Development of 4-[4-(Anilinomethyl)-3-phenyl-pyrazol-1-yl] Benzoic Acid Derivatives as Potent Anti-Staphylococci and Anti-Enterococci Agents
Abstract
:1. Introduction
2. Results and Discussion
2.1. MIC Studies
2.2. Bactericidal Properties and Time-Kill Assay
2.3. Activity against Biofilms
2.4. Real-Time Monitoring of S. aureus Biofilm
2.5. Activity against Persisters
2.6. Multistep Resistance Studies
2.7. Membrane Permeability Studies
3. Materials and Methods
3.1. Antimicrobial Compounds (1–11)
3.2. Minimum Inhibitory Concentration (MIC)
3.3. Minimum Bactericidal Concentration (MBC)
3.4. Time-Kill Assay
3.5. Biofilm Inhibition and Eradication Assays
3.6. Antibiofilm Studies Using Calgary Device
3.7. Real-Time Monitoring of S. aureus Biofilm
3.8. Persister Cell Killing Assay
3.9. Multi-Step Resistance Assay
3.10. Kinetic Fluorescence Measurements to Detect Membrane Permeabilization Using Propidium Iodide
3.11. Protein Leakage Assay
3.12. Flow Cytometry for Membrane Permeability
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Scholtz, V.; Vaňková, E.; Kašparová, P.; Premanath, R.; Karunasagar, I.; Julák, J. Non-thermal plasma treatment of ESKAPE pathogens: A review. Front Microbiol. 2021, 12, 737635. [Google Scholar] [CrossRef] [PubMed]
- Kavanagh, K.T. Control of MSSA and MRSA in the United States: Protocols, policies, risk adjustment and excuses. Antimicrob. Resist. Infect. Control 2019, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- CDC General Information about Staphylococcus Aureus. Available online: https://www.cdc.gov/hai/organisms/staph.html (accessed on 15 June 2022).
- Moisse, K. Antibiotic Resistance Could Bring “End of Modern Medicine”. Available online: http://abcnews.go.com/blogs/health/2012/03/16/antibiotic-resistance-could-bring-end-of-modern-medicine/ (accessed on 1 May 2020).
- Tal-Jasper, R.; Katz, D.E.; Amrami, N.; Ravid, D.; Avivi, D.; Zaidenstein, R.; Lazarovitch, T.; Dadon, M.; Kaye, K.S.; Marchaim, D. Clinical and epidemiological significance of carbapenem resistance in acinetobacter baumannii infections. Antimicrob. Agents Chemother. 2016, 60, 3127–3131. [Google Scholar] [CrossRef] [Green Version]
- Marsit, H.; Koubaa, M.; Gargouri, M.; Ben Jemaa, T.; Gaddour, H.; Kotti, F.; Sammoudi, A.; Turki, M.; Ben Jemaa, M. Hospital-acquired infections due to multidrug resistant acinetobacter baumannii: How challenging is the management? Fund Clin Pharmacol 2016, 30, 87. [Google Scholar]
- Farha, M.A.; Leung, A.; Sewell, E.W.; D’Elia, M.A.; Allison, S.E.; Ejim, L.; Pereira, P.M.; Pinho, M.G.; Wright, G.D.; Brown, E.D. Inhibition of WTA synthesis blocks the cooperative action of PBPs and sensitizes MRSA to beta-lactams. ACS Chem. Biol. 2013, 8, 226–233. [Google Scholar] [CrossRef]
- Mistry, T.L.; Truong, L.; Ghosh, A.K.; Johnson, M.E.; Mehboob, S. Benzimidazole-based fabi inhibitors: A promising novel scaffold for anti-staphylococcal drug development. ACS Infect. Dis. 2017, 3, 54–61. [Google Scholar] [CrossRef] [Green Version]
- Agudelo Higuita, N.I.; Huycke, M.M. Enterococcal disease, epidemiology, and implications for treatment. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- García-Solache, M.; Rice, L.B. The enterococcus: A model of adaptability to its environment. Clin. Microbiol. Rev. 2019, 32, e00058-18. [Google Scholar] [CrossRef] [Green Version]
- Said, M.S.; Tirthani, E.; Lesho, E. Enterococcus Infections; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- CDC Vancomycin-Resistant Enterococci (VRE). Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/vre-508.pdf (accessed on 23 May 2020).
