Status of Microbiota in Odontogenic Inflammatory Lesions and Dental Surgery Procedures Performed on an Outpatient Basis
Abstract
1. Introduction
2. Materials and Methods
- -
- Submucosal abscess requiring removal of the causative tooth as well as soft tissue incision and seton use, 15 patients, including 2 females and 13 males;
- -
- Periapical abscess requiring only removal of the causative tooth (drainage of the purulent content was obtained through the alveolus), 11 patients, including 3 females and 8 males.
2.1. Treatment Procedure—Study Group
2.2. Treatment Procedure—Control Group
2.3. Statistical Analysis
3. Results and Discussion
3.1. Characteristic of Examined Population
3.2. Microbiological Test Results
- -
- Gram-positive aerobic bacteria;
- -
- Gram-positive anaerobic bacteria;
- -
- Gram-negative aerobic bacteria;
- -
- Gram-negative anaerobic bacteria.
4. Discussion
5. Conclusions
- There were no statistically significant differences between the test and control groups in the number of bacterial strains cultured in either Test I or Test II. In each case, the highest number of cultured strains belonged to aerobic Gram-positive bacteria and the lowest to anaerobic Gram-negative bacteria;
- There were also no statistically significant differences between the test and control groups in the number of bacterial species cultured in either Test I or Test II. Gram-positive aerobic bacteria have the highest species diversity and Gram-negative aerobic bacteria the lowest;
- The highest percentage of potentially pathogenic strains was found for Gram-positive anaerobic bacteria and the lowest for Gram-positive aerobic bacteria;
- The highest percentage of potentially pathogenic species was found for Gram-negative aerobic bacteria and the lowest for Gram-positive aerobic bacteria. In the group of Gram-negative aerobic microorganisms, the only non-pathogenic bacteria were strains of Neisseria spp. The only pathogenic species found in the Gram-positive aerobic microbial group were Staphylococcus aureus, Staphylococcus epidermidis, and Streptococcus pneumoniae.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kilian, M.; Chapple, I.L.C.; Hannig, M.; Marsh, P.D.; Meuric, V.; Pedersen, A.M.L.; Tonetti, M.S.; Wade, W.G.; Zaura, E. The oral microbiome—An update for oral healthcare professionals. Br. Dent. J. 2016, 221, 657–666. [Google Scholar] [CrossRef]
