Tetracycline, Macrolide and Lincosamide Resistance in Streptococcus canis Strains from Companion Animals and Its Genetic Determinants
Abstract
1. Introduction
2. Results
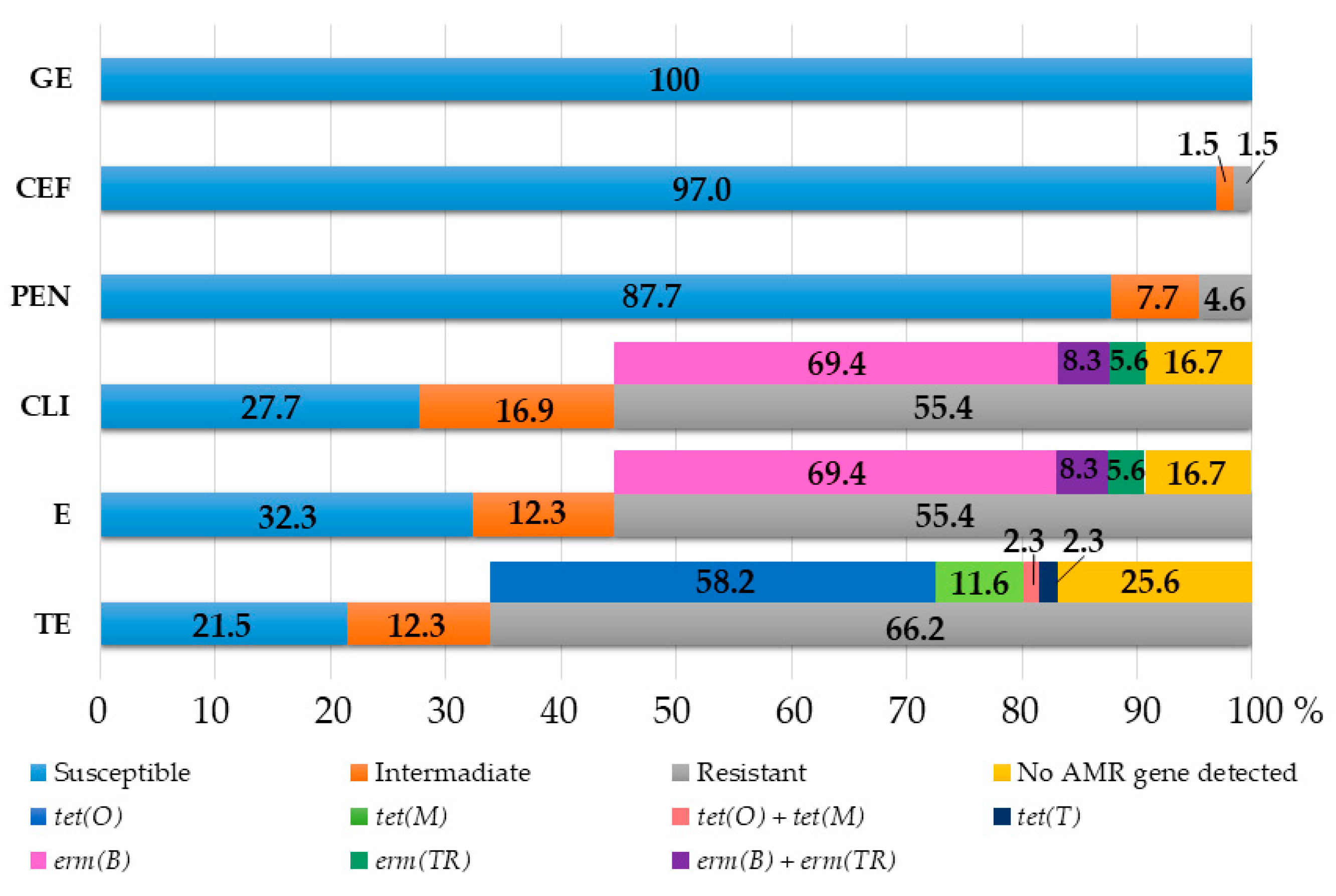
2.1. Antimicrobial Susceptibility Testing
2.2. Detection of Tetracycline, Macrolide and Lincosamide Resistance Genetic Determinants
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Antimicrobial Susceptibility Testing
4.3. Detection of Selected Resistance Genetic Determinants
4.4. Development of New Primers for Tet(T) Detection
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fulde, M.; Valentin-Weigand, P. Epidemiology and Pathogenicity of Zoonotic Streptococci. Curr. Top. Microbiol. Immunol. 2013, 368, 49–81. [Google Scholar] [CrossRef] [PubMed]
- Cheong, B.M.; Lim, A.Y. Sharing a microbe with man's best friend: A case of canine streptococcal infection in a diabetic patient. Med. J. Malaysia 2015, 70, 318–319. [Google Scholar] [PubMed]
- Frymus, T.; Addie, D.D.; Boucraut-Baralon, C.; Egberink, H.; Gruffydd-Jones, T.; Hartmann, K.; Horzinek, M.C.; Hosie, M.J.; Lloret, A.; Lutz, H.; et al. Streptococcal infections in cats: ABCD guidelines on prevention and management. J. Feline Med. Surg. 2015, 17, 620–625. [Google Scholar] [CrossRef] [PubMed]
- Timoney, J.F.; Velineni, S.; Ulrich, B.; Blanchard, P. Biotypes and ScM types of isolates of Streptococcus canis from diseased and healthy cats. Vet. Rec. 2017, 180, 358. [Google Scholar] [CrossRef]
- Chalmers, G.; McLean, J.; Hunter, D.B.; Brash, M.; Slavic, D.; Pearl, D.L.; Boerlin, P. Staphylococcus spp., Streptococcus canis, and Arcanobacterium phocae of healthy Canadian farmed mink and mink with pododermatitis. Can. J. Vet. Res. 2015, 79, 129–135. [Google Scholar]
- Hassan, A.A.; Akineden, O.; Usleber, E. Identification of Streptococcus canis Isolated from Milk of Dairy Cows with Subclinical Mastitis. J. Clin. Microbiol. 2005, 43, 1234–1238. [Google Scholar] [CrossRef]
- DeWinter, L.M.; Prescott, J.F. Relatedness of Streptococcus canis from canine streptococcal toxic shock syndrome and necrotizing fasciitis. Can. J. Vet. Res. 1999, 63, 90–95. [Google Scholar]
- Kulendra, E.; Corr, S. Necrotising fasciitis with sub–periosteal Streptococcus canis infection in two puppies. Vet. Comp. Orthop. Traumatol. 2008, 21, 474–477. [Google Scholar] [CrossRef]
- Lamm, C.G.; Ferguson, A.C.; Lehenbauer, T.W.; Love, B.C. Streptococcal Infection in Dogs: A retrospective study of 393 cases. Vet. Pathol. 2010, 47, 387–395. [Google Scholar] [CrossRef]
- Pinho, M.D.; Matos, S.C.; Pomba, C.; Lübke–Becker, A.; Wieler, L.H.; Preziuso, S.; Melo-Cristino, J.; Ramirez, M. Multilocus Sequence Analysis of Streptococcus canis Confirms the Zoonotic Origin of Human Infections and Reveals Genetic Exchange with Streptococcus dysgalactiae subsp. equisimilis. J. Clin. Microbiol. 2013, 51, 1099–1109. [Google Scholar] [CrossRef]
- Guerrero, A.E.; Stornelli, M.C.; Jurado, S.B.; Giacoboni, G.; Sguazza, G.H.; de la Sota, R.L.; Stornelli, M.A. Vaginal isolation of beta-haemolytic Streptococcus from bitches with and without neonatal deaths in the litters. Reprod. Domest. Anim. 2018, 53, 609–616. [Google Scholar] [CrossRef]
