Predictive Score for Carbapenem-Resistant Gram-Negative Bacilli Sepsis: Single-Center Prospective Cohort Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients, Setting, Study Design, and Methodology
2.2. Database, General Statistical Analyses, and Risk Score Development
3. Results
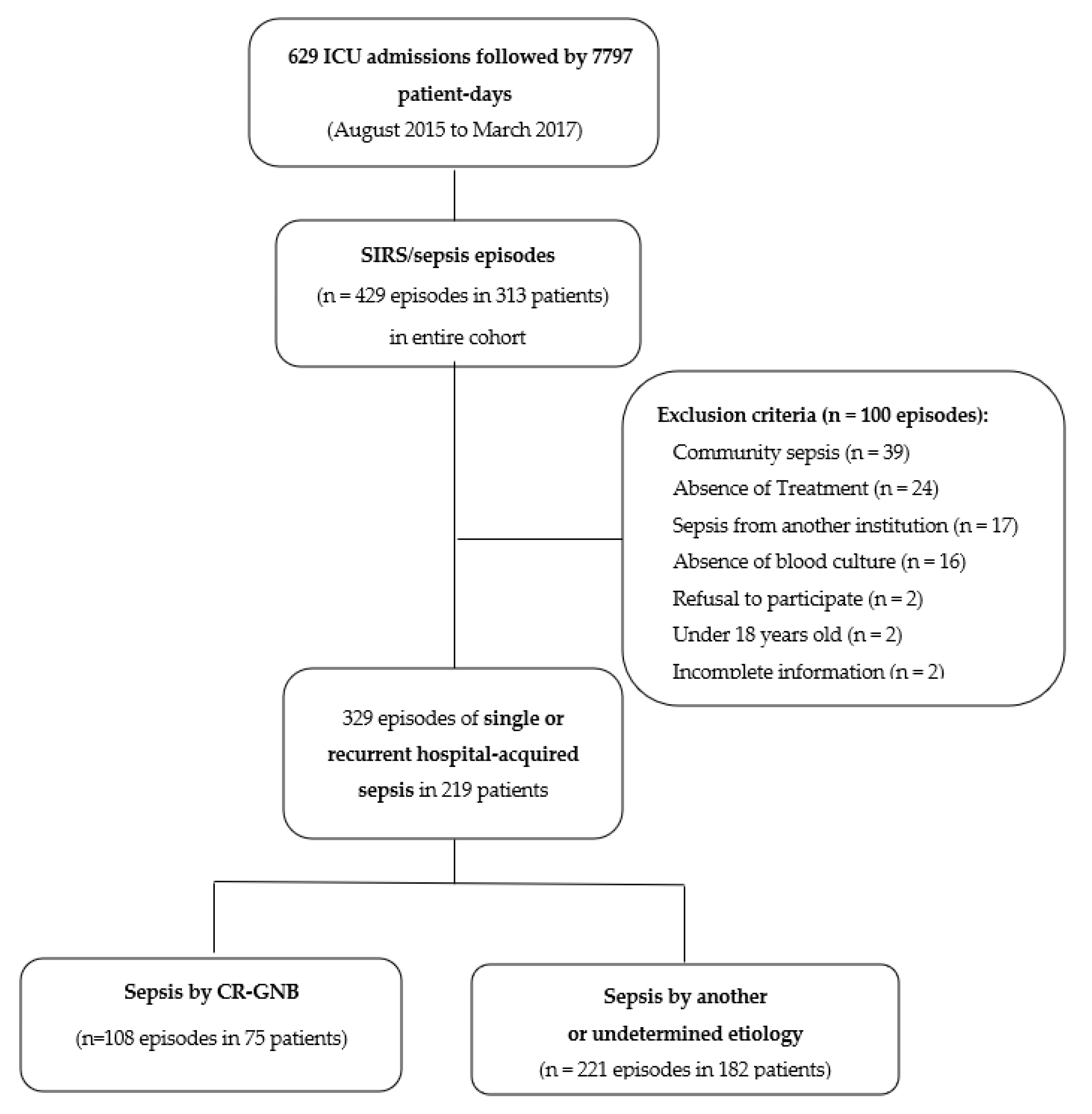
3.1. Patients and Episodes of Sepsis
3.2. Etiology and Infectious Focus of Sepsis
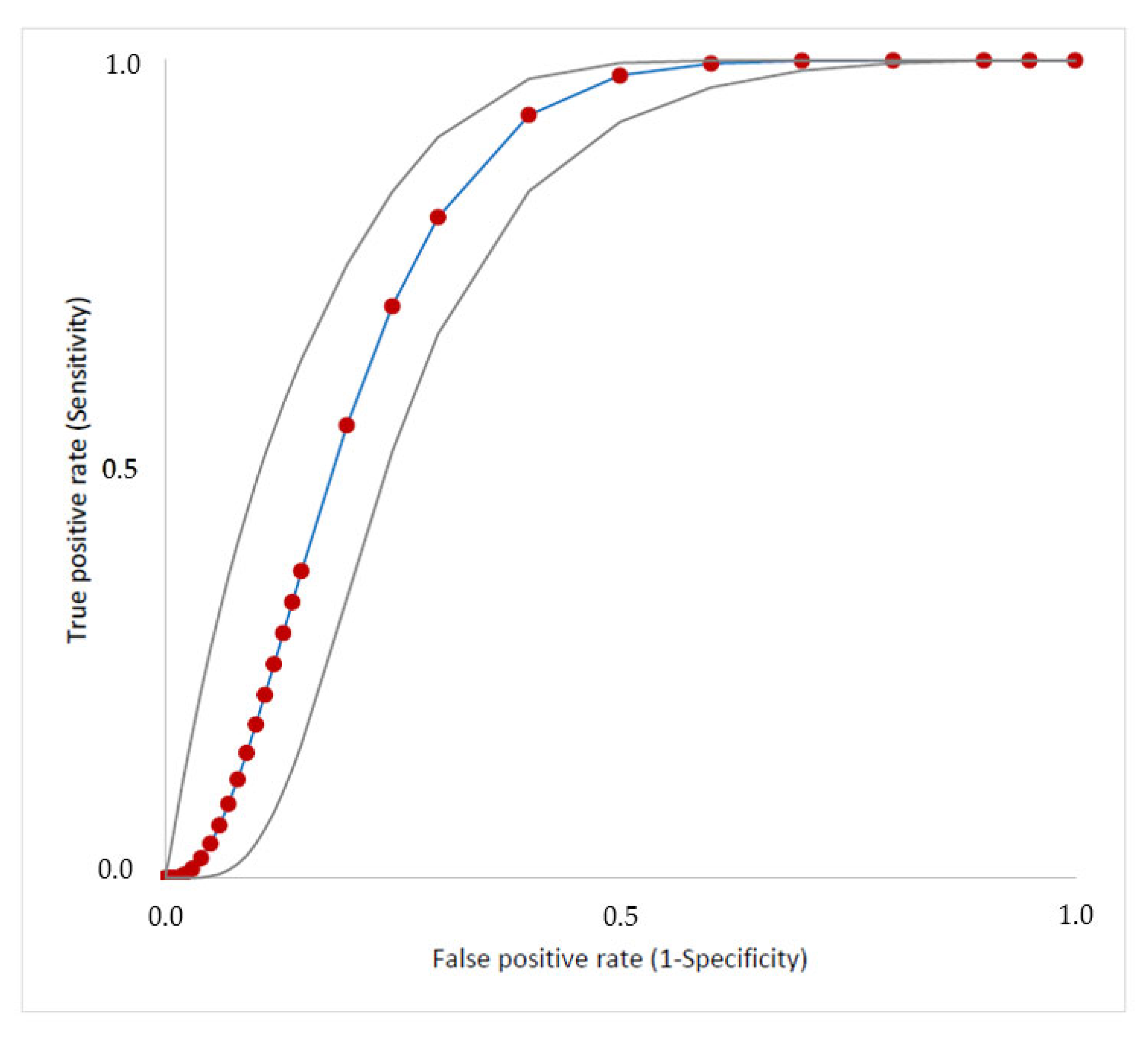
3.3. Performance of the Clinical–Epidemiological Score to Guide EAT
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Evans, L.; Rhodes, A.; Alhazzani, W.; Antonelli, M.; Coopersmith, C.M.; French, C.; Machado, F.R.; Mcintyre, L.; Ostermann, M.; Prescott, H.C.; et al. Surviving sepsis campaign: International guidelines for management of sepsis and septic shock 2021. Intensive Care Med. 2021, 47, 1181–1247. [Google Scholar] [CrossRef]
- Niederman, M.S.; Baron, R.M.; Bouadma, L.; Calandra, T.; Daneman, N.; DeWaele, J.; Kollef, M.H.; Lipman, J.; Nair, G.B. Initial antimicrobial management of sepsis. Crit. Care 2021, 25, 307. [Google Scholar] [CrossRef]
- Bassetti, M.; Peghin, M.; Vena, A.; Giacobbe, D.R. Treatment of Infections Due to MDR Gram-Negative Bacteria. Front. Med. 2019, 6, 74. [Google Scholar] [CrossRef]
- Doi, Y. Treatment Options for Carbapenem-resistant Gram-negative Bacterial Infections. Clin. Infect. Dis. 2019, 69, S565–S575. [Google Scholar] [CrossRef]