- Lemon, K.P.; Klepac-Ceraj, V.; Schiffer, H.K.; Brodie, E.L.; Lynch, S.V.; Kolter, R. Comparative Analyses of the Bacterial Microbiota of the Human Nostril and Oropharynx. mBio 2010, 1, e00129–10. [Google Scholar] [CrossRef]
- Verma, D.; Garg, P.K.; Dubey, A.K. Insights into the human oral microbiome. Arch. Microbiol. 2018, 200, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Cope, A.L.; Francis, N.; Wood, F.; Chestnutt, I.G. Systemic antibiotics for symptomatic apical periodontitis and acute apical abscess in adults. Cochrane Database Syst. Rev. 2018, 2018, CD010136. [Google Scholar] [CrossRef]
- Bartkowski, S.; Kurek, M.; Panaś, M.; Podziorny, H.; Serwatka, F.; Stypułkow-ska, J.; Zapała, J. Chirurgia Szczękowo-Twarzowa; Collegium Medicum UJ: Kraków, Poland, 1996. [Google Scholar]
- Ellison, S.J. An outcome audit of three day antimicrobial prescribing for the acute dentoalveolar abscess. Br. Dent. J. 2011, 211, 591–594. [Google Scholar] [CrossRef]
- Orzechowska-Wylęgała, B.; Wylęgała, A.; Buliński, M.; Niedzielska, I. Antibiotic therapies in maxillofacial surgery in the context of prophylaxis. BioMed Res. Int. 2015, 2015, 819086. [Google Scholar] [CrossRef] [PubMed]
- Curtis, M.A.; Diaz, P.I.; Van Dyke, T.E. The role of the microbiota in periodontal disease. Periodontology 2000 2020, 83, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Li, H.; Ni, C.; Du, Z.; Yan, F. Human oral microbiota and its modulation for oral health. Biomed. Pharmacother. 2018, 99, 883–893. [Google Scholar] [CrossRef] [PubMed]
- Plum, A.W.; Mortelliti, A.J.; Walsh, R.E. Microbial Flora and Antibiotic Resistance in Odontogenic Abscesses in Upstate New York. Ear Nose Throat J. 2018, 97, E27–E31. [Google Scholar] [CrossRef] [PubMed]
- Taub, D.; Yampolsky, A.; Diecidue, R.; Gold, L. Controversies in the Management of Oral and Maxillofacial Infections. Oral Maxillofac. Surg. Clin. N. Am. 2017, 29, 465–473. [Google Scholar] [CrossRef] [PubMed]
- AlRahabi, M.K.; Abuong, Z.A. Antibiotic abuse during endodontic treatment in private dental centers. Saudi Med. J. 2017, 38, 852–856. [Google Scholar] [CrossRef]
- Hossaini-Zadeh, M. Current Concepts of Prophylactic Antibiotics for Dental Patients. Dent. Clin. N. Am. 2016, 60, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Bansal, R.; Jain, A.; Goyal, M.; Singh, T.; Sood, H.; Malviya, H.S. Antibiotic abuse during endodontic treatment: A contributing factor to antibiotic resistance. J. Fam. Med. Prim. Care 2019, 8, 3518–3524. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.A.; Tamer, T.M.; Rageh, A.A.; Abou-Zeid, A.M.; Abd El-Zaher, E.H.F.; Kenawy, E.-R. Insight into multidrug-resistant microorganisms from microbial infected diabetic foot ulcers. Diabetes Metab. Syndr. 2019, 13, 1261–1270. [Google Scholar] [CrossRef]