- Ch’ng, J.-H.; Chong, K.K.L.; Lam, L.N.; Wong, J.J.; Kline, K.A. Biofilm-associated infection by enterococci. Nat. Rev. Microbiol. 2019, 17, 82–94. [Google Scholar] [CrossRef]
- Alkhaibari, I.S.; Raj, K.C.H.; Alnufaie, R.; Gilmore, D.; Alam, M.A. Synthesis of chimeric thiazolo-nootkatone derivatives as potent antimicrobial agents. ChemMedChem 2021, 16, 2628–2637. [Google Scholar] [CrossRef]
- Delancey, E.; Allison, D.; Kc, H.R.; Gilmore, D.F.; Fite, T.; Basnakian, A.G.; Alam, M.A. Synthesis of 4,4′-(4-formyl-1h-pyrazole-1,3-diyl) dibenzoic acid derivatives as narrow spectrum antibiotics for the potential treatment of acinetobacter baumannii infections. Antibiotics 2020, 9, 650. [Google Scholar] [CrossRef] [PubMed]
- Allison, D.; Delancey, E.; Ramey, H.; Williams, C.; Alsharif, Z.A.; Al-khattabi, H.; Ontko, A.; Gilmore, D.; Alam, M.A. Synthesis and antimicrobial studies of novel derivatives of 4-(4-formyl-3-phenyl-1H-pyrazol-1-yl) benzoic acid as potent anti-Acinetobacter baumannii agents. Bioorg. Med. Chem. Lett. 2017, 27, 387–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brider, J.; Rowe, T.; Gibler, D.J.; Gottsponer, A.; Delancey, E.; Branscum, M.D.; Ontko, A.; Gilmore, D.; Alam, M.A. Synthesis and antimicrobial studies of azomethine and N-arylamine derivatives of 4-(4-formyl-3-phenyl-1H-pyrazol-1-yl) benzoic acid as potent anti-methicillin-resistant Staphylococcus aureus agents. Med. Chem. Res. 2016, 25, 2691–2697. [Google Scholar] [CrossRef]