- Lima, E.M.; Cid, P.A.; Beck, D.S.; Pinheiro, L.H.Z.; Tonhá, J.P.S.; Alves, M.Z.O.; Lourenço, N.D.; Santos, R.Q.; Asensi, M.D.; Marques, J.A.; et al. Predictive factors for sepsis by carbapenem resistant Gram-negative bacilli in adult critical patients in Rio de Janeiro: A case-case-control design in a prospective cohort study. Antimicrob. Resist. Infect. Contol. 2020, 9, 132. [Google Scholar] [CrossRef]
- Nordmann, P.; Naas, T.; Poirel, L. Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2011, 17, 1791–1798. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clin. Infect. Dis. 2019, 69, S521–S528. [Google Scholar] [CrossRef]
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorlí, L.; Luque, S.; Gómez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and Treatment of Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa Infections. Clin. Microbiol. Rev. 2019, 32. [Google Scholar] [CrossRef]
- Kakoullis, L.; Papachristodoulou, E.; Chra, P.; Panos, G. Mechanisms of Antibiotic Resistance in Important Gram-Positive and Gram-Negative Pathogens and Novel Antibiotic Solutions. Antibiotics 2021, 10, 415. [Google Scholar] [CrossRef]
- Chen, I.L.; Lee, C.H.; Ting, S.W.; Wang, L.Y. Prediction of imipenem-resistant microorganisms among the nosocomial critically ill patients with Gram-negative bacilli septicemia: A simple risk score. Infect. Drug Resist. 2018, 11, 283–293. [Google Scholar] [CrossRef]
- Lambregts, M.M.C.; Hendriks, B.J.C.; Visser, L.G.; Bernards, S.T.; de Boer, M.G.J. Using local clinical and microbiological data to develop an institution specific carbapenem-sparing strategy in sepsis: A nested case-control study. Antimicrob. Resist. Infect. Control. 2019, 8, 19. [Google Scholar] [CrossRef]