- Pesavento, P.A.; Bannasch, M.J.; Bachmann, R.; Byrne, B.A.; Hurley, K.F. Fatal Streptococcus canis Infections in Intensively Housed Shelter Cats. Vet. Pathol. 2007, 44, 218–221. [Google Scholar] [CrossRef]
- Galpérine, T.; Cazorla, C.; Blanchard, E.; Boineau, F.; Ragnaud, J.-M.; Neau, D. Streptococcus canis infections in humans: Retrospective study of 54 patients. J. Infect. 2007, 55, 23–26. [Google Scholar] [CrossRef]
- Mališová, B.; Šantavý, P.; Lovečková, Y.; Hladký, B.; Kotásková, I.; Pol, J.; Lonský, V.; Němec, P.; Freiberger, T. Human native endocarditis caused byStreptococcus canis—A case report. APMIS 2019, 127, 41–44. [Google Scholar] [CrossRef]
- Taniyama, D.; Abe, Y.; Sakai, T.; Kikuchi, T.; Takahashi, T. Human case of bacteremia caused by Streptococcus canis sequence type 9 harboring the scm gene. IDCases 2017, 7, 48–52. [Google Scholar] [CrossRef]
- Khan, A.J.; Evans, H.E.; Macabuhay, M.R.; Lee, Y.; Werner, R. Primary peritonitis due to group G Streptococcus: A case report. Pediatrics 1975, 56, 1078–1079. [Google Scholar] [CrossRef]
- Lacave, G.; Coutard, A.; Troché, G.; Augusto, S.; Pons, S.; Zuber, B.; Laurent, V.; Amara, M.; Couzon, B.; Bédos, J.-P.; et al. Endocarditis caused by Streptococcus canis: An emerging zoonosis? Infection 2016, 44, 111–114. [Google Scholar] [CrossRef]
- Jacobs, J.A.; De Krom, M.C.; Kellens, J.T.; Stobberingh, E.E. Meningitis and sepsis due to group G streptococcus. Eur. J. Clin. Microbiol. Infect. Dis. 1993, 12, 224–225. [Google Scholar] [CrossRef]
- Bert, F.; Lambert-Zechovsky, N. Septicemia caused by Streptococcus canis in a human. J. Clin. Microbiol. 1997, 35, 777–779. [Google Scholar] [CrossRef]
- Zaidi, S.M.H.; Eranki, A. Streptococcus canis Bacteremia in a Renal Transplant Recipient. J. Investig. Med. High Impact Case Rep. 2019, 7, 2324709619834592. [Google Scholar] [CrossRef]
- Facklam, R. What Happened to the Streptococci: Overview of Taxonomic and Nomenclature Changes. Clin. Microbiol. Rev. 2002, 15, 613–630. [Google Scholar] [CrossRef] [PubMed]
- Tsuyuki, Y.; Kurita, G.; Murata, Y.; Goto, M.; Takahashi, T. Identification of Group G Streptococcal Isolates from Companion Animals in Japan and Their Antimicrobial Resistance Patterns. Jpn. J. Infect. Dis. 2017, 70, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Institute of Medicine (IOM). A15 antibiotic resistance-linking human and animal health. In Improving Food Safety through a One Health Approach: Workshop Summary; Wegner, H.C., Ed.; The National Academies Press: Washington, DC, USA, 2012; pp. 331–349. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Haenni, M.; Lupo, A.; Madec, J.-Y. Antimicrobial Resistance in Streptococcus spp. Microbiol. Spectr. 2018, 6, 1–25. [Google Scholar] [CrossRef]
- Salgado-Caxito, M.; Benavides, J.A.; Adell, A.D.; Paes, A.C.; Moreno-Switt, A.I. Global prevalence and molecular characterization of extended-spectrum β-lactamase producing-Escherichia coli in dogs and cats—A scoping review and meta-analysis. One Health 2021, 12, 100236. [Google Scholar] [CrossRef]
