Azithromycin Susceptibility Testing and Molecular Investigation of Neisseria gonorrhoeae Isolates Collected in Russia, 2020–2021
Abstract
1. Introduction
2. Results
2.1. Phenotypic Azithromycin Susceptibility and Analysis of Genetic Determinants Associated with Azithromycin Resistance
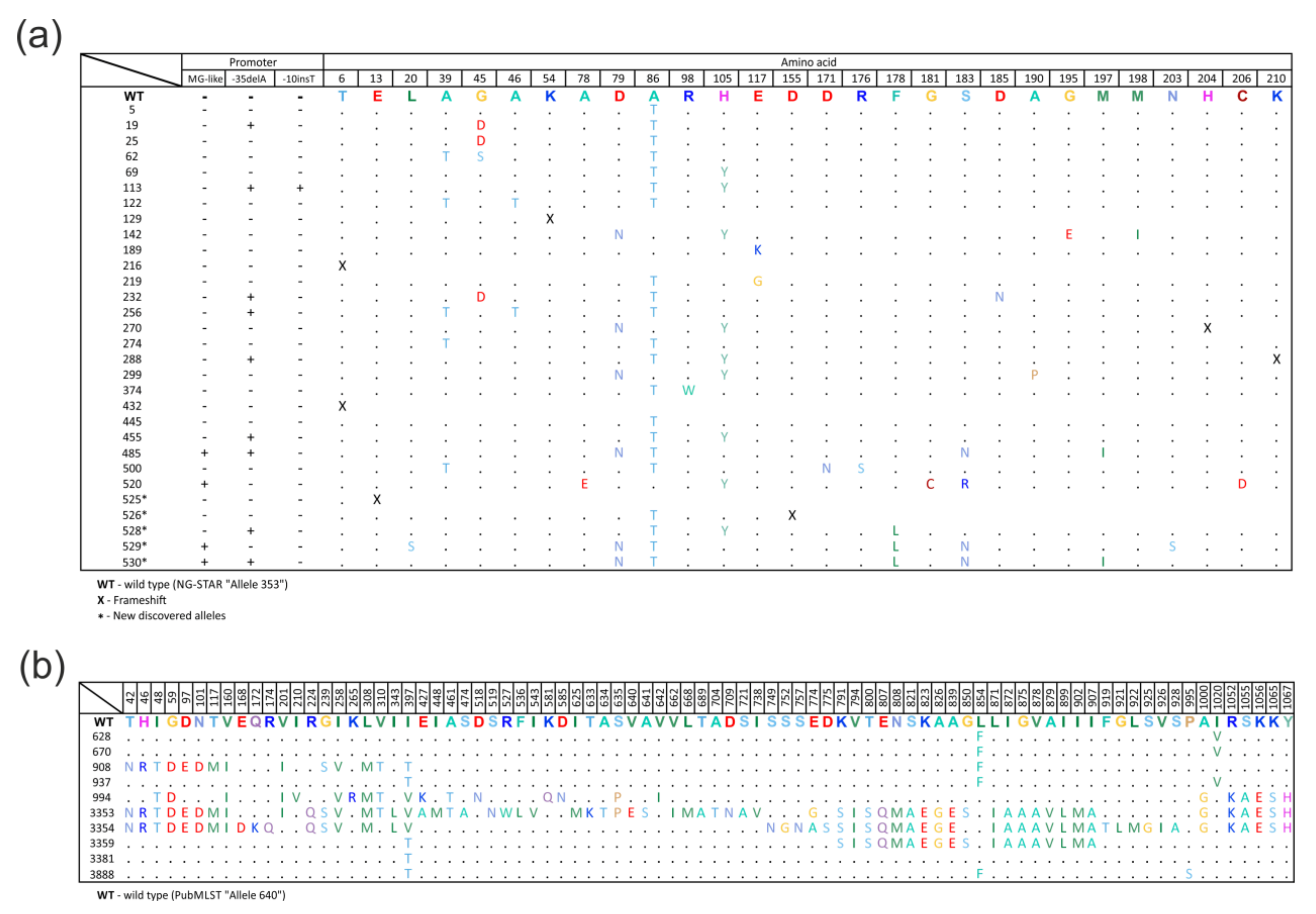
2.2. Diversity of the mtrR Gene Alleles
- allele 525—insertion of nucleotide A at position 38, resulting in a frameshift;
- allele 526—deletion of two GA nucleotides at position 465 of the gene, resulting in a frameshift;
- allele 528—T→G substitution at position 531, which does not lead to amino acid substitution;
- allele 529—T→C substitution at position 132, resulting in the Cys44Arg amino acid substitution;
- allele 530—T→G substitution at position 531, which does not lead to amino acid substitution, and T→C substitution at position 535, resulting in the Phe178Leu amino acid substitution.