- Hansa, R.K.; Khan, M.M.K.; Frangie, M.M.; Gilmore, D.F.; Shelton, R.S.; Savenka, A.V.; Basnakian, A.G.; Shuttleworth, S.L.; Smeltzer, M.S.; Alam, M.A. 4–4-(Anilinomethyl)-3-[4-(trifluoromethyl)phenyl]-1H-pyrazol-1-ylbenzoic acid derivatives as potent anti-gram-positive bacterial agents. Eur. J. Med. Chem. 2021, 219, 113402. [Google Scholar] [CrossRef] [PubMed]
- Pankey, G.A.; Sabath, L.D. Clinical relevance of bacteriostatic versus bactericidal mechanisms of action in the treatment of gram-positive bacterial infections. Clin. Infect. Dis. 2004, 38, 864–870. [Google Scholar] [CrossRef] [Green Version]
- Ayrapetyan, M.; Williams, T.; Oliver, J.D. Relationship between the viable but nonculturable state and antibiotic persister cells. J. Bacteriol. 2018, 200, e00249-18. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, M.; Wozniak, D.J.; Stoodley, P.; Hall-Stoodley, L. Prevention and treatment of Staphylococcus aureus biofilms. Expert Rev. Anti. Infect. Ther. 2015, 13, 1499–1516. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.-X.; Bai, B.; Lin, Z.-W.; Pu, Z.-Y.; Yao, W.-M.; Chen, Z.; Li, D.-Y.; Deng, X.-B.; Deng, Q.-W.; Yu, Z.-J. Characterization of biofilm formation by enterococcus faecalis isolates derived from urinary tract infections in China. J. Med. Microbiol. 2018, 67, 60–67. [Google Scholar] [CrossRef]
- Costerton, W.; Veeh, R.; Shirtliff, M.; Pasmore, M.; Post, C.; Ehrlich, G. The application of biofilm science to the study and control of chronic bacterial infections. J. Clin. Investig. 2003, 112, 1466–1477. [Google Scholar] [CrossRef] [Green Version]
- Hall-Stoodley, L.; Stoodley, P. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 2005, 13, 7–10. [Google Scholar] [CrossRef]
