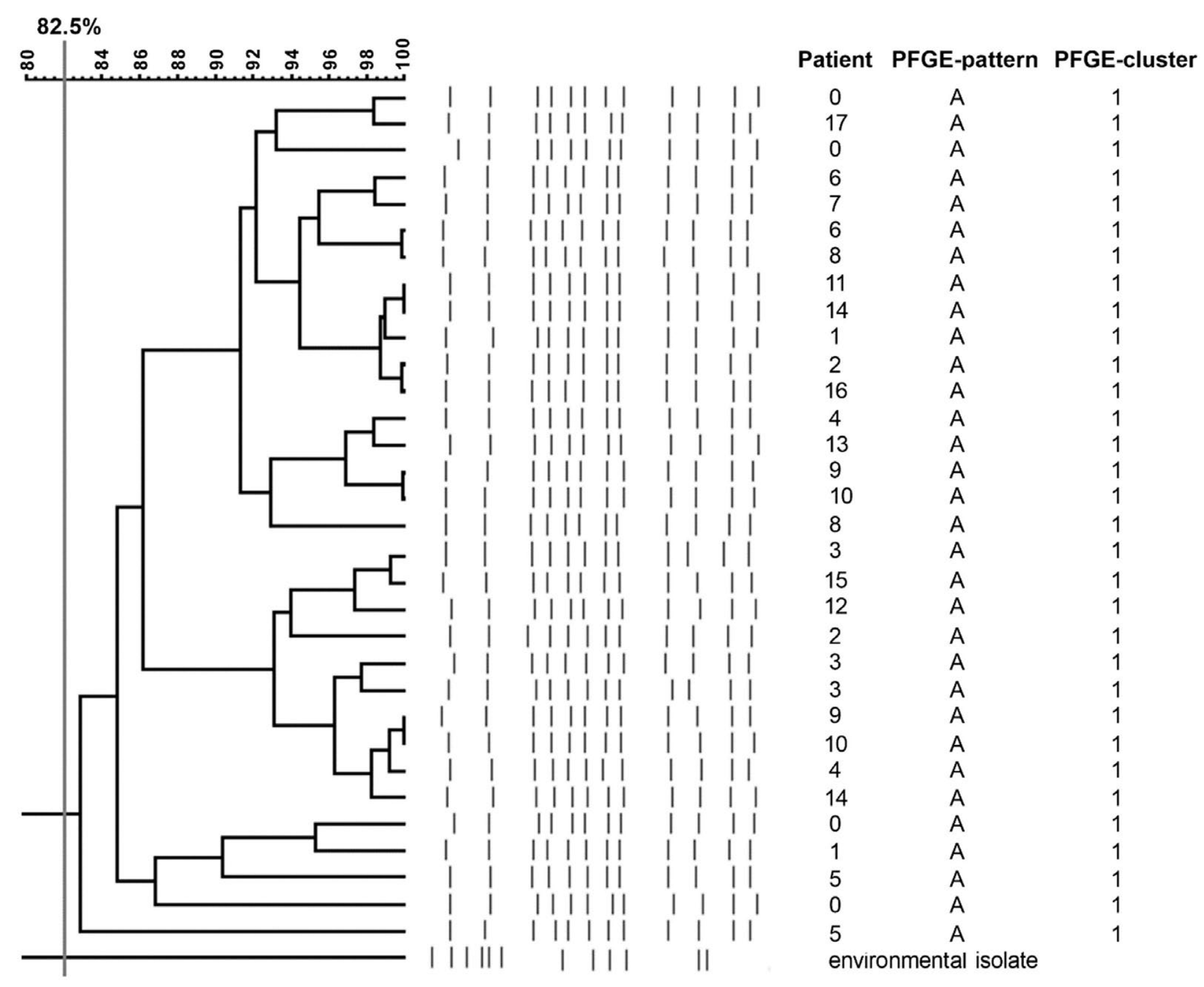
| Patient No. | Date of Birth | Date of 1. ESBL-KP Isolation | Sex | Pregnancy | Childbirth | HBD | Apgar Score | Birth Weight (g) | Assisted Ventilation | Antibiotic Therapy | Colonization Specimen | Infection Specimen | Clinical Diagnosis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| „0” | 9 Sep | 10 Oct | M | C | CS | 38 | 10,10,10 | 2930 | no | yes | OC,R | - | PNA |
| 1. | 20 Oct | 26 Oct | M | N | CS | 38 | 6,7,7 | 4200 | yes | yes | R | CON | CONS, PNA |
| 2. | 21 Oct | 26 Oct | M | N | NAT | 37 | 9,9,9 | 3130 | yes | yes | R | B,U,CSF | PNA, UROSEP, MEN |
| 3. | 27 Oct | 2 Nov | M | N | CS | 35 | 6,8,9 | 1880 | no | yes | R | B | SEP |
| 4. | 29 Oct | 3 Nov | F | C | CS | 33 | 8,8,8 | 2230 | no | yes | OC,R | U | UTI |
| 5. | 28 Oct | 4 Nov | F | N | CS | 37 | 8,8,8 | 2000 | no | no | OC,R | - | COL |
| 6. | 21 Oct | 7 Nov | F | C | NAT | 30 | 7,8,9 | 1460 | yes | yes | R | B | SEP,PNA |
| 7. | 3 Nov | 8 Nov | M | C | CS | 35 | 10,10,10 | 1700 | no | yes | OC,R | - | PNA |
| 8. | 4 Nov | 8 Nov | M | C | CS | 39 | 10,10,10 | 3400 | yes | yes | R | - | PNA |
| 9. | 4 Nov | 8 Nov | F | C | CS | 35 | 9,9,10 | 2450 | no | no | OC,R | - | COL |
| 10. | 13 Oct | 13 Nov | F | C | CS | 28 | 2,3,5 | 1080 | yes | yes | R | - | PNA |
| 11. | 14 Oct | 13 Nov | M | N | CS | 29 | 6,6,7 | 1250 | yes | yes | R | - | PNA |
| 12. | 12 Nov | 21 Nov | F | C | NAT | 40 | 10,10,10 | 3470 | no | no | R | - | COL |
| 13. | 19 Nov | 21 Nov | F | C | CS | 39 | 10,10,10 | 2300 | no | no | R | - | COL |
| 14. | 8 Nov | 21 Nov | M | C | CS | 35 | 9,9,9 | 3080 | yes | yes | OC,R | - | PNA |
| 15. | 21 Nov | 23 Nov | M | C | CS | 39 | 8,9,10 | 2800 | no | no | R | - | COL |
| 16. | 21 Oct | 23 Nov | M | C | CS | 39 | 8,9,10 | 4110 | no | no | R | - | COL |
| 17. | 4 Nov | 7 Dec | M | N | NAT | 38 | 8,8,8 | 3180 | no | yes | R | U | UTI |