2.3. Diversity of the mtrD Gene Alleles
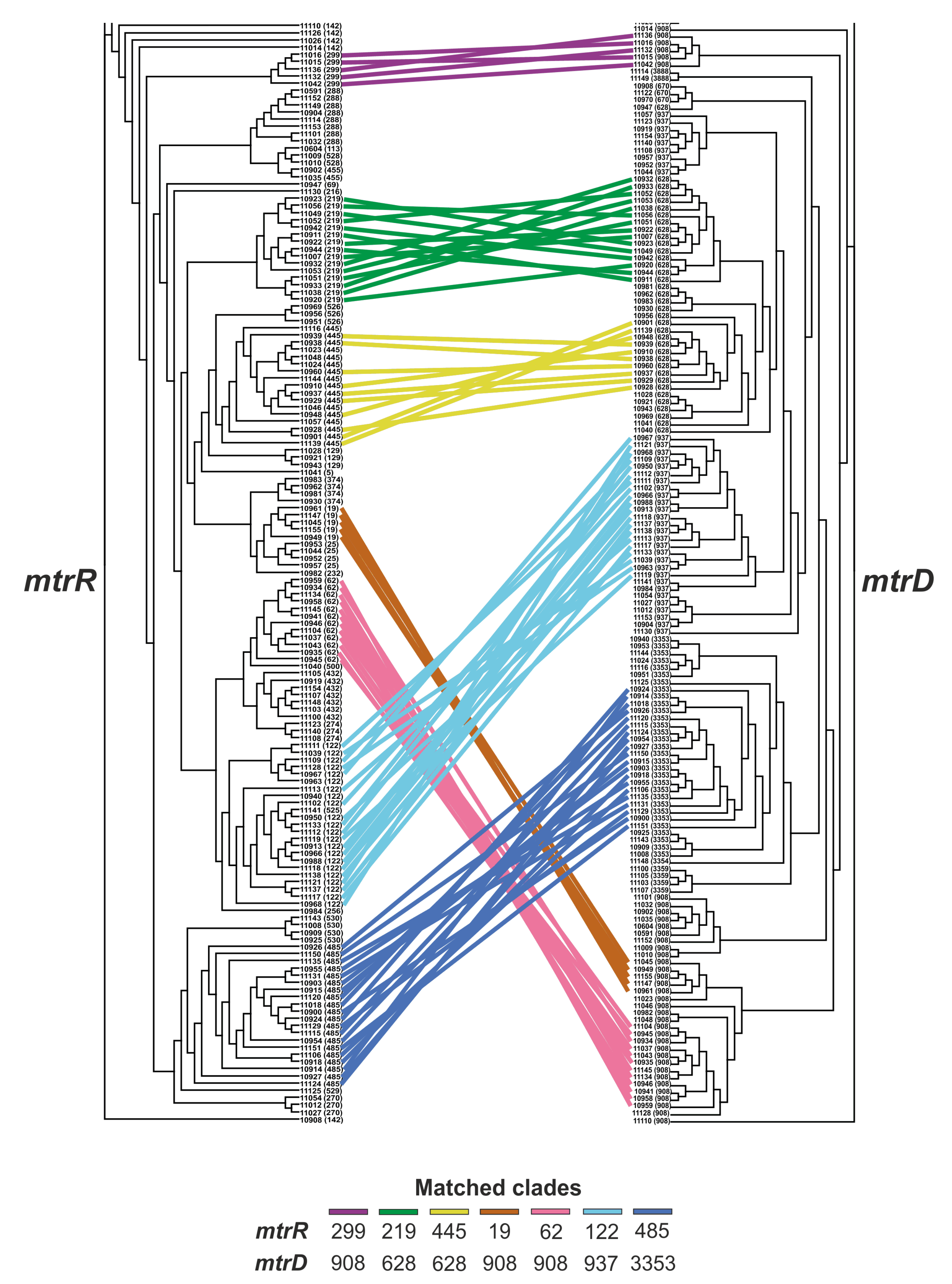
2.4. Relationship between the mtrR and mtrD Alleles
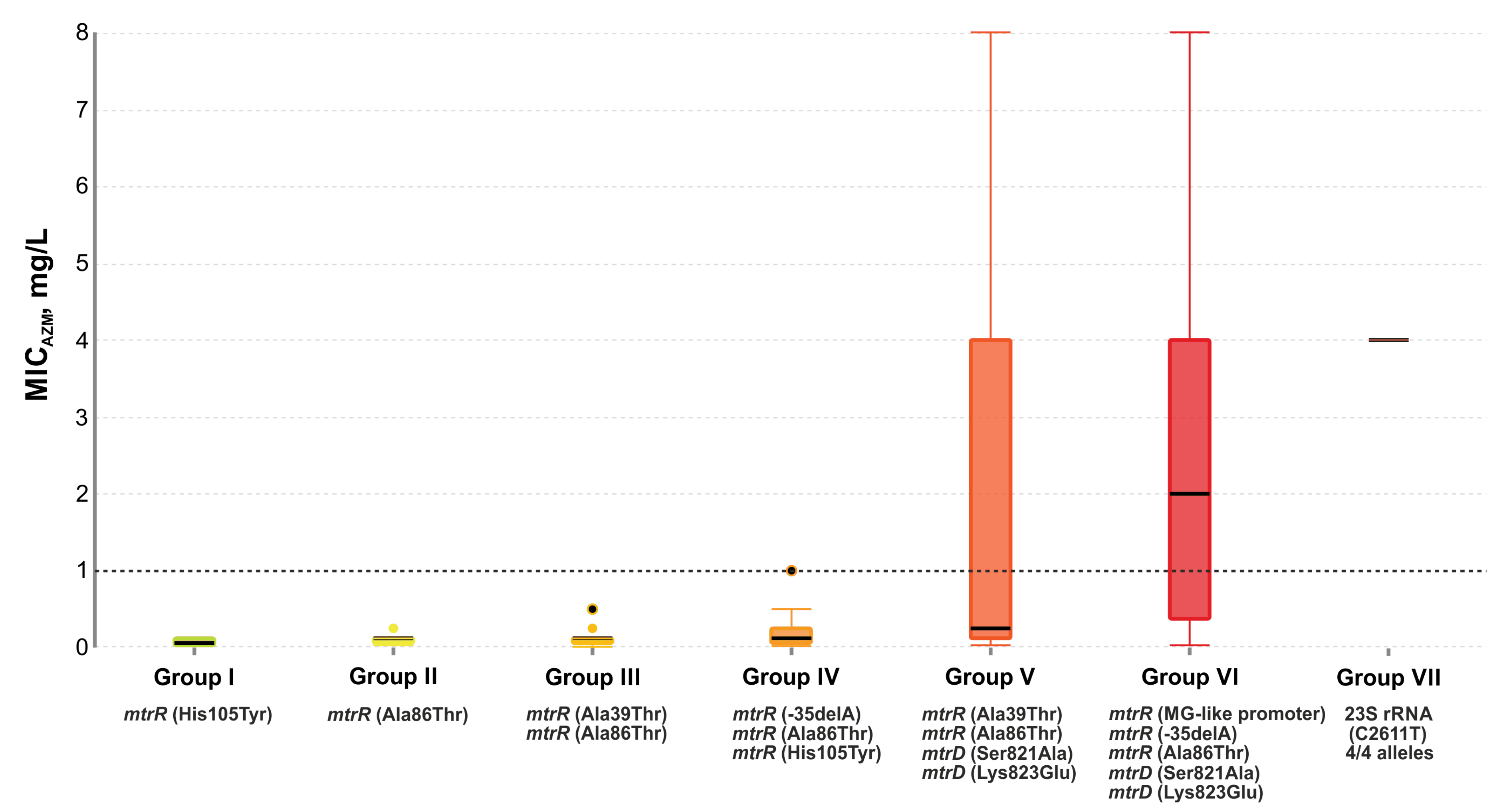
2.5. Effect of Genetic Profile on MICAzm
- group I: 12 isolates with the single His105Tyr substitution in the mtrR gene;
- group II: 47 isolates, 98% of which had the Ala86Thr mutation in the mtrR gene;
- group III: 35 isolates with His105Tyr and Ala39Thr substitutions in the mtrR gene. Several isolates carried the Gly45Ser mutation;
- group IV: 23 isolates with the His105Tyr substitution (100%) and the -35delA mutation in the promoter region of the mtrR gene (83%). There were also the -10insT, Ala39Thr, and Gly45Asp mutations;
- group V: 13 isolates with the His105Tyr (100%) and Ala39Thr (54%) substitutions in the mtrR gene and the Ser821Ala, Lys823Glu substitutions in the mtrD gene associated with mosaic structure (92%);
- group VI: 23 isolates with nucleotide sequences derived from N. meningitides both in the mtrR gene and its promoter (MG-like promoter), and in the mtrD gene (Ser821Ala and Lys823Glu mutations). There was also the -35delA mutation in the promoter region of the mtrR gene and the Ala86Thr mutation in the mtrR gene;
- group VII: two isolates with the C2611T mutations in all four alleles of the 23S rRNA gene. One isolate contained an MG-like promoter, the -35delA, Ala39Thr, Ala86Thr mutations in the mtrR gene, and the Ser821Ala, Lys823Glu mutations in the mtrD gene;
- ungrouped (7 isolates).