- Sousa, E.L.R.; Gomes, B.P.; Jacinto, R.; Zaia, A.A.; Ferraz, C.C.R. Microbiological profile and antimicrobial susceptibility pattern of infected root canals associated with periapical abscesses. Eur. J. Clin. Microbiol. 2012, 32, 573–580. [Google Scholar] [CrossRef]
- Łysakowska, M.E.; Ciebiada-Adamiec, A.; Sienkiewicz, M.; SokoŁowski, J.; Banaszek, K. The cultivable microbiota of primary and secondary infected root canals, their susceptibility to antibiotics and association with the signs and symptoms of infection. Int. Endod. J. 2016, 49, 422–430. [Google Scholar] [CrossRef]
- Loyola-Rodriguez, J.P.; Garcia-Cortes, J.O.; Martinez-Martinez, R.E.; Patiño-Marin, N.; Martinez-Castanon, G.-A.; Zavala-Alonso, N.V.; Amano, A. Molecular identification and antibiotic resistant bacteria isolated from primary dentition infections. Aust. Dent. J. 2014, 59, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Götz, C.; Reinhart, E.; Wolff, K.D.; Kolk, A. Oral soft tissue infections: Causes, therapeutic approaches and microbiological spectrum with focus on antibiotic treatment. J. Cranio-Maxillofac. Surg. 2015, 43, 1849–1854. [Google Scholar] [CrossRef]
- LaMonte, M.J.; Andrews, C.A.; Hovey, K.M.; Buck, M.J.; Li, L.; McSkimming, D.I.; Banack, H.R.; Rotterman, J.; Sun, Y.; Kirkwood, K.L.; et al. Subgingival microbiome is associated with alveolar bone loss measured 5 years later in postmenopausal women. J. Periodontol. 2020, 92, 648–661. [Google Scholar] [CrossRef]
- Fernández-Ferro, M.; Fernández-Sanromán, J.; Costas-López, A.; López-Betancourt, A.; Casañas-Villalba, N.; López-Fernández, P. Complex Dentoalveolar Fractures: Main Clinical Variables Description and Analysis. J. Craniofacial Surg. 2020, 31, e761–e765. [Google Scholar] [CrossRef]
- Aragon-Martinez, O.H.; Isiordia-Espinoza, M.A.; Nava, F.J.T.; Romo, S.A. Dental Care Professionals Should Avoid the Administration of Amoxicillin in Healthy Patients During Third Molar Surgery: Is Antibiotic Resistence the Only Problem? J. Oral Maxillofac. Surg. 2016, 74, 1512–1513. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Isiordia-Espinoza, M.A.; Aragon-Martinez, O.H.; Martínez-Morales, J.F.; Zapata-Morales, J.R. Risk of wound infection and safety profile of amoxicillin in healthy patients which required third molar surgery: A systematic review and meta-analysis. Br. J. Oral Maxillofac. Surg. 2015, 53, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Kanno, S.; Nosho, K.; Sukawa, Y.; Mitsuhashi, K.; Kurihara, H.; Igarashi, H.; Takahashi, T.; Tachibana, M.; Takahashi, H.; et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int. J. Cancer 2015, 137, 1258–1268. [Google Scholar] [CrossRef] [PubMed]