- Formenti, N.; Grassi, A.; Parisio, G.; Romeo, C.; Guarneri, F.; Birbes, L.; Pitozzi, A.; Scali, F.; Maisano, A.M.; Boniotti, M.B.; et al. Extended-Spectrum-β-Lactamase- and AmpC-Producing Escherichia coli in Domestic Dogs: Spread, Characterisation and Associated Risk Factors. Antibiotics 2021, 10, 1251. [Google Scholar] [CrossRef]
- Garcia-Fierro, R.; Drapeau, A.; Dazas, M.; Saras, E.; Rodrigues, C.; Brisse, S.; Madec, J.-Y.; Haenni, M. Comparative phylogenomics of ESBL-, AmpC- and carbapenemase-producing Klebsiella pneumoniae originating from companion animals and humans. J. Antimicrob. Chemother. 2022, 77, 1263–1271. [Google Scholar] [CrossRef]
- Pires dos Santos, T.; Damborg, P.; Moodley, A.; Guardabassi, L. Systematic Review on Global Epidemiology of Methicillin-Resistant Staphylococcus pseudintermedius: Inference of Population Structure from Multilocus Sequence Typing Data. Front. Microbiol. 2016, 7, 1599. [Google Scholar] [CrossRef]
- Rzewuska, M.; Stefańska, I.; Kizerwetter-Świda, M.; Chrobak-Chmiel, D.; Szczygielska, P.; Leśniak, M.; Binek, M. Characterization of Extended-Spectrum-β-Lactamases Produced by Escherichia coli Strains Isolated from Dogs in Poland. Pol. J. Microbiol. 2015, 64, 285–288. [Google Scholar] [CrossRef]
- Kizerwetter-Świda, M.; Chrobak-Chmiel, D.; Rzewuska, M.; Binek, M. Changes in the population structure of canine methicillin-resistant Staphylococcus pseudintermedius in Poland. Vet. Microbiol. 2017, 208, 106–109. [Google Scholar] [CrossRef]
- Moyaert, H.; Morrissey, I.; De Jong, A.; El Garch, F.; Klein, U.; Ludwig, C.; Thiry, J.; Youala, M. Antimicrobial Susceptibility Monitoring of Bacterial Pathogens Isolated from Urinary Tract Infections in Dogs and Cats Across Europe: ComPath Results. Microb. Drug Resist. 2017, 23, 391–403. [Google Scholar] [CrossRef]
- Fukushima, Y.; Tsuyuki, Y.; Goto, M.; Yoshida, H.; Takahashi, T. Species Identification of β-Hemolytic Streptococci from Diseased Companion Animals and Their Antimicrobial Resistance Data in Japan (2017). Jpn. J. Infect. Dis. 2019, 72, 94–98. [Google Scholar] [CrossRef]
- Kurita, G.; Tsuyuki, Y.; Shibata, S.; Itoh, M.; Goto, M.; Yoshida, H.; Takahashi, T. Species identification of β-hemolytic streptococci from diseased companion animals and their antimicrobial resistance patterns in Japan (2021). Jpn. J. Vet. Res. 2022, 70, 19–28. [Google Scholar] [CrossRef]
- Moyaert, H.; De Graef, E.M.; Haesebrouck, F.; Decostere, A. Acquired antimicrobial resistance in the intestinal microbiota of diverse cat populations. Res. Vet. Sci. 2006, 81, 1–7. [Google Scholar] [CrossRef]
- Pedersen, K.; Jensen, H.; Finster, K.; Jensen, V.F.; Heuer, O.E. Occurrence of antimicrobial resistance in bacteria from diagnostic samples from dogs. J. Antimicrob. Chemother. 2007, 60, 775–781. [Google Scholar] [CrossRef]
- Awji, E.G.; Damte, D.; Lee, S.-J.; Lee, J.-S.; Kim, Y.-H.; Park, S.-C. The In Vitro Activity of 15 Antimicrobial Agents against Bacterial Isolates from Dogs. J. Vet. Med. Sci. 2012, 74, 1091–1094. [Google Scholar] [CrossRef]