- Falcone, M.; Tiseo, G.; Dentali, F.; La Regina, M.; Foglia, E.; Gambacorta, M.; Garagiola, E.; Bonardi, G.; Clerici, P.; Colombo, F.; et al. Predicting resistant etiology in hospitalized patients with blood cultures positive for Gram-negative bacilli. Eur. J. Inter. Med. 2018, 53, 21–28. [Google Scholar] [CrossRef]
- Levy, M.M.; Fink, M.P.; Marshall, J.C.; Abraham, E.; Angus, D.; Cook, D.; Cohen, J.; Opal, S.M.; Jean-Louis Vincent, J.-L.; Ramsay, G.; et al. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit. Care Med. 2003, 31, 1250–1256. [Google Scholar] [CrossRef]
- von Elm, E.; Altman, D.G.; Egger, M.; Pocock, S.J.; Gøtzsche, P.C.; Vandenbroucke, J.P.; STROBE Initiative. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: Guidelines for reporting observational studies. J. Clin. Epidemiol. 2008, 4, e296. [Google Scholar] [CrossRef]
- Singer, M.; Deutschman, C.S.; Seymour, C.M.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.-D.; Craig, M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef]
- Klein Klouwenberg, P.M.; Ong, D.S.Y.; Bos, L.D.J.; de Beer, F.M.; van Hooijdonk, R.T.M.; Huson, M.A.; Straat, M.; van Vught, L.A.; Wieske, L.; Horn, J.; et al. Interobserver agreement of Centers for Disease Control and Prevention criteria for classifying infections in critically ill patients. Crit. Care Med. 2013, 41, 2373–2378. [Google Scholar] [CrossRef]
- CDC. National Healthcare Safety Network (NHSN) Patient Safety Component Manual. Available online: https://www.cdc.gov/nhsn/pdfs/pscmanual/pcsmanual_current.pdf (accessed on 18 June 2018).