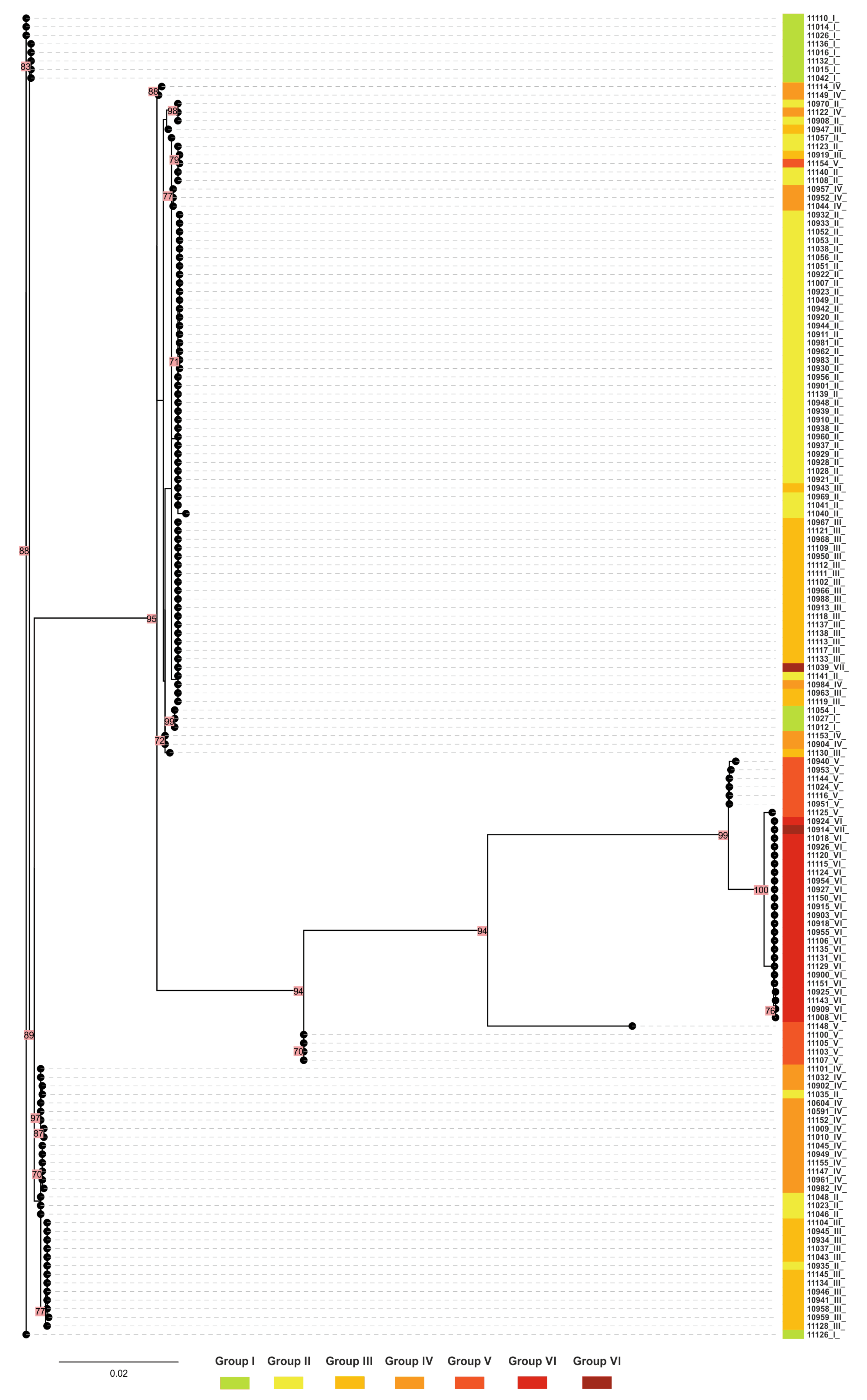
2.6. Phylogenetic Tree for mtrR and mtrD Genes
2.7. MtrR and mtrD Alleles in Isolates Belonging to NG-MAST 228 and 807
3. Discussion
4. Materials and Methods
4.1. Collection and Characterization of N. gonorrhoeae Isolates
4.2. Testing of N. gonorrhoeae Susceptibility for Azithromycin
4.3. Study of Genetic Determinants of N. gonorrhoeae Resistance to Azithromycin
4.4. Determination of mtrR and mtrD Allele Types
4.5. NG-MAST Typing
4.6. Construction of Phylogenetic Trees
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Global progress report on HIV, viral hepatitis and sexually transmitted infections, 2021. In The Global Health Sector Strategies 2016–2021: Actions for Impact; World Health Organization: Geneva, Switzerland, 2021.