- Conlon, B.P.; Rowe, S.E.; Gandt, A.B.; Nuxoll, A.S.; Donegan, N.P.; Zalis, E.A.; Clair, G.; Adkins, J.N.; Cheung, A.L.; Lewis, K. Persister formation in staphylococcus aureus is associated with ATP depletion. Nat. Microbiol. 2016, 1, 16051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrader, S.M.; Botella, H.; Jansen, R.; Ehrt, S.; Rhee, K.; Nathan, C.; Vaubourgeix, J. Multiform antimicrobial resistance from a metabolic mutation. Sci. Adv. 2021, 7, eabh2037. [Google Scholar] [CrossRef] [PubMed]
- Igler, C.; Rolff, J.; Regoes, R. Multi-step vs. single-step resistance evolution under different drugs, pharmacokinetics, and treatment regimens. eLife 2021, 10, e64116. [Google Scholar] [CrossRef] [PubMed]
- Terstappen, G.C.; Schlüpen, C.; Raggiaschi, R.; Gaviraghi, G. Target deconvolution strategies in drug discovery. Nat. Rev. Drug Discov. 2007, 6, 891–903. [Google Scholar] [CrossRef]
- Shi, Y.-G.; Zhang, R.-R.; Zhu, C.-M.; Xu, M.-F.; Gu, Q.; Ettelaie, R.; Lin, S.; Wang, Y.-F.; Leng, X.-Y. Antimicrobial mechanism of alkyl gallates against escherichia coli and staphylococcus aureus and its combined effect with electrospun nanofibers on Chinese Taihu icefish preservation. Food Chem. 2021, 346, 128949. [Google Scholar] [CrossRef]
- Sun, H.; Huang, S.-Y.; Jeyakkumar, P.; Cai, G.-X.; Fang, B.; Zhou, C.-H. Natural berberine-derived azolyl ethanols as new structural antibacterial agents against drug-resistant escherichia coli. J. Med. Chem. 2022, 65, 436–459. [Google Scholar] [CrossRef]