- Hussan, H.; Clinton, S.K.; Roberts, K.; Bailey, M.T. Fusobacterium’s link to colorectal neoplasia sequenced: A systematic review and future insights. World J. Gastroenterol. 2017, 23, 8626–8650. [Google Scholar] [CrossRef]
- Zhou, Y.; He, H.; Xu, H.; Li, Y.; Li, Z.; Du, Y.; He, J.; Zhou, Y.; Wang, H.; Nie, Y. Association of oncogenic bacteria with colorectal cancer in South China. Oncotarget 2016, 7, 80794–80802. [Google Scholar] [CrossRef] [PubMed]
- Viljoen, K.S.; Dakshinamurthy, A.; Goldberg, P.; Blackburn, J.M. Quantitative profiling of colorectal cancer-associated bacteria reveals associations between Fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PLoS ONE 2015, 10, e0119462. [Google Scholar] [CrossRef]
- Piekarz, T.; Mertas, A.; Wiatrak, K.; Rój, R.; Kownacki, P.; Śmieszek-Wilczewska, J.; Kopczyńska, E.; Wrzoł, M.; Cisowska, M.; Szliszka, E.; et al. The Influence of Toothpaste Containing Australian Melaleuca alternifolia Oil and Ethanolic Extract of Polish Propolis on Oral Hygiene and Microbiome in Patients Requiring Conservative Procedures. Molecules 2017, 22, 1957. [Google Scholar] [CrossRef]
- Wiatrak, K.; Morawiec, T.; Rój, R.; Kownacki, P.; Nitecka-Buchta, A.; Niedzielski, D.; Wychowański, P.; Machorowska-Pieniążek, A.; Cholewka, A.; Baldi, D.; et al. Article evaluation of effectiveness of a toothpaste containing tea tree oil and ethanolic extract of propolis on the improvement of oral health in patients using removable partial dentures. Molecules 2021, 26, 4071. [Google Scholar] [CrossRef]
- Bogacz, M.; Morawiec, T.; Śmieszek-Wilczewska, J.; Janowska-Bogacz, K.; Bubiłek-Bogacz, A.; Rój, R.; Pinocy, K.; Mertas, A. Evaluation of Drug Susceptibility of Microorganisms in Odontogenic Inflammations and Dental Surgery Procedures Performed on an Outpatient Basis. BioMed Res. Int. 2019, 2019, 2010453. [Google Scholar] [CrossRef]
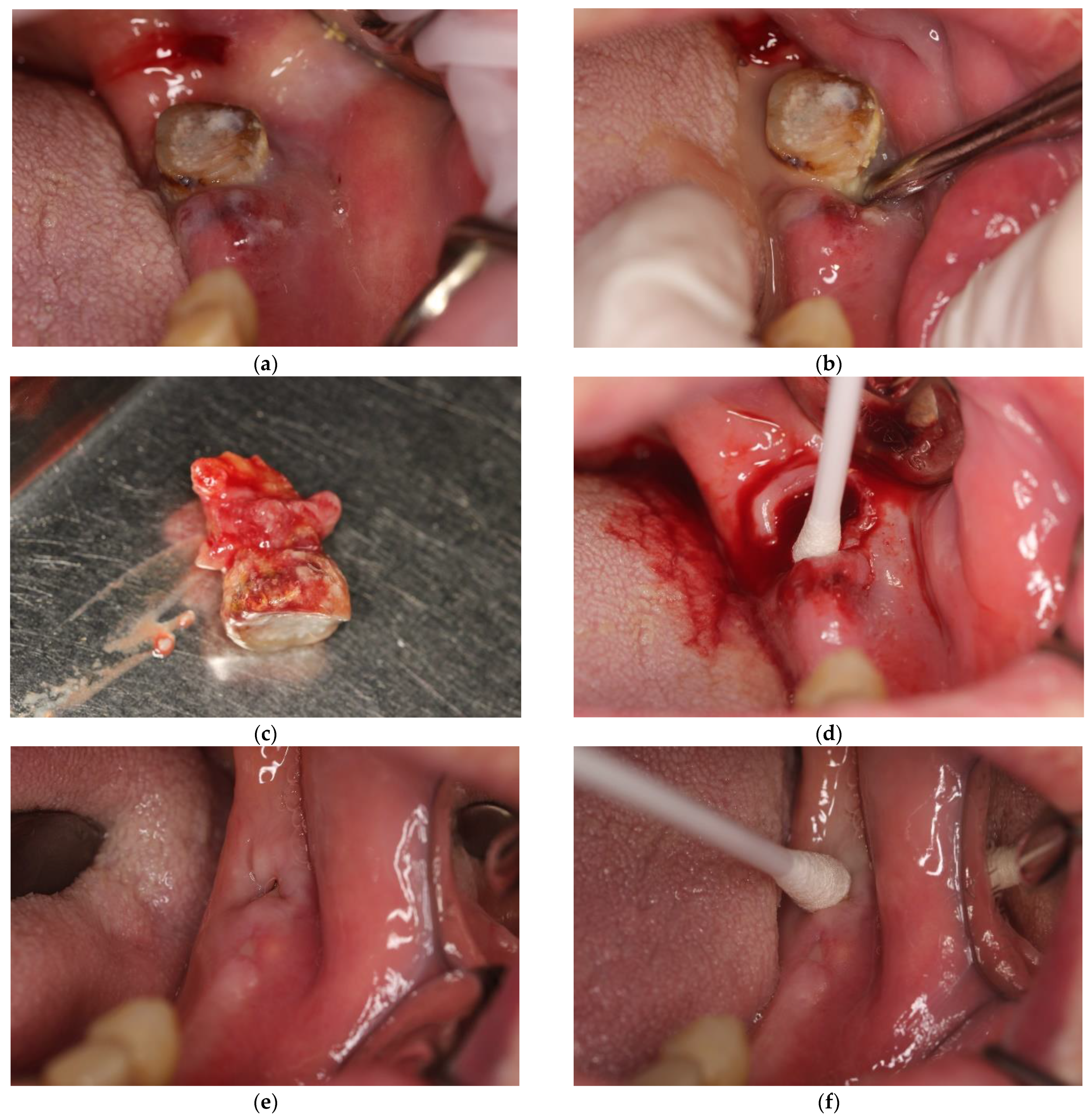
| Study Group (Mean ± SD) | Control Group (Mean ± SD) | p | |
|---|---|---|---|
| Age of patients (years) | 47.5 ± 14.5 | 33.0 ± 16.8 | 0.0017 |
| Bacterial Strains | Study Group % (Nbact/∑bact) | Control Group % (Nbact/∑bact) | p | |