- Golparian, D.; Unemo, M. Antimicrobial resistance prediction in Neisseria gonorrhoeae: Current status and future prospects. Expert Rev. Mol. Diagn. 2022, 22, 29–48. [Google Scholar] [CrossRef]
- Shaskolskiy, B.; Kandinov, I.; Dementieva, E.; Gryadunov, D. Antibiotic resistance in Neisseria gonorrhoeae: Challenges in research and treatment. Microorganisms 2022, 10, 1699. [Google Scholar] [CrossRef]
- Barbee, L.A.; St Cyr, S.B. Management of Neisseria gonorrhoeae in the United States: Summary of evidence from the development of the 2020 gonorrhea treatment recommendations and the 2021 Centers for Disease Control and Prevention sexually transmitted infection treatment guidelines. Clin. Infect. Dis. 2022, 74 (Suppl. S2), S95–S111. [Google Scholar] [CrossRef]
- WHO Guidelines for the Treatment of Neisseria gonorrhoeae; World Health Organization: Geneva, Switzerland, 2016.
- Unemo, M.; Ross, J.; Serwin, A.B.; Gomberg, M.; Cusini, M.; Jensen, J.S. 2020 European guideline for the diagnosis and treatment of gonorrhoea in adults. Int. J. STD AIDS 2020, 956462420949126. [Google Scholar] [CrossRef] [PubMed]
- Derbie, A.; Mekonnen, D.; Woldeamanuel, Y.; Abebe, T. Azithromycin resistant gonococci: A literature review. Antimicrob. Resist. Infect. Control 2020, 9, 138. [Google Scholar] [CrossRef] [PubMed]
- George, C.R.R.; Enriquez, R.P.; Gatus, B.J.; Whiley, D.M.; Lo, Y.R.; Ishikawa, N.; Wi, T.; Lahra, M.M. Systematic review and survey of Neisseria gonorrhoeae ceftriaxone and azithromycin susceptibility data in the Asia Pacific, 2011 to 2016. PLoS ONE 2019, 14, e0213312. [Google Scholar] [CrossRef]
- Shimuta, K.; Lee, K.; Yasuda, M.; Furubayashi, K.; Uchida, C.; Nakayama, S.I.; Takahashi, H.; Ohnishi, M. Characterization of 2 Neisseria gonorrhoeae strains with high-level azithromycin resistance isolated in 2015 and 2018 in Japan. Sex Transm. Dis. 2021, 48, e85–e87. [Google Scholar] [CrossRef] [PubMed]
- Mlynarczyk-Bonikowska, B.; Kowalewski, C.; Krolak-Ulinska, A.; Marusza, W. Molecular mechanisms of drug resistance and epidemiology of multidrug-resistant variants of Neisseria gonorrhoeae. Int. J. Mol. Sci. 2022, 23, 10499. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, S.A.; Dave, J.; Ison, C.A. High-level azithromycin resistance occurs in Neisseria gonorrhoeae as a result of a single point mutation in the 23S rRNA genes. Antimicrob. Agents Chemother. 2010, 54, 3812–3816. [Google Scholar] [CrossRef] [PubMed]
- Whiley, D.M.; Kundu, R.L.; Jennison, A.V.; Buckley, C.; Limnios, A.; Hogan, T.; Enriquez, R.; El Nasser, J.; George, C.R.; Lahra, M.M. Azithromycin-resistant Neisseria gonorrhoeae spreading amongst men who have sex with men (MSM) and heterosexuals in New South Wales, Australia, 2017. J. Antimicrob. Chemother. 2018, 73, 1242–1246. [Google Scholar] [CrossRef]
- Zhou, Q.; Liu, J.; Chen, S.; Xu, W.; Han, Y.; Yin, Y. The accuracy of molecular detection targeting the mutation C2611T for detecting moderate-level azithromycin resistance in Neisseria gonorrhoeae: A systematic review and meta-analysis. Antibiotics 2021, 10, 1027. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12.0. 2022. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 1 September 2022).