- Saleh, I.; Kc, H.R.; Roy, S.; Abugazleh, M.K.; Ali, H.; Gilmore, D.; Alam, M.A. Design, synthesis, and antibacterial activity of N-(trifluoromethyl)phenyl substituted pyrazole derivatives. RSC Med. Chem. 2021, 12, 1690–1697. [Google Scholar] [CrossRef]
- Alkhaibari, I.S.; Kc, H.R.; Roy, S.; Abu-gazleh, M.K.; Gilmore, D.F.; Alam, M.A. Synthesis of 3,5-Bis(trifluoromethyl)phenyl-substituted pyrazole derivatives as potent growth inhibitors of drug-resistant bacteria. Molecules 2021, 26, 5083. [Google Scholar] [CrossRef]
- Elkhatib, W.; Noreddin, A. In vitro antibiofilm efficacies of different antibiotic combinations with zinc sulfate against pseudomonas aeruginosa recovered from hospitalized patients with urinary tract infection. Antibiotics 2014, 3, 64–84. [Google Scholar] [CrossRef] [Green Version]
- Boix-Lemonche, G.; Lekka, M.; Skerlavaj, B. A rapid fluorescence-based microplate assay to investigate the interaction of membrane active antimicrobial peptides with whole gram-positive bacteria. Antibiotics 2020, 9, 92. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.-P.; Sangaraiah, N.; Meng, J.-P.; Zhou, C.-H. Unique carbazole-oxadiazole derivatives as new potential antibiotics for combating gram-positive and negative bacteria. J. Med. Chem. 2022, 65, 6171–6190. [Google Scholar] [CrossRef] [PubMed]
- Alkhaibari, I.; Kc, H.R.; Angappulige, D.H.; Gilmore, D.; Alam, M.A. Novel pyrazoles as potent growth inhibitors of staphylococci, enterococci and acinetobacter baumannii bacteria. Future Med. Chem. 2022, 14, 233–244. [Google Scholar] [CrossRef] [PubMed]

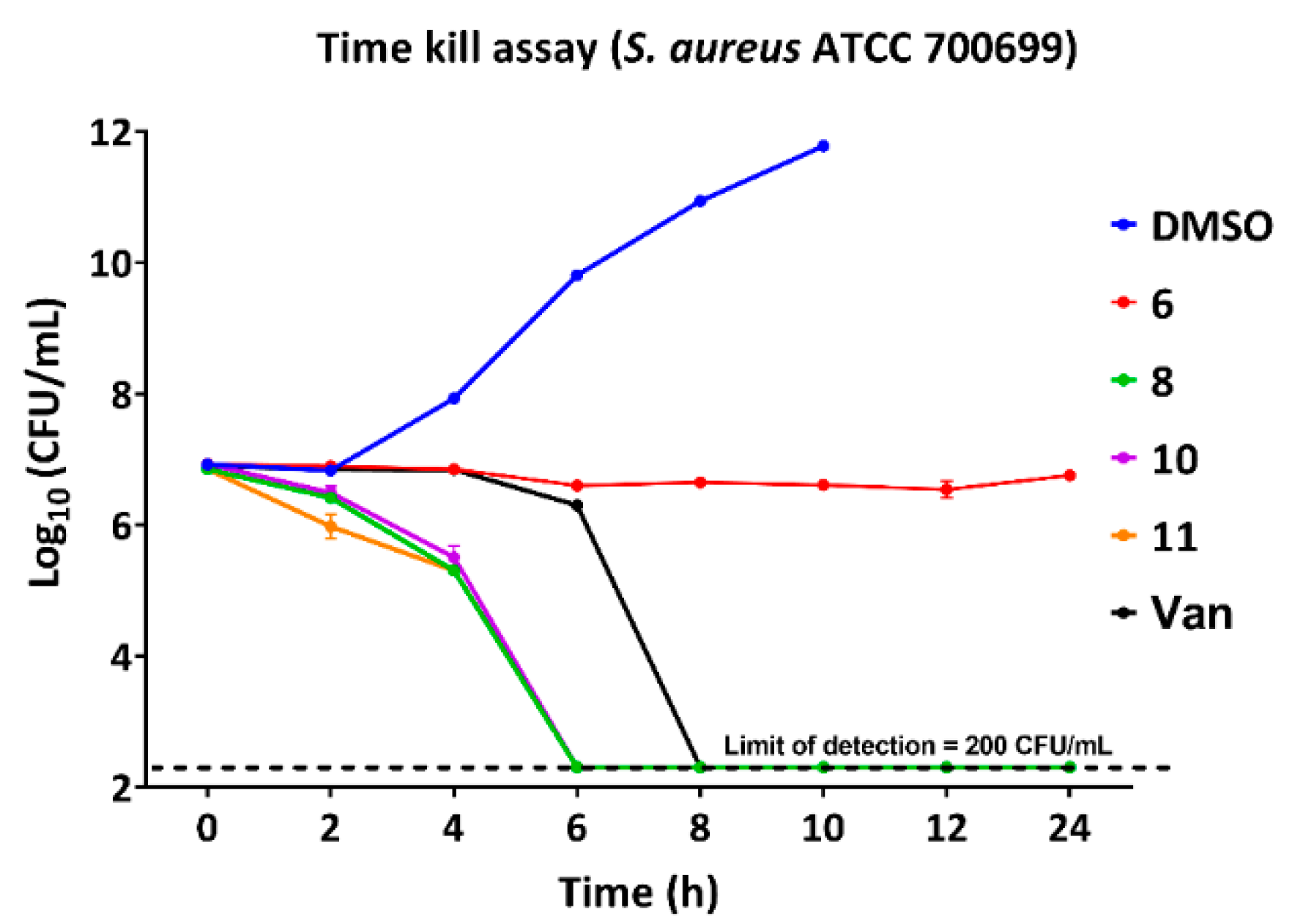


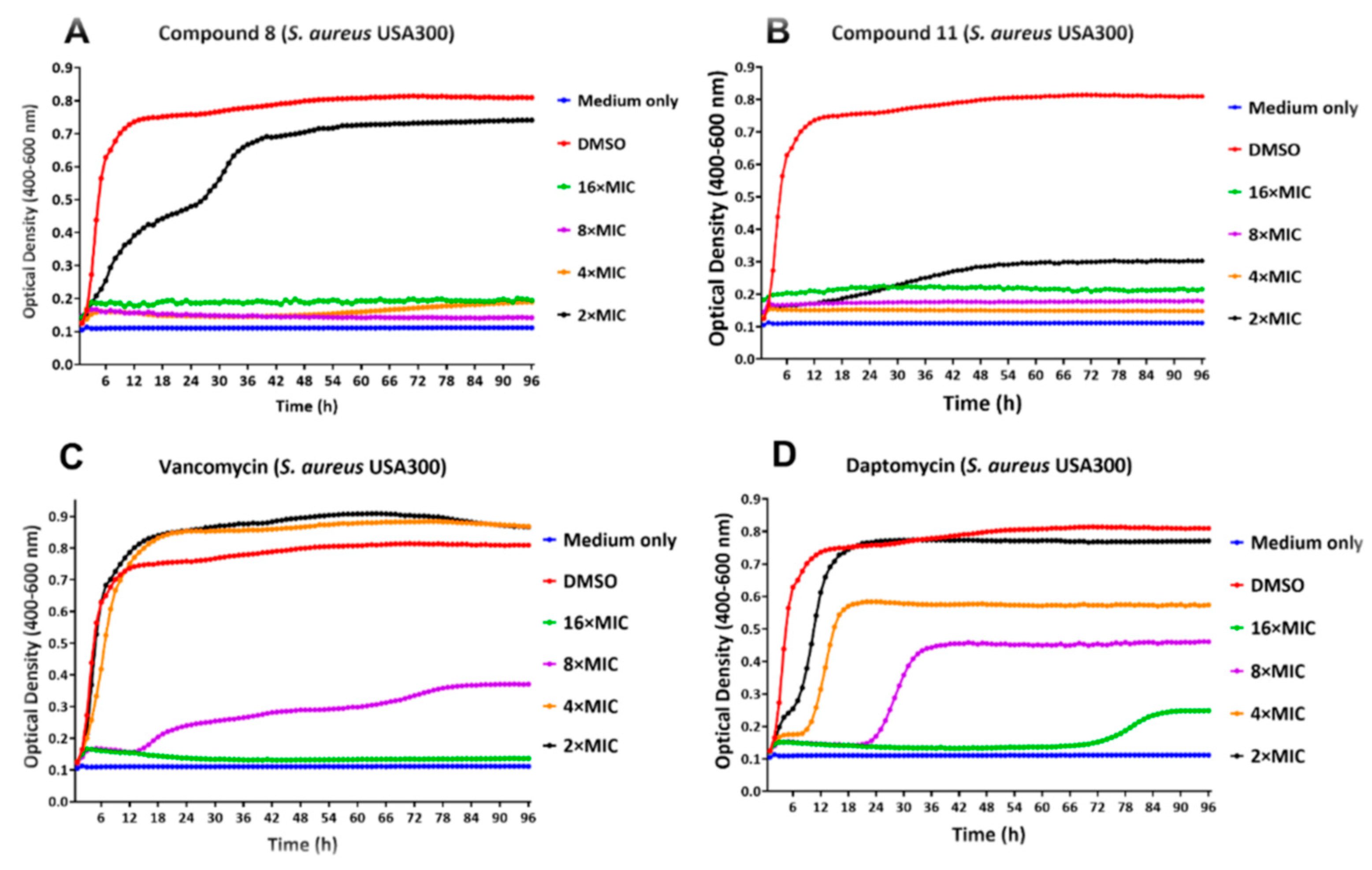