|---|---|---|---|---|
| Anaerobic Gram-positive | Test I | 23.71% (23/97) | 17.43% (19/109) | 0.2641 |
| Test II | 23.66% (22/93) | 17.35% (17/98) | 0.2796 | |
| p | 0.9928 | 0.9873 | – | |
| Anaerobic Gram-negative | Test I | 10.31% (10/97) | 16.51% (18/109) | 0.0787 |
| Test II | 9.68% (9/93) | 12.24% (12/98) | 0.5708 | |
| p | 0.7915 | 0.3837 | – | |
| Aerobic Gram-positive | Test I | 41.24% (40/97) | 42.20% (46/109) | 0.8885 |
| Test II | 41.94% (39/93) | 41.84% (41/98) | 0.9890 | |
| p | 0.9222 | 0.9576 | – | |
| Aerobic Gram-negative | Test I | 24.74% (24/97) | 23.85% (26/109) | 0.8819 |
| Test II | 24.73 (23/93) | 28.57% (28/98) | 0.5488 | |
| p | 0.9986 | 0.4402 | – | |
| SPP | Study Group % (NSPP/Nbact) | Control Group % (NSPP/Nbact) | p | |
|---|---|---|---|---|
| Anaerobic Gram-positive | Test I | 47.83% (11/23) | 42.11% (8/19) | 0.7141 |
| Test II | 59.09% (13/22) | 58.82% (10/17) | 0.7555 | |
| p | 0.4490 | 0.5043 | – | |
| Anaerobic Gram-negative | Test I | 10.00% (1/10) | 12.50% (1/18) | 0.5952 |
| Test II | 0.00% (0/9) | 0.00% (0/12) | 1.0000 | |
| p | 0.3704 | 0.6000 | – | |
| Aerobic Gram-positive | Test I | 2.50% (1/40) | 4.35% (2/46) | 0.9019 |
| Test II | 5.13% (2/39) | 2.44% (1/41) | 0.9648 | |
| p | 0.9822 | 0.9192 | – | |
| Aerobic Gram-negative | Test I | 16.67% (4/24) | 11.54% (3/26) | 0.9091 |
| Test II | 17.39% (4/23) | 17.86% (5/28) | 0.7447 | |
| p | 0.7474 | 0.7874 | – | |
| Bacterial Species | Study Group % (Nsp/∑sp) | Control Group % (Nsp/∑sp) | p | |
|---|---|---|---|---|
| Anaerobic Gram-positive | Test I | 35.00% (14/40) | 28.95% (11/38) | 0.5670 |
| Test II | 34.21% (13/38) | 25.71% (9/35) | 0.4293 | |
| p | 0.9416 | 0.7386 | – | |
| Anaerobic Gram-negative | Test I | 17.50% (7/40) | 18.42% (7/38) | 0.9162 |
| Test II | 15.79% (6/38) | 17.14% (6/35) | 0.8770 | |
| p | 0.8405 | 0.8874 | – | |
| Aerobic Gram-positive | Test I | 37.50% (15/40) | 39.47% (15/38) | 0.8579 |
| Test II | 34.21% (13/38) | 40.00% (14/35) | 0.6087 | |
| p | 0.7621 | 0.9634 | – | |
| Aerobic Gram-negative | Test I | 9.76% (4/40) | 13.16% (5/38) | 0.9348 |
| Test II | 15.79% (6/38) | 17.14% (6/35) | 0.8770 | |
| p | 0.6704 | 0.6368 | – | |
| GPP | Study Group % (NGPP/Nsp) | Control Group % (NGPP/Nsp) | p | |
|---|---|---|---|---|
| Anaerobic Gram-positive | Test I | 42.86% (6/14) | 54.55% (6/11) | 0.8592 |
| Test II | 53.85% (7/13) | 33.33% (3/9) | 0.3050 | |
| p | 0.8528 | 0.3110 | – | |
| Anaerobic Gram-negative | Test I | 14.29% (1/7) | 14.29% (1/7) | 0.7692 |