- Beggs, G.A.; Ayala, J.C.; Kavanaugh, L.G.; Read, T.D.; Hooks, G.M.; Schumacher, M.A.; Shaferm, W.M.; Brennan, R.G. Structures of Neisseria gonorrhoeae MtrR-operator complexes reveal molecular mechanisms of DNA recognition and antibiotic resistance-conferring clinical mutations. Nucl. Acids Res. 2021, 49, 4155–4170. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.J.; Thomas, J.C.; Schmerer, M.W.; Cartee, J.C.; St Cyr, S.; Schlanger, K.; Kersh, E.N.; Raphael, B.H.; Gernert, K.M. Global emergence and dissemination of Neisseria gonorrhoeae ST-9363 isolates with reduced susceptibility to azithromycin. Genome Biol. Evol. 2022, 14, evab287. [Google Scholar] [CrossRef] [PubMed]
- Rouquette-Loughlin, C.E.; Reimche, J.L.; Balthazar, J.T.; Dhulipala, V.; Gernert, K.M.; Kersh, E.N.; Pham, C.D.; Pettus, K.; Abrams, A.J.; Trees, D.L.; et al. Mechanistic basis for decreased antimicrobial susceptibility in a clinical isolate of Neisseria gonorrhoeae possessing a mosaic-like mtr efflux pump locus. mBio 2018, 9, e02281-18. [Google Scholar] [CrossRef]
- Demczuk, W.; Martin, I.; Peterson, S.; Bharat, A.; Van Domselaar, G.; Graham, M.; Lefebvre, B.; Allen, V.; Hoang, L.; Tyrrell, G.; et al. Genomic epidemiology and molecular resistance mechanisms of azithromycin-resistant Neisseria gonorrhoeae in Canada from 1997 to 2014. J. Clin. Microbiol. 2016, 54, 1304–1313. [Google Scholar] [CrossRef]
- Trembizki, E.; Doyle, C.; Jennison, A.; Smith, H.; Bates, J.; Lahra, M.; Whiley, D. A Neisseria gonorrhoeae strain with a meningococcal mtrR sequence. J. Med. Microbiol. 2014, 63, 1113–1115. [Google Scholar] [CrossRef]
- Cousin, S., Jr.; Whittington, W.L.; Roberts, M.C. Acquired macrolide resistance genes in pathogenic Neisseria spp. isolated between 1940 and 1987. Antimicrob. Agents Chemother. 2003, 47, 3877–3880. [Google Scholar] [CrossRef]
- Luna, V.A.; Cousin, S., Jr.; Whittington, W.L.; Roberts, M.C. Identification of the conjugative mef gene in clinical Acinetobacter junii and Neisseria gonorrhoeae isolates. Antimicrob. Agents Chemother. 2000, 44, 2503–2506. [Google Scholar] [CrossRef]
- Belkacem, A.; Jacquier, H.; Goubard, A.; Mougari, F.; La Ruche, G.; Patey, O.; Micaelo, M.; Semaille, C.; Cambau, E.; Bercot, B. Molecular epidemiology and mechanisms of resistance of azithromycin-resistant Neisseria gonorrhoeae isolated in France during 2013-14. J. Antimicrob. Chemother. 2016, 71, 2471–2478. [Google Scholar] [CrossRef]
- Kubanov, A.; Solomka, V.; Plakhova, X.; Chestkov, A.; Petrova, N.; Shaskolskiy, B.; Dementieva, E.; Leinsoo, A.; Gryadunov, D.; Deryabin, D. Summary and trends of the Russian Gonococcal Antimicrobial Surveillance Programme, 2005 to 2016. J. Clin. Microbiol. 2019, 57, e02024-18. [Google Scholar] [CrossRef]
- Shaskolskiy, B.; Dementieva, E.; Kandinov, I.; Chestkov, A.; Kubanov, A.; Deryabin, D.; Gryadunov, D. Genetic diversity of Neisseria gonorrhoeae multi-antigen sequence types in Russia and Europe. Int. J. Infect. Dis. 2020, 93, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Aniskevich, A.; Shimanskaya, I.; Boiko, I.; Golubovskaya, T.; Golparian, D.; Stanislavova, I.; Jacobsson, S.; Adaskevich, A.; Unemo, M. Antimicrobial resistance in Neisseria gonorrhoeae isolates and gonorrhoea treatment in the Republic of Belarus, Eastern Europe, 2009-2019. BMC Infect. Dis. 2021, 21, 520. [Google Scholar] [CrossRef] [PubMed]