- Nikolaisen, N.K.; Lassen, D.C.K.; Chriél, M.; Larsen, G.; Jensen, V.F.; Pedersen, K. Antimicrobial resistance among pathogenic bacteria from mink (Neovison vison) in Denmark. Acta Vet. Scand. 2017, 59, 60. [Google Scholar] [CrossRef]
- Hewitt, J.S.; Allbaugh, R.A.; Kenne, D.E.; Sebbag, L. Prevalence and Antibiotic Susceptibility of Bacterial Isolates from Dogs With Ulcerative Keratitis in Midwestern United States. Front. Vet. Sci. 2020, 7, 583965. [Google Scholar] [CrossRef]
- Haenni, M.; Hourquet, C.; Saras, E.; Madec, J.-Y. Genetic determinants of antimicrobial resistance in Streptococcus canis in France. J. Glob. Antimicrob. Resist. 2015, 3, 142–143. [Google Scholar] [CrossRef]
- Fukushima, Y.; Tsuyuki, Y.; Goto, M.; Yoshida, H.; Takahashi, T. Novel Quinolone Nonsusceptible Streptococcus canis Strains with Point Mutations in Quinolone Resistance-Determining Regions and Their Related Factors. Jpn. J. Infect. Dis. 2020, 73, 242–249. [Google Scholar] [CrossRef]
- Haenni, M.; Saras, E.; Bertin, S.; Leblond, P.; Madec, J.-Y.; Payot, S. Diversity and Mobility of Integrative and Conjugative Elements in Bovine Isolates of S treptococcus agalactiae, S. dysgalactiae subsp. dysgalactiae, and S. uberis. Appl. Environ. Microbiol. 2010, 76, 7957–7965. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.G.; Genteluci, G.L.; De Mattos, M.C.; Glatthardt, T.; Figueiredo, A.M.S.; Ferreira-Carvalho, B.T. Group C Streptococcus dysgalactiae subsp. equisimilis in south-east Brazil: Genetic diversity, resistance profile and the first report of human and equine isolates belonging to the same multilocus sequence typing lineage. J. Med. Microbiol. 2015, 64, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Ciszewski, M.; Zegarski, K.; Szewczyk, E.M. Streptococcus dysgalactiae subsp. equisimilis Isolated from Infections in Dogs and Humans: Are Current Subspecies Identification Criteria accurate? Curr. Microbiol. 2016, 73, 684–688. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.Y.; Zheng, N.; Han, R.W.; Ho, H.; Wang, J.; Wang, Y.T.; Wang, S.Q.; Li, H.G.; Liu, H.W.; Yu, Z.N. Antimicrobial resistance and virulence genes of Streptococcus isolated from dairy cows with mastitis in China. Microb. Pathog. 2019, 131, 33–39. [Google Scholar] [CrossRef]
- Oh, S.I.; Kim, J.W.; Jung, J.Y.; Chae, M.; Lee, Y.R.; Kim, J.H.; So, B.; Kim, H.Y. Pathologic and molecular characterization ofStreptococcus dysgalactiaesubsp.equisimilisinfection in neonatal piglets. J. Vet. Sci. 2018, 19, 313–317. [Google Scholar] [CrossRef]
- Alves-Barroco, C.; Caço, J.; Roma-Rodrigues, C.; Fernandes, A.R.; Bexiga, R.; Oliveira, M.; Chambel, L.; Tenreiro, R.; Mato, R.; Santos-Sanches, I. New Insights on Streptococcus dysgalactiae subsp. dysgalactiae Isolates. Front. Microbiol. 2021, 12, 686413. [Google Scholar] [CrossRef]
- Shen, J.; Wu, X.; Yang, Y.; Lv, Y.; Li, X.; Ding, X.; Wang, S.; Yan, Z.; Yan, Y.; Yang, F.; et al. Antimicrobial Resistance and Virulence Factor of Streptococcus dysgalactiae Isolated from Clinical Bovine Mastitis Cases in Northwest China. Infect. Drug Resist. 2021, 14, 3519–3530. [Google Scholar] [CrossRef]