| Variable | Descriptive Statistic | Colonized (n = 6) | Infected (n = 12) | p-Value (Univariate) |
|---|---|---|---|---|
| Gender (male) | n (%) | 2 (33.3) | 9 (75.0) | 0.232 |
| Birth weight (g) | median (IQR) | 2625 (2338–3303) | 2580 (1640–3143) | 0.437 |
| Gestational age (week) | median (IQR) | 39 (37.5–39) | 35 (32–38) | 0.022 |
| Apgar score | median (IQR) | 9.5 (8–10) | 8 (7–9) | 0.016 |
| Gestational course (complicated) | n (%) | 5 (83.3) | 7 (58.3) | 0.596 |
| Cesarean section (yes) | n (%) | 5 (83.3) | 9 (75.0) | 0.841 |
| Assisted ventilation | n (%) | 0 (0.0) | 7 (58.3) | 0.060 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pruss, A.; Kwiatkowski, P.; Masiuk, H.; Bilska, I.; Giedrys-Kalemba, S.; Dołęgowska, B. Epidemiological Analysis of Extended-Spectrum β-Lactamase-Producing Klebsiella pneumoniae Outbreak in a Neonatal Clinic in Poland. Antibiotics 2023, 12, 50. https://doi.org/10.3390/antibiotics12010050
Pruss A, Kwiatkowski P, Masiuk H, Bilska I, Giedrys-Kalemba S, Dołęgowska B. Epidemiological Analysis of Extended-Spectrum β-Lactamase-Producing Klebsiella pneumoniae Outbreak in a Neonatal Clinic in Poland. Antibiotics. 2023; 12(1):50. https://doi.org/10.3390/antibiotics12010050
Chicago/Turabian StylePruss, Agata, Paweł Kwiatkowski, Helena Masiuk, Iwona Bilska, Stefania Giedrys-Kalemba, and Barbara Dołęgowska. 2023. "Epidemiological Analysis of Extended-Spectrum β-Lactamase-Producing Klebsiella pneumoniae Outbreak in a Neonatal Clinic in Poland" Antibiotics 12, no. 1: 50. https://doi.org/10.3390/antibiotics12010050
APA StylePruss, A., Kwiatkowski, P., Masiuk, H., Bilska, I., Giedrys-Kalemba, S., & Dołęgowska, B. (2023). Epidemiological Analysis of Extended-Spectrum β-Lactamase-Producing Klebsiella pneumoniae Outbreak in a Neonatal Clinic in Poland. Antibiotics, 12(1), 50. https://doi.org/10.3390/antibiotics12010050