| Test II | 0.00% (0/6) | 0.00% (0/6) | 1.0000 | |
| p | 0.5385 | 0.5385 | – | |
| Aerobic Gram-positive | Test I | 6.67% (1/15) | 13.33% (2/15) | 0.5000 |
| Test II | 15.38% (2/13) | 7.14% (1/14) | 0.4711 | |
| p | 0.4444 | 0.5268 | – | |
| Aerobic Gram-negative | Test I | 75.00% (3/4) | 40.00% (2/5) | 0.3571 |
| Test II | 66.67% (4/6) | 83.33% (5/6) | 0.5000 | |
| p | 0.6667 | 0.1970 | – | |
| Aerobic | Anaerobic | p | |
|---|---|---|---|
| Gram-positive | 4.82% (8/166) | 51.85% (42/81) | <0.0001 |
| Gram-negative | 15.84% (16/101) | 4.08% (2/49) | 0.0383 |
| p | 0.0023 | <0.0001 | – |
| Aerobic | Anaerobic | p | |
|---|---|---|---|
| Gram-positive | 10.00% (3/30) | 48.15% (13/27) | 0.0015 |
| Gram-negative | 73.33% (11/15) | 15.38% (2/13) | 0.0072 |
| p | <0.0001 | 0.0458 | – |
| Anaerobic Gram-Positive | ||||||
|---|---|---|---|---|---|---|
| Bacterial Species | Study Group | Control Group | ||||
| Test I | Test II | Total | Test I | Test II | Total | |
| Actinomyces naeslundii | 4 | 4 | 8 | 2 | 7 | 9 |
| Bifidobacterium odolescentis | 0 | 4 | 4 | 3 | 1 | 4 |
| Atopobium minutum | 3 | 1 | 4 | 0 | 1 | 1 |
| Propionibacterium propionicum | 0 | 4 | 4 | 0 | 0 | 0 |
| Bifidobacterium dentium | 2 | 1 | 3 | 3 | 2 | 5 |
| Clostridium perfringens | 1 | 1 | 2 | 1 | 0 | 1 |
| Actinomyces israelii | 2 | 0 | 2 | 1 | 0 | 1 |
| Peptococcus niger | 2 | 0 | 2 | 0 | 0 | 0 |
| Actinomyces meyeri | 2 | 0 | 2 | 0 | 0 | 0 |
| Bifidobacterium breve | 1 | 0 | 1 | 3 | 1 | 4 |
| Clostridium sporogenes | 0 | 1 | 1 | 0 | 1 | 1 |
| Faclamia sourekii | 0 | 1 | 1 | 0 | 1 | 1 |
| Clostridium novyi biovar A | 1 | 0 | 1 | 1 | 0 | 1 |
| Abiotrophia odicens | 1 | 0 | 1 | 0 | 0 | 0 |
| Anaerococcus prevotii | 0 | 1 | 1 | 0 | 0 | 0 |
| Clostridium butyricum | 0 | 1 | 1 | 0 | 0 | 0 |
| Clostridium chauvoei | 0 | 1 | 1 | 0 | 0 | 0 |
| Clostridium tertium | 1 | 0 | 1 | 0 | 0 | 0 |
| Pseudoramibacter alactolyticus | 1 | 0 | 1 | 0 | 0 | 0 |
| Actinomyces viscosus | 0 | 1 | 1 | 0 | 0 | 0 |
| Lactobacillus acidophilus | 0 | 1 | 1 | 0 | 0 | 0 |
| Lactobacillus catenaformis | 1 | 0 | 1 | 0 | 0 | 0 |
| Lactobacillus jensenii | 1 | 0 | 1 | 0 | 0 | 0 |
| Actinomyces odontolyticus | 0 | 0 | 0 | 2 | 2 | 4 |
| Bifidobacterium longum | 0 | 0 | 0 | 1 | 1 | 2 |
| Clostridium novyi | 0 | 0 | 0 | 1 | 0 | 1 |
| Eubacterium limosum | 0 | 0 | 0 | 1 | 0 | 1 |
| Total | 23 | 22 | 45 | 19 | 17 | 36 |
| Anaerobic Gram-Negative | ||||||
|---|---|---|---|---|---|---|