- Harris, P.A.; Taylor, R.; Minor, B.L.; Elliott, V.; Fernandez, M.; O’Neal, L.; McLeod, L.; Delacqua, G.; Delacqua, F.; Kirby, J.; et al. The REDCap consortium: Building an international community of software platform partners. J. Biomed. Inform. 2019, 95, 103208. [Google Scholar] [CrossRef]
- Rossner, B. Fundamentals of Biostatistics, 8th ed.; Mason Cengage Learning: Belmont, CA, USA, 2015. [Google Scholar]
- Lasko, T.A.; Bhagwat, J.G.; Zou, K.H.; Ohno-Machado, L. The use of receiver operating characteristic curves in biomedical informatics. J. Biomed. Inform. 2005, 38, 404–415. [Google Scholar] [CrossRef]
- Harris, A.D.; Karchmer, T.B.; Carmeli, Y.; Samore, M.H. Methodological principles of case-control studies that analyzed risk factors for antibiotic resistance: A systematic review. Clin. Infect. Dis. 2001, 32, 1055–1061. [Google Scholar] [CrossRef]
- Mehta, H.B.; Mehta, V.; Girman, C.J.; Adhikari, D.; Johnson, M.L. Regression coefficient-based scoring system should be used to assign weights to the risk index. J. Clin. Epidemiol. 2016, 79, 22–28. [Google Scholar] [CrossRef]
- Mendes, D.; Alves, C.; Batel-Marques, F. Number needed to treat (NNT) in clinical literature: An appraisal. BMC Med. 2017, 15, 112. [Google Scholar] [CrossRef]
- CDC. Antibiotic Resistance Threats in the United States, 2019; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019. Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed on 10 August 2022).
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance. 2014. Available online: https://apps.who.int/iris/handle/10665/112642. (accessed on 10 August 2022).
- World Bank. Drug-Resistant Infections: A Threat to Our Economic Future; World Bank: Washington, DC, USA, 2017; Available online: https://openknowledge.worldbank.org/handle/10986/26707 (accessed on 10 August 2022).
- Kumar, A.; Roberts, D.; Wood, K.E.; Light, B.; Parrillo, J.E.; Sharma, S.; Suppes, R.; Feinstein, D.; Zanotti, S.; Taiberg, L.; et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 2006, 34, 1589–1596. [Google Scholar] [CrossRef]
- Seymour, C.W.; Liu, V.X.; Iwashyna, T.J.; Brunkhorst, F.M.; Rea, T.D.; Scherag, A.; Rubenfeld, G.; Kahn, J.M.; Shankar-Hari, M.; Singer, M.; et al. Assessment of Clinical Criteria for Sepsis: For the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 762–774. [Google Scholar] [CrossRef]
- Giamarellos-Bourboulis, E.J.; Tsaganos, T.; Tsangaris, I.; M Lada, M.; Routsi, C.; Sinapidis, D.; Koupetori, M.; Bristianou, M.; Adamis, G.; Mandragos, K.; et al. Validation of the new Sepsis-3 definitions: Proposal for improvement in early risk identification. Clin. Microbiol. Infect. 2017, 23, 104–109. [Google Scholar] [CrossRef]
- Ferreira-Coimbra, J.; Ferreira-Coimbra, J.; Ardanuy, C.; Diaz, E.; Leone, M.; De Pascale, G.; Póvoa, P.; Prat-Aymerich, C.; Serrano-Garcia, R.; Solé-Violan, J.; et al. Ventilator-associated pneumonia diagnosis: A prioritization exercise based on multi-criteria decision analysis. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 281–286. [Google Scholar] [CrossRef]
- Rea-Neto, A.; Youssef, N.C.M.; Tuche, F.; Brunkhorst, F.; Marco Ranieri, V.; Reinhart, K.; Sakr, Y. Diagnosis of ventilator-associated pneumonia: A systematic review of the literature. Crit. Care 2008, 12, R56. [Google Scholar] [CrossRef]