- Fukushima, Y.; Tsuyuki, Y.; Goto, M.; Yoshida, H.; Takahashi, T. Biofilm Production Ability and Other Microbiological Features of Streptococcus canis. Jpn. J. Infect. Dis. 2022, 75, 63–69. [Google Scholar] [CrossRef]
- Roberts, M.C. Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 2005, 245, 195–203. [Google Scholar] [CrossRef]
- Dechêne-Tempier, M.; Marois-Créhan, C.; Libante, V.; Jouy, E.; Leblond-Bourget, N.; Payot, S. Update on the Mechanisms of Antibiotic Resistance and the Mobile Resistome in the Emerging Zoonotic Pathogen Streptococcus suis. Microorganisms 2021, 9, 1765. [Google Scholar] [CrossRef]
- Hadjirin, N.F.; Miller, E.L.; Murray, G.G.R.; Yen, P.L.K.; Phuc, H.D.; Wileman, T.M.; Hernandez-Garcia, J.; Williamson, S.M.; Parkhill, J.; Maskell, D.J.; et al. Large-scale genomic analysis of antimicrobial resistance in the zoonotic pathogen Streptococcus suis. BMC Biol. 2021, 19, 191. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R.I.; Garrigues-Jeanjean, N.; Mackie, R.I. Molecular Ecology of Tetracycline Resistance: Development and Validation of Primers for Detection of Tetracycline Resistance Genes Encoding Ribosomal Protection Proteins. Appl. Environ. Microbiol. 2001, 67, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Clermont, D.; Chesneau, O.; De Cespédès, G.; Horaud, T. New tetracycline resistance determinants coding for ribosomal protection in streptococci and nucleotide sequence of tet(T) isolated from Streptococcus pyogenes A498. Antimicrob. Agents Chemother. 1997, 41, 112–116. [Google Scholar] [CrossRef]
- Rojo-Bezares, B.; Toca, L.; Azcona-Gutiérrez, J.M.; Ortega-Unanue, N.; Toledano, P.; Sáenz, Y. Streptococcus dysgalactiae subsp. equisimilis from invasive and non-invasive infections in Spain: Combining epidemiology, molecular characterization, and genetic diversity. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1013–1021. [Google Scholar] [CrossRef]
- Nishimoto, Y.; Kobayashi, N.; Alam, M.M.; Ishino, M.; Uehara, N.; Watanabe, N. Analysis of the Prevalence of Tetracycline Resistance Genes in Clinical Isolates ofEnterococcus faecalisandEnterococcus faeciumin a Japanese Hospital. Microb. Drug Resist. 2005, 11, 146–153. [Google Scholar] [CrossRef]
- Katsarou, E.I.; Chatzopoulos, D.C.; Giannoulis, T.; Ioannidi, K.S.; Katsafadou, A.I.; Kontou, P.I.; Lianou, D.T.; Mamuris, Z.; Mavrogianni, V.S.; Michael, C.K.; et al. MLST-Based Analysis and Antimicrobial Resistance of Staphylococcus epidermidis from Cases of Sheep Mastitis in Greece. Biology 2021, 10, 170. [Google Scholar] [CrossRef]
- Hraoui, M.; Boubaker, I.B.-B.; Rachdi, M.; Slim, A.; Ben Redjeb, S. Macrolide and tetracycline resistance in clinical strains of Streptococcus agalactiae isolated in Tunisia. J. Med. Microbiol. 2012, 61, 1109–1113. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Q. Relationship between tetracycline antibiotic susceptibility and genotype in oral cavity Lactobacilli clinical isolates. Antimicrob. Resist. Infect. Control 2019, 8, 1–8. [Google Scholar] [CrossRef]
- Roberts, M.C. Mechanism of Resistance for Characterized Tet and Otr Genes, Last Modified 20 April 2021. Available online: https://faculty.washington.edu/marilynr/tetweb1.pdf (accessed on 30 June 2022).