| Bacterial Species | Study Group | Control Group | ||||
| Test I | Test II | Total | Test I | Test II | Total | |
| Campylobacter gracilis | 1 | 3 | 4 | 3 | 2 | 5 |
| Campylobacter ureolyticus | 2 | 2 | 4 | 1 | 1 | 2 |
| Pseudoflavonifractor capillosus | 3 | 0 | 3 | 0 | 1 | 1 |
| Veillonella parvula | 1 | 1 | 2 | 4 | 0 | 4 |
| Mitsuokella multacida | 1 | 0 | 1 | 6 | 5 | 11 |
| Prevotella oralis | 1 | 0 | 1 | 0 | 1 | 1 |
| Fusobacterium mortiferum | 0 | 1 | 1 | 0 | 0 | 0 |
| Fusobacterium nucleatum | 1 | 0 | 1 | 0 | 0 | 0 |
| Prevotella intermedia | 0 | 1 | 1 | 0 | 0 | 0 |
| Parabacteroides distasonis | 0 | 1 | 1 | 0 | 0 | 0 |
| Capnocytophaga ochracea | 0 | 0 | 0 | 2 | 2 | 4 |
| Bacteroides ovatus | 0 | 0 | 0 | 1 | 0 | 1 |
| Prevotella melaninogenica | 0 | 0 | 0 | 1 | 0 | 1 |
| Total | 10 | 9 | 19 | 18 | 12 | 30 |
| Aerobic Gram-Positive | ||||||
|---|---|---|---|---|---|---|
| Bacterial Species | Study Group | Control Group | ||||
| Test I | Test II | Total | Test I | Test II | Total | |
| Streptococcus mitis | 12 | 14 | 26 | 12 | 13 | 25 |
| Streptococcus sanguinis | 7 | 7 | 14 | 8 | 6 | 14 |
| Streptococcus salivarius | 5 | 6 | 11 | 12 | 8 | 20 |
| Sarcina spp. | 2 | 1 | 3 | 0 | 1 | 1 |
| Staphylococcus epidermidis MSCNS | 2 | 1 | 3 | 0 | 0 | 0 |
| Streptococcus acidominimus | 2 | 0 | 2 | 1 | 2 | 3 |
| Staphylococcus lentus | 2 | 0 | 2 | 1 | 0 | 1 |
| Streptococcus anginosus | 0 | 2 | 2 | 0 | 1 | 1 |
| Streptococcus pneumoniae | 0 | 2 | 2 | 1 | 0 | 1 |
| Streptococcus suis | 1 | 1 | 2 | 0 | 0 | 0 |
| Streptococcus intermedius | 1 | 0 | 1 | 2 | 1 | 3 |
| Streptococcus mutans | 1 | 0 | 1 | 2 | 0 | 2 |
| Aerococcus viridans | 1 | 0 | 1 | 0 | 1 | 1 |
| Enterococcus malodoratus | 0 | 1 | 1 | 1 | 0 | 1 |
| Gemella spp. | 1 | 0 | 1 | 0 | 1 | 1 |
| Enterococcus columbae | 1 | 0 | 1 | 0 | 0 | 0 |
| Enterococcus hirae | 0 | 1 | 1 | 0 | 0 | 0 |
| Staphylococcus caprae | 0 | 1 | 1 | 0 | 0 | 0 |
| Staphylococcus chromogenes | 1 | 0 | 1 | 0 | 0 | 0 |
| Staphylococcus hominis | 1 | 0 | 1 | 0 | 0 | 0 |
| Staphylococcus sciuri | 0 | 1 | 1 | 0 | 0 | 0 |
| Staphylococcus warneri | 0 | 1 | 1 | 0 | 0 | 0 |
| Streptococcus uberis | 0 | 0 | 0 | 1 | 2 | 3 |
| Enterococcus gallinarum | 0 | 0 | 0 | 0 | 2 | 2 |
| Staphylococcus aureus MSSA | 0 | 0 | 0 | 1 | 1 | 2 |
| Streptococcus canis | 0 | 0 | 0 | 1 | 1 | 2 |
| Enterococcus casseliflavus | 0 | 0 | 0 | 0 | 1 | 1 |
| Globicatella sanquinis | 0 | 0 | 0 | 1 | 0 | 1 |
| Lactococcus lactis | 0 | 0 | 0 | 1 | 0 | 1 |
| Streptococcus bovis biovar I | 0 | 0 | 0 | 1 | 0 | 1 |