- Dantas, L.F.; Dalmas, B.; Andrade, R.M.; Hamacher, S.; Bozza, F.A. Predicting acquisition of carbapenem-resistant Gram-negative pathogens in intensive care units. J. Hosp. Infect. 2013, 103, 121–127. [Google Scholar] [CrossRef]
- Lee, C.-H.; Su, T.-Y.; Ye, J.-J.; Hsu, P.-C.; Kuo, A.J.; Chia, J.H.; Lee, M.-H. Risk factors and clinical significance of bacteremia caused by Pseudomonas aeruginosa resistant only to carbapenems. J. Microbiol. Immunol. Infect. 2017, 50, 677–683. [Google Scholar] [CrossRef]
- Nicolas-Chanoine, M.H.; Vigan, M.; Laouénan, C.; Robert, J. Risk factors for carbapenem-resistant Enterobacteriaceae infections: A French case-control-control study. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 383–393. [Google Scholar] [CrossRef]
- Palacios-Baena, Z.R.; Giannella, M.; Manissero, D.; Rodríguez-Baño, J.; Viale, P.; Lopes, S.; Wilson, K.; McCool, R.; Longshaw, C. Risk factors for carbapenem-resistant Gram-negative bacterial infections: A systematic review. Clin. Microbiol. Infect. 2021, 27, 228–235. [Google Scholar] [CrossRef]
- Trung, N.T.; Thau, N.S.; Bang, M.H.; Song, L.H. PCR-based Sepsis@Quick test is superior in comparison with blood culture for identification of sepsis-causative pathogens. Sci. Rep. 2019, 9, 13663. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Girardello, R.; Cury, A.P.; Di Gioia, T.S.R.; de Almeida , J.N., Jr.; da Silva Duarte, A.J. Emergence of colistin resistance in the largest university hospital complex of Sao Paulo, Brazil, over five years. Braz. J. Infect. Dis. 2017, 21, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Vena, A.; Giacobbe, D.R.; Castaldo, N.; Cattelan, A.; Mussini, C.; Luzzati, R.; De Rosa, F.G.; Del Puente, F.; Mastroianni, C.M.; Cascio, A.; et al. Clinical Experience with Ceftazidime-Avibactam for the Treatment of Infections due to Multidrug-Resistant Gram-Negative Bacteria Other than Carbapenem-Resistant Enterobacterales. Antibiotics 2020, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Monti, G.L.G.; Taddeo, D.; Isella, F.; Zangrillo, A. Clinical Aspects of Sepsis: An Overview. In Sepsis Diagnositic Methods and Protocols; Walker, J., Ed.; Humana Press—Springer Science: London, UK, 2015; pp. 17–33. [Google Scholar]
- Lima, E.M. Fatores Preditivos para Sepse por Bacilos Gram-Negativos Resistentes aos Carbapenemas em Pacientes Adultos Críticos do Rio de Janeiro; Master Science, Instituto Oswaldo Cruz, Fundação Oswaldo Cruz: Rio de Janeiro, Brazil, 2017. [Google Scholar]

| CR-GNB Sepsis (n = 75) | Non-CR-GNB Sepsis (n = 182) | |
|---|---|---|
| Demographic data | ||
| Age in years, median (range) | 62 (23–91) | 63 (19–92) |
| Male, n (%) | 39 (52) | 72 (40) |
| Prior ICU admission, n (%) | 17 (23) | 30 (16) |
| ICU admission reason, n (%) | ||
| Infection | 53 (71) | 94 (52) |
| Sepsis | 41 (55) | 68 (37) |
| Respiratory failure | 32 (43) | 58 (32) |
| Septic shock | 29 (39) | 59 (32) |
| Respiratory disease | 20 (27) | 48 (26) |
| Surgery | 17 (23) | 60 (33) |
| Elective surgery | 11 (15) | 35 (19) |
| Urgent surgery | 6 (8) | 25 (14) |