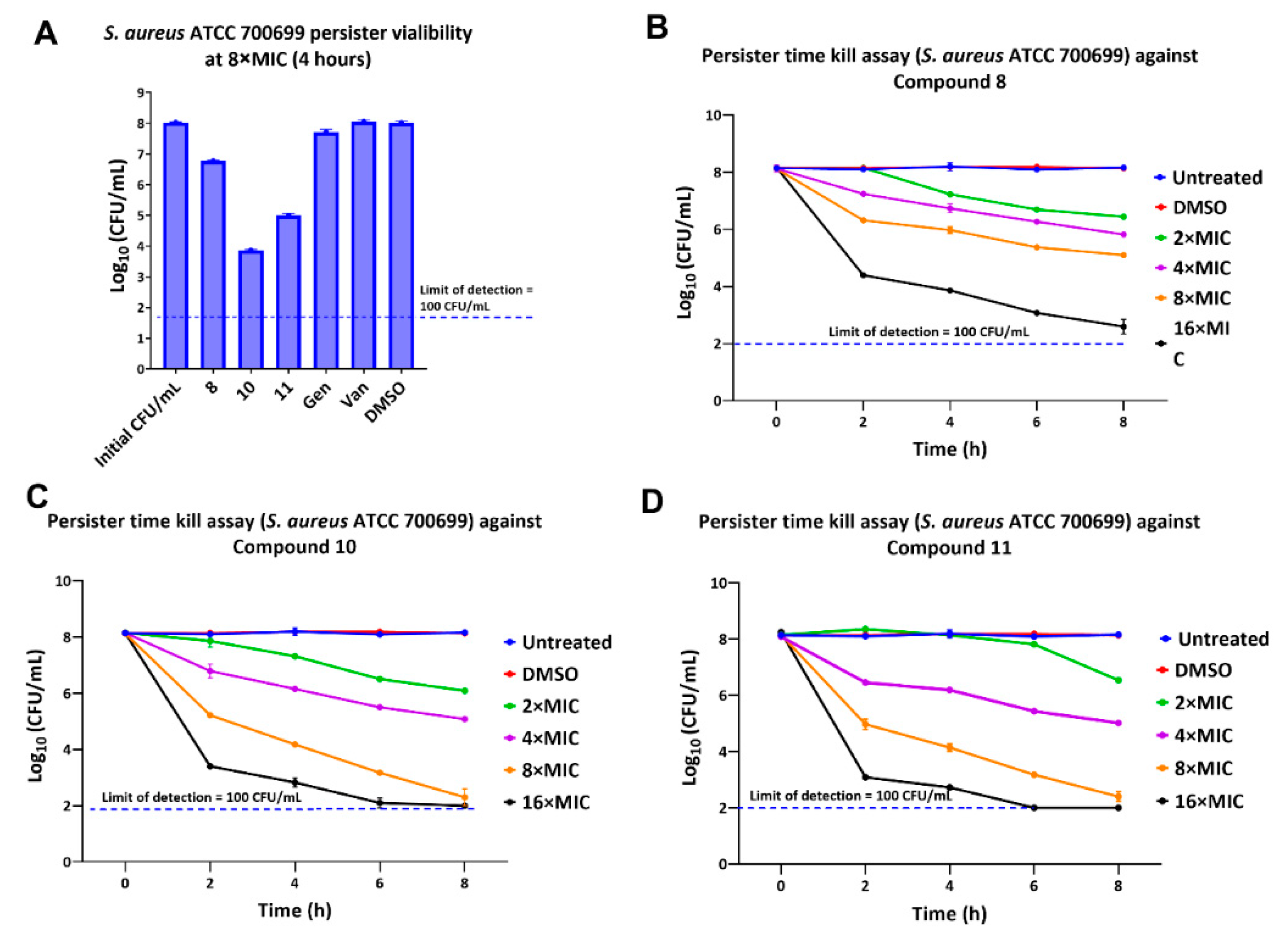



| Comp | Sa23 | Sa99 | Sa12 | Sa92 | Sa91 | SaN | Sa00 | Sa1 | Ef12 | Ef21 | Ef99 | Bs | Se |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 25 | 25 | 25 | 50 | 25 | >50 | >50 | >50 | >50 | 50 | >50 | 25 | 50 |
| 2 | 12.5 | 12.5 | 12.5 | 25 | 12.5 | 12.5 | 12.5 | 12.5 | 12.5 | 50 | 25 | 6.25 | 25 |
| 3 | 12.5 | 12.5 | 12.5 | 25 | 12.5 | 12.5 | 12.5 | 12.5 | >50 | 50 | 25 | 6.25 | 25 |
| 4 | 12.5 | 12.5 | 12.5 | 12.5 | 12.5 | 6.25 | 12.5 | 12.5 | 12.5 | 25 | 25 | 6.25 | 25 |
| 5 | 25 | 25 | 50 | 50 | 25 | >50 | >50 | >50 | 50 | >50 | 25 | 12.5 | 50 |
| 6 | 6.25 | 3.12 | 6.25 | 6.25 | 3.12 | 3.12 | 6.25 | 6.25 | 6.25 | 12.5 | 6.25 | 3.12 | 12.5 |
| 7 | 3.12 | 6.25 | 6.25 | 12.5 | 6.25 | 3.12 | 6.25 | 6.25 | 6.25 | 12.5 | 6.25 | 3.12 | 12.5 |
| 8 | 1.56 | 1.56 | 0.78 | 3.12 | 0.78 | 1.56 | 1.56 | 1.56 | 3.12 | 3.12 | 3.12 | 0.78 | 6.25 |
| 9 | 6.25 | 12.5 | 12.5 | 12.5 | 6.25 | 3.12 | 12.5 | 12.5 | 6.25 | 25 | 25 | 3.12 | 25 |
| 10 | 3.12 | 3.12 | 3.12 | 3.12 | 3.12 | 3.12 | 3.12 | 3.12 | 3.12 | 6.25 | 6.25 | 1.56 | 6.25 |
| 11 | 3.12 | 3.12 | 3.12 | 3.12 | 3.12 | 3.12 | 3.12 | 3.12 | 3.12 | 6.25 | 6.25 | 1.56 | 6.25 |
| Van | 0.78 | 3.12 | 0.78 | 1.56 | 1.56 | 0.78 | 0.78 | 0.78 | 3.12 | >50 | >50 | 0.19 | 3.12 |
| Dap | 1.56 | 6.25 | 0.78 | 3.12 | 6.25 | 3.12 | 3.12 | 3.12 | 12.5 | 12.5 | 12.5 | 0.78 | 0.78 |
| Comp | Bs | Sa12 | Sa23 | Sa99 | Sa91 | Sa92 | Se |
|---|---|---|---|---|---|---|---|
| 6 | 12.5 | 50 | 50 | 50 | 12.5 | 50 | 50 |
| 7 | 12.5 | 50 | 25 | 25 | 12.5 | 50 | 50 |
| 8 | 3.12 | 3.12 | 12.5 | 6.25 | 3.12 | 12.5 | 12.5 |