| Total | 40 | 39 | 79 | 46 | 41 | 87 |
| Aerobic Gram-Negative | ||||||
|---|---|---|---|---|---|---|
| Bacterial Species | Study Group | Control Group | ||||
| Test I | Test II | Total | Test I | Test II | Total | |
| Neisseria subflava | 20 | 18 | 38 | 21 | 23 | 44 |
| Enterobacter cloacae | 2 | 0 | 2 | 0 | 0 | 0 |
| Escherichia coli | 1 | 1 | 2 | 0 | 0 | 0 |
| Klebsiella pneumoniae | 0 | 1 | 1 | 0 | 1 | 1 |
| Enterobacter kobeii | 0 | 1 | 1 | 0 | 0 | 0 |
| Hafnia alvei | 0 | 1 | 1 | 0 | 0 | 0 |
| Neisseria caprae | 0 | 1 | 1 | 0 | 0 | 0 |
| Serratia odorifera | 1 | 0 | 1 | 0 | 0 | 0 |
| Klebsiella oxytoca | 0 | 0 | 0 | 2 | 1 | 3 |
| Burkholderia cepacia | 0 | 0 | 0 | 0 | 1 | 1 |
| Chryseobacterium indologenes | 0 | 0 | 0 | 1 | 0 | 1 |
| Enterobacter aerogenes | 0 | 0 | 0 | 0 | 1 | 1 |
| Neisseria flavescens | 0 | 0 | 0 | 1 | 0 | 1 |
| Neisseria sicca | 0 | 0 | 0 | 1 | 0 | 1 |
| Providencia rustigianii | 0 | 0 | 0 | 0 | 1 | 1 |
| Total | 24 | 23 | 47 | 26 | 28 | 54 |
| Bacterial Species | Study Group | Control Group | ||||
|---|---|---|---|---|---|---|
| Test I | Test II | Total | Test I | Test II | Total | |
| Candida albicans | 5 | 5 | 10 | 4 | 3 | 7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morawiec, T.; Śmieszek-Wilczewska, J.; Bogacz, M.; Jędrusik-Pawłowska, M.; Bubiłek-Bogacz, A.; Mertas, A. Status of Microbiota in Odontogenic Inflammatory Lesions and Dental Surgery Procedures Performed on an Outpatient Basis. Antibiotics 2022, 11, 1025. https://doi.org/10.3390/antibiotics11081025
Morawiec T, Śmieszek-Wilczewska J, Bogacz M, Jędrusik-Pawłowska M, Bubiłek-Bogacz A, Mertas A. Status of Microbiota in Odontogenic Inflammatory Lesions and Dental Surgery Procedures Performed on an Outpatient Basis. Antibiotics. 2022; 11(8):1025. https://doi.org/10.3390/antibiotics11081025
Chicago/Turabian StyleMorawiec, Tadeusz, Joanna Śmieszek-Wilczewska, Mateusz Bogacz, Magdalena Jędrusik-Pawłowska, Anna Bubiłek-Bogacz, and Anna Mertas. 2022. "Status of Microbiota in Odontogenic Inflammatory Lesions and Dental Surgery Procedures Performed on an Outpatient Basis" Antibiotics 11, no. 8: 1025. https://doi.org/10.3390/antibiotics11081025
APA StyleMorawiec, T., Śmieszek-Wilczewska, J., Bogacz, M., Jędrusik-Pawłowska, M., Bubiłek-Bogacz, A., & Mertas, A. (2022). Status of Microbiota in Odontogenic Inflammatory Lesions and Dental Surgery Procedures Performed on an Outpatient Basis. Antibiotics, 11(8), 1025. https://doi.org/10.3390/antibiotics11081025