| Cardiovascular disease | 12 (16) | 23 (13) |
| Renal and urinary tract disease | 10 (13) | 23 (13) |
| Gastrointestinal or intrabdominal disease | 9 (12) | 20 (11) |
| Neurological disease | 3 (4) | 19 (10) |
| Charlson comorbidity index a, median (range) | 2 (0–19) | 2 (0–11) |
| Total SOFA score a, median (range) | 7 (1–17) | 7 (0–18) |
| SOFA parameters ≥2 points, % (n/valid)) | ||
| Cardiovascular | 56 (42/75) | 51 (92/181) |
| Respiratory | 45 (61/74) | 50 (89/178) |
| Renal | 38 (28/74) | 39 (70/180) |
| Neurological | 34 (21/61) | 28 (42/152) |
| Liver | 15 (10/68) | 16 (24/149) |
| Coagulation | 13 (10/75) | 16 (28/178) |
| SAPS 3 score a, median (range) | 64 (24-103) | 62 (24–114) |
| Mortality rate b, % (n) | ||
| 30 day mortality | 66 (42/64) | 44 (68/154) |
| Hospital mortality | 70 (45/64) | 55 (85/154) |
| Clinical Factors | CR-GNB Sepsis (n = 108) | Non-CR-GNB Sepsis (n = 221) | p-Value i |
|---|---|---|---|
| Comorbidity a, n (%) | |||
| Previous infection | 101 (94) | 117 (53) | <0.001 * |
| Renal failure | 57 (53) | 87 (39) | 0.02 * |
| CR-GNB Infection/Colonization | 52 (48) | 43 (20) | <0.001 * |
| Previous surgeries | 50 (46) | 122 (55) | 0.13 |
| Diabetes mellitus | 49 (45) | 87 (39) | 0.16 |
| Hemodialysis | 46 (43) | 57 (26) | 0.002 * |
| Nosocomial diarrhea b | 31 (29) | 21 (10) | <0.001 * |
| Neoplasia | 28 (26) | 79 (36) | 0.74 |
| Pulmonary disease | 27 (25) | 44 (20) | 0.29 |
| Gastrointestinal disease | 27 (25) | 51 (23) | 0.70 |
| Immunosuppressive condition c | 26 (24) | 34 (15) | 0.06 |
| Genitourinary disease | 9 (8) | 21 (10) | 0.73 |
| Neutropenia d | 6 (6) | 6 (3) | 0.20 |
| Chronic liver disease | 5 (5) | 25 (11) | 0.06 |
| AIDS or chronic infectious disease | 4 (4) | 11 (5) | 0.60 |
| Pregnancy | 4 (4) | 5 (2) | 0.45 |
| Invasive devices a, n (%) | |||
| Central vascular catheter | 108 (100) | 192 (87) | <0.001 * |
| Mechanical ventilation | 104 (96) | 149 (67) | <0.001 * |
| Indwelling urinary catheter | 97 (90) | 179 (81) | 0.04 * |
| Nutrition, n (%) | |||
| Enteral nutrition | 93 (87) | 147 (67) | <0.001 * |
| Parenteral nutrition | 69 (64) | 116 (53) | 0.05 |
| Length of hospital stay e, median (range) | 54 (4–292) | 27 (0–340) | <0.001 * |
| Length of ICUl stay e, median (range) | 33 (0–291) | 12 (0–339) | <0.001 |
| Previous antimicrobial use, n (%) | 108 (100) | 180 (81) | <0.001 * |
| Previous carbapenem use, n (%) | 95 (88) | 93 (42) | <0.001 * |
| Previous polymyxin use, n (%) | 62 (57) | 51 (23) | <0.001 * |
| SIRS f, n (%) | 106 (98) | 217 (99) | 0.34 |
| Total SOFA score g, n (%) | 8 (0-20) | 8 (0–18) | 0.60 |
| SOFA parameters ≥ 2 points, % (n/valid)) | |||
| Cardiovascular | 62 (66/107) | 60 (130/217) | 0.76 |
| Respiratory | 53 (56/106) | 53 (114/215) | 0,97 |
| Renal | 49 (52/107) | 45 (98/216) | 0.59 |
| Neurological | 32 (27/85) | 31 (55/180) | 0.83 |
| Liver | 20 (15/76) | 18 (30/163) | 0.80 |
| Coagulation | 15 (16/106) | 17 (37/212) | 0.61 |
| Delta SOFA h, n (%) | 46 (43) | 83 (39) | 0.64 |
| Type of Infection, n (%) | |||
| Ventilator-associated pneumonia | 51 (47) | 22 (10) | 0.001 * |
| Vascular catheter infection | 18 (17) | 31 (14) | 0.52 |
| Surgical infection | 17 (16) | 42 (19) | 0.47 |