- Chopra, I.; Roberts, M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef]
- Rice, L.B. Tn 916 Family Conjugative Transposons and Dissemination of Antimicrobial Resistance Determinants. Antimicrob. Agents Chemother. 1998, 42, 1871–1877. [Google Scholar] [CrossRef]
- Jurado-Rabadán, S.; de la Fuente, R.; Ruiz-Santa-Quiteria, J.A.; Orden, J.A.; de Vries, L.E.; Agersø, Y. Detection and linkage to mobile genetic elements of tetracycline resistance gene tet(M) in Escherichia coliisolates from pigs. BMC Vet. Res. 2014, 10, 155. [Google Scholar] [CrossRef]
- Ludwig, C.; De Jong, A.; Moyaert, H.; El Garch, F.; Janes, R.; Klein, U.; Morrissey, I.; Thiry, J.; Youala, M. Antimicrobial susceptibility monitoring of dermatological bacterial pathogens isolated from diseased dogs and cats across Europe (ComPath results). J. Appl. Microbiol. 2016, 121, 1254–1267. [Google Scholar] [CrossRef]
- Roberts, M.C. Mechanisms of MLS Resistance-Last Modified 2 May 2022. Available online: https://faculty.washington.edu/marilynr/ermwebA.pdf (accessed on 30 June 2022).
- van Hoek, A.H.A.M.; Mevius, D.; Guerra, B.; Mullany, P.; Roberts, A.P.; Aarts, H.J.M. Acquired Antibiotic Resistance Genes: An Overview. Front. Microbiol. 2011, 2, 203. [Google Scholar] [CrossRef]
- Mingoia, M.; Morici, E.; Marini, E.; Brenciani, A.; Giovanetti, E.; Varaldo, P.E. Macrolide resistance geneerm(TR) anderm(TR)-carrying genetic elements inStreptococcus agalactiae: Characterization of ICESagTR7, a new composite element containing IMESp2907. J. Antimicrob. Chemother. 2016, 71, 593–600. [Google Scholar] [CrossRef]
- Amezaga, M.R.; McKenzie, H. Molecular epidemiology of macrolide resistance in β-haemolytic streptococci of Lancefield groups A, B, C and G and evidence for a new mef element in group G streptococci that carries allelic variants of mef and msr(D). J. Antimicrob. Chemother. 2006, 57, 443–449. [Google Scholar] [CrossRef]
- Kataja, J.; Huovinen, P.; Seppala, H. Erythromycin resistance genes in group A streptococci of different geographical origins: The Macrolide Resistance Study Group. J. Antimicrob. Chemother. 2000, 46, 789–792. [Google Scholar] [CrossRef][Green Version]
- Varaldo, P.E.; Montanari, M.P.; Giovanetti, E. Genetic Elements Responsible for Erythromycin Resistance in Streptococci. Antimicrob. Agents Chemother. 2009, 53, 343–353. [Google Scholar] [CrossRef]
- Kaczorek, E.; Małaczewska, J.; Wójcik, R.; Rękawek, W.; Siwicki, A.K. Phenotypic and genotypic antimicrobial susceptibility pattern of Streptococcus spp. isolated from cases of clinical mastitis in dairy cattle in Poland. J. Dairy Sci. 2017, 100, 6442–6453. [Google Scholar] [CrossRef]
- Baracco, G.J. Infections Caused by Group C and G Streptococcus (Streptococcus dysgalactiae subsp. equisimilis and Others): Epidemiological and Clinical Aspects. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Samir, A.; Abdel-Moein, K.A.; Zaher, H.M. Emergence of penicillin-macrolide-resistant Streptococcus pyogenes among pet animals: An ongoing public health threat. Comp. Immunol. Microbiol. Infect. Dis. 2020, 68, 101390. [Google Scholar] [CrossRef] [PubMed]
- Bonofiglio, L.; Gagetti, P.; Gabarrot, G.G.; Kaufman, S.; Mollerach, M.; Toresani, I.; Vigliarolo, L.; von Specht, M.; Lopardo, H.A. Susceptibility to β-lactams in β-hemolytic streptococci. Rev. Argent Microbiol. 2018, 50, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Bush, K. The ABCD’s of β-lactamase nomenclature. J. Infect. Chemother. 2013, 19, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Ruegg, P.L.; Oliveira, L.; Jin, W.; Okwumabua, O. Phenotypic antimicrobial susceptibility and occurrence of selected resistance genes in gram-positive mastitis pathogens isolated from Wisconsin dairy cows. J. Dairy Sci. 2015, 98, 4521–4534. [Google Scholar] [CrossRef]
- CA-SFM. Comité de l’antibiogramme de la Société Française de Microbiologie. Antibiogram Committee of the French Society of Microbiology Guidelines: Recommandations Vétérinaires 2021. 2021. Available online: https://www.sfm-microbiologie.org/wp-content/uploads/2021/12/CASFM_VET2021.pdf (accessed on 30 June 2022). (In French).
- Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial disk and dilution susceptibility test for bacteria isolated from animals. In CLSI Supplement VET08, 4th ed.; CLSI: Wayne, PA, USA, 2018. [Google Scholar]
- Pang, Y.; Bosch, T.; Roberts, M.C. Single polymerase chain reaction for the detection of tetracycline-resistant determinants Tet K and Tet L. Mol. Cell. Probes 1994, 8, 417–422. [Google Scholar] [CrossRef]
- Trzcinski, K.; Cooper, B.S.; Hryniewicz, W.; Dowson, C.G. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 2000, 45, 763–770. [Google Scholar] [CrossRef]
- Nawaz, M.; Wang, J.; Zhou, A.; Ma, C.; Wu, X.; Moore, J.E.; Millar, B.C.; Xu, J. Characterization and Transfer of Antibiotic Resistance in Lactic Acid Bacteria from Fermented Food Products. Curr. Microbiol. 2011, 62, 1081–1089. [Google Scholar] [CrossRef]
- Gibreel, A.; Tracz, D.M.; Nonaka, L.; Ngo, T.M.; Connell, S.R.; Taylor, D.E. Incidence of Antibiotic Resistance in Campylobacter jejuni Isolated in Alberta, Canada, from 1999 to 2002, with Special Reference to tet (O)-Mediated Tetracycline Resistance. Antimicrob. Agents Chemother. 2004, 48, 3442–3450. [Google Scholar] [CrossRef]
- Agersø, Y.; Pedersen, A.G.; Aarestrup, F.M. Identification of Tn5397-like and Tn916-like transposons and diversity of the tetracycline resistance gene tet(M) in enterococci from humans, pigs and poultry. J. Antimicrob. Chemother. 2006, 57, 832–839. [Google Scholar] [CrossRef]
- de Vries, L.E.; Christensen, H.; Skov, R.L.; Aarestrup, F.M.; Agersø, Y. Diversity of the tetracycline resistance gene tet(M) and identification of Tn916- and Tn5801-like (Tn6014) transposons in Staphylococcus aureus from humans and animals. J. Antimicrob. Chemother. 2009, 64, 490–500. [Google Scholar] [CrossRef]
- Toomey, N.; Bolton, D.; Fanning, S. Characterisation and transferability of antibiotic resistance genes from lactic acid bacteria isolated from Irish pork and beef abattoirs. Res. Microbiol. 2010, 161, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Pihlajamäki, M.; Kataja, J.; Seppälä, H.; Elliot, J.; Leinonen, M.; Huovinen, P.; Jalava, J. Ribosomal Mutations in Streptococcus pneumoniae Clinical Isolates. Antimicrob. Agents Chemother. 2002, 46, 654–658. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kohn, M.A.; Senyak, J. Sample Size Calculators. UCSF CTSI. 20 December 2021. Available online: https://www.sample-size.net/ (accessed on 25 July 2022).