- Karymbaeva, S.; Boiko, I.; Jacobsson, S.; Mamaeva, G.; Ibraeva, A.; Usupova, D.; Golparian, D.; Unemo, M. Antimicrobial resistance and molecular epidemiological typing of Neisseria gonorrhoeae isolates from Kyrgyzstan in Central Asia, 2012 and 2017. BMC Infect. Dis. 2021, 21, 559. [Google Scholar] [CrossRef] [PubMed]
- Kandinov, I.; Dementieva, E.; Kravtsov, D.; Chestkov, A.; Kubanov, A.; Solomka, V.; Deryabin, D.; Gryadunov, D.; Shaskolskiy, B. Molecular typing of Neisseria gonorrhoeae clinical isolates in Russia, 2018-2019: A link between penA alleles and NG-MAST types. Pathogens 2020, 9, 941. [Google Scholar] [CrossRef]
- Sanchez-Buso, L.; Cole, M.J.; Spiteri, G.; Day, M.; Jacobsson, S.; Golparian, D.; Sajedi, N.; Yeats, C.A.; Abudahab, K.; Underwood, A.; et al. Europe-wide expansion and eradication of multidrug-resistant Neisseria gonorrhoeae lineages: A genomic surveillance study. Lancet Microbe 2022, 3, e452–e463. [Google Scholar] [CrossRef]
- Tanaka, M.; Furuya, R.; Kobayashi, I.; Kanesaka, I.; Ohno, A.; Katsuse, A.K. Antimicrobial resistance and molecular characterisation of Neisseria gonorrhoeae isolates in Fukuoka, Japan, 1996-2016. J. Glob. Antimicrob. Resist. 2019, 17, 3–7. [Google Scholar] [CrossRef]
- Shaskolskiy, B.; Dementieva, E.; Kandinov, I.; Filippova, M.; Petrova, N.; Plakhova, X.; Chestkov, A.; Kubanov, A.; Deryabin, D.; Gryadunov, D. Resistance of Neisseria gonorrhoeae isolates to beta-lactams (benzylpenicillin and ceftriaxone) in Russia, 2015-2017. PLoS ONE 2019, 14, e0220339. [Google Scholar] [CrossRef]
- Mavroidi, A.; Tzouvelekis, L.S.; Kyriakis, K.P.; Avgerinou, H.; Daniilidou, M.; Tzelepi, E. Multidrug-resistant strains of Neisseria gonorrhoeae in Greece. Antimicrob. Agents Chemother. 2001, 45, 2651–2654. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; UGENE Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Hall, C.L.; Harrison, M.A.; Pond, M.J.; Chow, C.; Harding-Esch, E.M.; Sadiq, S.T. Genotypic determinants of fluoroquinolone and macrolide resistance in Neisseria gonorrhoeae. Sex Health 2019, 16, 479–487. [Google Scholar] [CrossRef]
- Martin, I.M.; Ison, C.A.; Aanensen, D.M.; Fenton, K.A.; Spratt, B.G. Rapid sequence-based identification of gonococcal transmission clusters in a large metropolitan area. J. Infect. Dis. 2004, 189, 1497–1505. [Google Scholar] [CrossRef] [PubMed]
| Group (Number of Isolates) | Median MICAzm (mg/L) | Genetic Profile mtrR | Genetic Profile mtrD | 23S rRNA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mtr Allele | MG-Like Promoter | -35delA | -10insT | Ala39 Thr | Gly45 Asp | Gly45 Ser | Ala86 Thr | His105 Tyr | mtrD Allele | Ser821 Ala | Lys823 Glu | C2611T (4 Alleles) | ||
| I (12) | 0.06 | 299 (42%) 142 (33%) 270 (25%) | – | – | – | – | – | – | – | 100% * | 908 (75%) 937 (25%) | – | – | – |