| Non-ventilator hospital-acquired pneumonia | 14 (13) | 51 (23) | 0.04 * |
| Tracheobronchitis | 14 (13) | 14 (6) | 0.05 |
| Catheter-associated UTI | 6 (6) | 6 (3) | 0.20 |
| Sinusitis | 6 (6) | 5 (2) | 0.14 |
| Soft tissue infection | 3 (3) | 10 (5) | 0.47 |
| Intra-abdominal infection | 3 (3) | 7 (3) | 0.85 |
| Endocarditis | 2 (2) | 2 (1) | 0.50 |
| Mastoiditis | 2 (2) | 5 (2) | 0.85 |
| Non-catheter associated UTI | 1 (1) | 9 (4) | 0.12 |
| Osteomyelitis | 1 (1) | 4 (2) | 0.54 |
| Undetermined focus | 0 (0) | 43 (20) | - |
| Others | 0 (0) | 4 (2) j | - |
| Concurrent infection, n (%) | |||
| Bacterial | 21 (20) | 24 (11) | 0.03 * |
| Viral | 3 (3) | 9 (4) | 0.55 |
| Fungal | 7 (7) | 6 (3) | 0.10 |
| Serum PCR g (mg/dL), median (range) | 4.8 (1–31) | 4 (1–31) | 0.25 |
| Cutoff | Sensibility % (95% CI) | Specificity % (95% CI) | False Negative (n) | False Positive (n) | NPV % (95% CI) | PPV % (95% CI) | Youden’s Index 6 | Rate of Accuracy % (95% CI) |
|---|---|---|---|---|---|---|---|---|
| ≥1 | 100 (97–100) | 15 (11–20) | 0 | 188 | 100 (90–100) | 37 (31–42) | 0.149 | 43 (38–48) |
| ≥2 | 99 (95–100) | 31 (26–37) | 1 | 136 | 99 (94–100) | 36 (31–42) | 0.302 | 50 (45–55) |
| ≥3 | 92 (85–96) | 65 (58–71) | 9 | 78 | 94 (89–97) | 56 (49–63) | 0.564 | 74 (69–78) |
| 4 | 66 (56–74) | 76 (70–81) | 37 | 53 | 82 (76–87) | 57 (49–66) | 0.418 | 73 (68–77) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomes, M.Z.R.; Braga, D.Q.; Pinheiro, D.O.B.P.; Verduc, R.C.A.S.; dos Reis, L.V.; de Lima, E.M.; Lourenço, N.D.; Cid, P.A.; Beck, D.S.; Pinheiro, L.H.Z.; et al. Predictive Score for Carbapenem-Resistant Gram-Negative Bacilli Sepsis: Single-Center Prospective Cohort Study. Antibiotics 2023, 12, 21. https://doi.org/10.3390/antibiotics12010021
Gomes MZR, Braga DQ, Pinheiro DOBP, Verduc RCAS, dos Reis LV, de Lima EM, Lourenço ND, Cid PA, Beck DS, Pinheiro LHZ, et al. Predictive Score for Carbapenem-Resistant Gram-Negative Bacilli Sepsis: Single-Center Prospective Cohort Study. Antibiotics. 2023; 12(1):21. https://doi.org/10.3390/antibiotics12010021
Chicago/Turabian StyleGomes, Marisa Zenaide Ribeiro, Douglas Quintanilha Braga, Debora Otero Britto Passos Pinheiro, Renata Cristina Amorim Silveira Verduc, Letícia Vellozo dos Reis, Elisangela Martins de Lima, Newton Dias Lourenço, Patrícia Aquen Cid, Debora Souza Beck, Luiz Henrique Zanata Pinheiro, and et al. 2023. "Predictive Score for Carbapenem-Resistant Gram-Negative Bacilli Sepsis: Single-Center Prospective Cohort Study" Antibiotics 12, no. 1: 21. https://doi.org/10.3390/antibiotics12010021
APA StyleGomes, M. Z. R., Braga, D. Q., Pinheiro, D. O. B. P., Verduc, R. C. A. S., dos Reis, L. V., de Lima, E. M., Lourenço, N. D., Cid, P. A., Beck, D. S., Pinheiro, L. H. Z., Tonhá, J. P. S., de Sousa, L. S., Dias, M. L. S., da Silva Machado, A. A., Castro, M. M., Dutra, V. P. R., de Mello, L. S., da Silva, M. C., Tozo, T. M., ... dos Santos Gomes Junior, S. C., on behalf of the Nucleus of Hospital Research Study Collaborators. (2023). Predictive Score for Carbapenem-Resistant Gram-Negative Bacilli Sepsis: Single-Center Prospective Cohort Study. Antibiotics, 12(1), 21. https://doi.org/10.3390/antibiotics12010021