| Animal | Antibiotic * | Number of Strains with MIC (μg/mL): | MIC50 | MIC90 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | ≥128 | ||||
| Cat | PEN | 9 | 1 | ≤0.25 | ≤0.25 | ||||||||
| CEF | 5 | 2 | 2 | 1 | ≤0.25 | 1 | |||||||
| GE | 1 | 4 | 5 | 8 | 16 | ||||||||
| TE | 1 | 2 | 7 | ≥128 | ≥128 | ||||||||
| E | 2 | 1 | 7 | ≥128 | ≥128 | ||||||||
| CLI | 3 | 7 | ≥128 | ≥128 | |||||||||
| Dog | PEN | 45 | 3 | 4 | 2 | 1 | ≤0.25 | 1 | |||||
| CEF | 28 | 18 | 6 | 2 | 1 | ≤0.25 | 1 | ||||||
| GE | 1 | 12 | 33 | 7 | 1 | 1 | 16 | 32 | |||||
| TE | 1 | 12 | 6 | 2 | 1 | 4 | 29 | ≥128 | ≥128 | ||||
| E | 15 | 4 | 4 | 1 | 2 | 1 | 28 | ≥128 | ≥128 | ||||
| CLI | 9 | 9 | 5 | 3 | 1 | 28 | ≥128 | ≥128 | |||||
| Strain | Resistance Phenotype 1 | Resistance Genes Detected |
|---|---|---|
| 12/16 | TE | n.d. 2 |
| 22/18 | n.d. | |
| 27/18 | tet(O) 3 | |
| 35/20 | n.d. | |
| 44/21 | tet(O) 3 | |
| 1/16 | tet(M) linked with Tn916-like transposon | |
| 3/16 | tet(M) linked with Tn916-like transposon | |
| 52/21 | tet(T) | |
| 31/20 | E-CLI | erm(B) |
| 14/16 | TE-E-CLI | tet(O), erm(B), erm(TR) |
| 15/16 | tet(O), erm(B), erm(TR) | |
| 18/16 | tet(M) 3, erm(B) | |
| 23/18 | erm(B) | |
| 24/18 | tet(O), erm(B) | |
| 32/20 | tet(O) 3, erm(B) | |
| 48/21 | tet(O) 3 | |
| 51/21 | tet(O), erm(B) | |
| 58/21 | erm(B) | |
| 60/21 | tet(O), erm(B) | |
| 2/16 | TE-E-CLI | tet(O), erm(B) |
| 4/16 | tet(O), erm(B) | |
| 5/16 | tet(O), erm(B) | |
| 6/16 | tet(O), erm(TR) | |
| 7/16 | tet(O), erm(B), erm(TR) | |
| 10/16 | tet(O), erm(B) | |
| 17/16 | tet(O), erm(B) | |
| 19/16 | n.d. | |
| 20/17 | tet(O), erm(B) | |
| 25/18 | tet(O) 3, erm(B) | |
| 39/21 | tet(O), erm(B) | |
| 47/21 | tet(O), erm(B) | |
| 49/21 | n.d. | |
| 50/21 | tet(M) linked with Tn916-like transposon | |
| 55/21 | n.d. | |
| 56/21 | erm(TR) | |
| 57/21 | erm(B) | |
| 61/21 | tet(O), erm(B) | |
| 62/21 | tet(O) 3, erm(B) | |
| 63/21 | tet(O), erm(B) | |
| 65/22 | tet(M) linked with Tn916-like transposon, tet(O), erm(B) | |
| 59/21 | TE-E-CLI-P | tet(O), erm(B) |
| 64/21 | n.d. | |
| 41/21 | tet(O), erm(B) | |
| 53/21 | TE-E-CLI-CEF | tet(M) linked with Tn916-like transposon, erm(B) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stefańska, I.; Kwiecień, E.; Kizerwetter-Świda, M.; Chrobak-Chmiel, D.; Rzewuska, M. Tetracycline, Macrolide and Lincosamide Resistance in Streptococcus canis Strains from Companion Animals and Its Genetic Determinants. Antibiotics 2022, 11, 1034. https://doi.org/10.3390/antibiotics11081034
Stefańska I, Kwiecień E, Kizerwetter-Świda M, Chrobak-Chmiel D, Rzewuska M. Tetracycline, Macrolide and Lincosamide Resistance in Streptococcus canis Strains from Companion Animals and Its Genetic Determinants. Antibiotics. 2022; 11(8):1034. https://doi.org/10.3390/antibiotics11081034
Chicago/Turabian StyleStefańska, Ilona, Ewelina Kwiecień, Magdalena Kizerwetter-Świda, Dorota Chrobak-Chmiel, and Magdalena Rzewuska. 2022. "Tetracycline, Macrolide and Lincosamide Resistance in Streptococcus canis Strains from Companion Animals and Its Genetic Determinants" Antibiotics 11, no. 8: 1034. https://doi.org/10.3390/antibiotics11081034
APA StyleStefańska, I., Kwiecień, E., Kizerwetter-Świda, M., Chrobak-Chmiel, D., & Rzewuska, M. (2022). Tetracycline, Macrolide and Lincosamide Resistance in Streptococcus canis Strains from Companion Animals and Its Genetic Determinants. Antibiotics, 11(8), 1034. https://doi.org/10.3390/antibiotics11081034