| II (47) | 0.12 | 219 (33%) 445 (30%) 374 (9%) 274 (6%) 129 (4%) 142 (4%) 526 (4%) 5 (2%) 62 (2%) 455 (2%) 500 (2%) 525 (2%) | – | – | – | 11% | – | 2% | 98% | – | 628 (74%) 908 (11%) 937 (11%) 670 (4%) | – | – | – |
| III (35) | 0.12 | 122 (57%) 62 (31%) 129 (3%) 69 (3%) 432 (3%) 216 (3%) | – | – | – | 94% | – | 31% | 100% | 6% | 937 (60%) 908 (34%) 628 (6%) | – | – | – |
| IV (23) | 0.12 | 288 (36%) 19 (22%) 25 (13%) 528 (9%) 113 (4%) 142 (4%) 232 (4%) 256 (4%) 455 (4%) | – | 83% | 4% | 4% | 39% | – | 100% | 52% | 908 (61%) 937 (26%) 3888 (9%) 670 (4%) | – | – | – |
| V (13) | 0.25 | 432 (45%) 445 (23%) 25 (8%) 122 (8%) 526 (8%) 529 (8%) | 8% | – | – | 54% | 8% | – | 100% | – | 3353 (54%) 3359 (30%) 937 (8%) 3354(8%) | 92% | 92% | – |
| VI (23) | 2.0 | 485 (83%) 530 (17%) | 100% | 100% | – | – | – | – | 100% | – | 3353 (100%) | 100% | 100% | – |
| VII (2) | 4.0 | 485 (50%) 122 (50%) | 50% | 50% | – | 50% | – | – | 100% | – | 3353 (50%) 937 (50%) | 50% | 50% | 100% |
| Ungrouped (7) | 1 | 129 (30%) 142 (14%) 299 (14%) 219 (14%) 189 (14%) 520 (14%) | – | – | – | – | – | – | 43% | 43% | 908 (33%) 628 (33%) 994 (17%) 3381 (17%) | – | – | – |
| NG-MAST | Number of Isolates | mtrR Allele | mtrD Allele | Comment |
|---|---|---|---|---|
| 228 | 4 | 219 | 628 | Alleles mtrR 5, 445, 526, 537 differ from allele mtrR 219 by one nucleotide |
| 4 | 445 | 628 | ||
| 2 | 5 | 628 | ||
| 3 | 537 | 628 | ||
| 1 | 62 | 908 | ||
| 807 | 14 | 219 | 628 | |
| 3 | 445 | 628 | ||
| 3 | 526 | 628 | ||
| 6 | 5 | 628 | ||
| 1 | 500 | 628 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kandinov, I.; Shaskolskiy, B.; Kravtsov, D.; Vinokurova, A.; Gorshkova, S.; Kubanov, A.; Solomka, V.; Shagabieva, J.; Deryabin, D.; Dementieva, E.; et al. Azithromycin Susceptibility Testing and Molecular Investigation of Neisseria gonorrhoeae Isolates Collected in Russia, 2020–2021. Antibiotics 2023, 12, 170. https://doi.org/10.3390/antibiotics12010170
Kandinov I, Shaskolskiy B, Kravtsov D, Vinokurova A, Gorshkova S, Kubanov A, Solomka V, Shagabieva J, Deryabin D, Dementieva E, et al. Azithromycin Susceptibility Testing and Molecular Investigation of Neisseria gonorrhoeae Isolates Collected in Russia, 2020–2021. Antibiotics. 2023; 12(1):170. https://doi.org/10.3390/antibiotics12010170
Chicago/Turabian StyleKandinov, Ilya, Boris Shaskolskiy, Dmitry Kravtsov, Alexandra Vinokurova, Sofya Gorshkova, Alexey Kubanov, Victoria Solomka, Julia Shagabieva, Dmitry Deryabin, Ekaterina Dementieva, and et al. 2023. "Azithromycin Susceptibility Testing and Molecular Investigation of Neisseria gonorrhoeae Isolates Collected in Russia, 2020–2021" Antibiotics 12, no. 1: 170. https://doi.org/10.3390/antibiotics12010170
APA StyleKandinov, I., Shaskolskiy, B., Kravtsov, D., Vinokurova, A., Gorshkova, S., Kubanov, A., Solomka, V., Shagabieva, J., Deryabin, D., Dementieva, E., & Gryadunov, D. (2023). Azithromycin Susceptibility Testing and Molecular Investigation of Neisseria gonorrhoeae Isolates Collected in Russia, 2020–2021. Antibiotics, 12(1), 170. https://doi.org/10.3390/antibiotics12010170