| 10 | 6.25 | 12.5 | 25 | 12.5 | 12.5 | 25 | 25 |
| 11 | 12.5 | 25 | 12.5 | 12.5 | 12.5 | 25 | 25 |
| Van | 1.56 | 1.56 | 12.5 | 6.25 | 6.25 | 6.25 | 3.12 |
| Comps | Sa23 | Sa00 | Efs12 | |||
|---|---|---|---|---|---|---|
| MIC | MBEC | MIC | MBEC | MIC | MBEC | |
| 8 | 1.56 | 3.12 | 1.56 | 12.5 | 3.12 | 6.25 |
| 10 | 3.12 | 6.25 | 3.12 | 50 | 3.12 | 12.5 |
| 11 | 3.12 | 6.25 | 3.12 | 25 | 3.12 | 6.25 |
| Van | 0.78 | >50 | 0.78 | >50 | 3.12 | >50 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raj KC, H.; Gilmore, D.F.; Alam, M.A. Development of 4-[4-(Anilinomethyl)-3-phenyl-pyrazol-1-yl] Benzoic Acid Derivatives as Potent Anti-Staphylococci and Anti-Enterococci Agents. Antibiotics 2022, 11, 939. https://doi.org/10.3390/antibiotics11070939
Raj KC H, Gilmore DF, Alam MA. Development of 4-[4-(Anilinomethyl)-3-phenyl-pyrazol-1-yl] Benzoic Acid Derivatives as Potent Anti-Staphylococci and Anti-Enterococci Agents. Antibiotics. 2022; 11(7):939. https://doi.org/10.3390/antibiotics11070939
Chicago/Turabian StyleRaj KC, Hansa, David F. Gilmore, and Mohammad A. Alam. 2022. "Development of 4-[4-(Anilinomethyl)-3-phenyl-pyrazol-1-yl] Benzoic Acid Derivatives as Potent Anti-Staphylococci and Anti-Enterococci Agents" Antibiotics 11, no. 7: 939. https://doi.org/10.3390/antibiotics11070939
APA StyleRaj KC, H., Gilmore, D. F., & Alam, M. A. (2022). Development of 4-[4-(Anilinomethyl)-3-phenyl-pyrazol-1-yl] Benzoic Acid Derivatives as Potent Anti-Staphylococci and Anti-Enterococci Agents. Antibiotics, 11(7), 939. https://doi.org/10.3390/antibiotics11070939







